Introduction
Cancer is the leading cause of morbidity and
mortality globally (1). Brain and
other central nervous system tumor types are common amongst adults
and children, ranking 17th in incidence among all cancer types
globally (2) and is the 2nd leading
cause of mortality in the pediatric age group (3).
Brain tumor types are either primary or secondary
(4). Currently, there exists a World
Health Organization (WHO) classification for the prognostication of
gliomas based on their morphological characteristics.
Oligodendrogliomas are divided into low-grade (WHO Grade II) and
anaplastic (WHO Grade III) tumor types. Astrocytic tumor types are
classified into low-grade diffuse astrocytoma (WHO Grade II),
anaplastic astrocytoma (WHO Grade III) and glioblastoma multiforme
(GBM) and its variants (WHO Grade IV) (4). GBM is the most malignant and aggressive
of these tumor types (4).
Conventionally, brain tumor types are diagnosed by
imaging. In certain cases, imaging may not be adequate to reach a
diagnosis, so a biopsy and histopathological examination of the
tissue may be required (5). The
molecular genetics of tumor cells serve a notable role in the
pathogenesis of brain tumor types. This is particularly the case
for oligodendrogliomas that have their own unique set of
alterations, with the most notable being the loss of heterozygosity
(LOH) on chromosomal arms 1p/19q. LOH 1p/19q has emerged as an
independent predictive marker of an improved response to radio- and
chemotherapy in addition to prolonged overall survival in
anaplastic oligodendrglioma (6–11).
Combined with the status of isocitrate dehydrogenase 1 and 2 and
tumor protein p53 (TP53) mutations, the prognosis and diagnosis of
distinct subgroups of gliomas has become dependent on these genetic
alterations due to their functions in these tumor types (12,13).
Over the years, fluorescence in situ
hybridization (FISH) analysis has become a useful diagnostic test
to determine the molecular profile of oligodendrogliomas, along
with other techniques including microsatellite markers and array
comparative genomic hybridization (aCGH) (14). There are several disadvantages to
using FISH, one of which is the decreasing intensity of the signals
over time. Other limitations include the need to optimize the
control probes for each locus, which may be time-consuming and
costly (14).
Using aCGH as a clinical tool has proven to be more
useful and reliable compared with FISH as it is able to detect a
wide and extensive number of genetic changes in one screening of
the human genome. It is a highly sensitive tool that is able to
distinguish even the smallest genomic regions and subtlest DNA copy
number difference (15). A study by
Mohapatra et al (15),
demonstrated this by using formalin-fixed, paraffin-embedded
oligodendroglioma samples in aCGH analysis with a novel labelling
technique to generate high-quality hybridization probes. It was
hypothesized that aCGH was able to detect single copy losses and
50% more DNA losses in the cells when compared with FISH analysis
(15).
In more recent years, a method to analyze the entire
genome has been identified through the use of next generation
sequencing (NGS) technologies (16,17). There
are a number of techniques that fall under the definition of NGS
including whole genome, whole exome and RNA sequencing. NGS is a
method that is able to provide in-depth information regarding the
functionalities of the whole genome including its genomic
alterations, transcriptional changes, gene fusions and alternative
splicing (16,17). RNA sequencing (RNA-Seq) in particular,
is a high-throughput whole transcriptome sequencing technology that
utilizes isolated RNA for deep sequencing for profiling and
generates comprehensive transcriptomic information of the cells.
Previous studies have been performed using NGS to provide an
RNA-Seq profiling dataset and identify biomarkers for diagnosis and
prognosis with the potential to be candidate genes for glioma
therapy (18–20). The main objective of the present study
was to capture the transcriptomic landscape along the pathogenesis
of oligodendrogliomas from low-grade to high-grade tumor
progression. Using aCGH and NGS, the present study also aimed to
identify differentially expressed genes, genetic aberrations and
novel markers involved in the pathogenesis of
oligodendrogliomas.
Materials and methods
Clinical patient data
Patient samples of 6 patients were collected from
the respective hospitals for a period of 3 years between 1st
January 2014 and 31st December 2016. A select number of archived
samples were also obtained from the Brain Tumor Tissue Bank (Brain
Tumor Foundation of Canada, London, ON, Canada) due to the limited
availability of samples obtained from patients. Upon admission to
the hospital, demographic data and a brief clinical history were
elicited from the patients. This included the age and sex of the
patient and the state in which the patient was domiciled. The final
selection of 6 patients consisted of 3 low-grade oligodendroglioma
and 3 anaplastic oligodendroglioma cases (Table I). Written informed consent was
obtained prior to brain tumor removal from the patient during
surgery. Subsequent to obtaining consent, surgery was performed the
next day, typically within 12–24 h. In addition, the present study
received ethical approval from the Medical Research Ethics
Committee of the Ministry of Health (Putrajaya, Malaysia).
 | Table I.Patient demographics and
histopathology of tumor types. |
Table I.
Patient demographics and
histopathology of tumor types.
| Patient | Sample no. | Age (years) | Sex | Histopathology | Grade |
|---|
| 1 | 88 | 23 | Male | Low grade
oligodendroglioma | II |
| 2 | 1578 | 34 | Female | Low grade
oligodendroglioma | II |
| 3 | 1090 | 37 | Female | Low grade
oligodendroglioma | II |
| 4 | 1385 | 35 | Female | Anaplastic
oligodendroglioma | III |
| 5 | 1395 | 44 | Female | Anaplastic
oligodendrglioma | III |
| 6 | 1430 | 45 | Female | Anaplastic
oligodendrglioma | III |
Tissue sample collection
The tumor samples were collected and then inserted
into a PAXgene™ Tissue Container (Qiagen GmbH, Hilden,
Germany). The tissue specimens were placed first into a standard
tissue cassette. The cap of the PAXgene Tissue Container was then
unscrewed and lifted up to remove the screw cap-rack assembly. The
tissue cassette was attached to the rack by inserting the lower
edge of the tissue cassette into the bottom edge of the rack and
pressed to secure it under the rack upper release tab. Then, the
rack holding the tissue cassette was placed into chamber 1
containing the PAXgene Tissue Fix solution and capped into place.
Fixation of the tissue was performed at room temperature for 2–4 h.
Following fixation, the cap was unscrewed and lifted up to remove
the screw cap-rack assembly with the tissue cassette from chamber
1. The rack-tissue cassette was then placed directly above chamber
2 which contained the PAXgene Tissue Stabilizer and submerged with
the cap screw tightened. The PAXgene Tissue Container was then
stored at −80°C until required for further experimentation.
DNA and RNA extraction
Approximately 20 mg tissue specimens were harvested
by placing them in 350 µl buffer cytotoxic T-lymphocyte with
β-mercaptoethanol (GeneAll Biotechnology Co., Ltd., Seoul, South
Korea) followed by complete homogenizing with a handheld
rotor-stator TissueRuptor (Qiagen GmbH, Hilden, Germany) for 30
sec. The lysates were left to incubate for 10 min at room
temperature, allowing the nucleoprotein complexes to completely
dissociate. The lysates were then centrifuged at 10,000 × g for 3
min and the supernatant transferred to a mini spin column. The mini
spin column containing the supernatant was then centrifuged at
≥10,000 × g for 30 sec at room temperature. The pass-through was
used for total RNA purification while the spin column with the DNA
bound to the membrane was transferred to a new collection tube and
used for DNA purification. The pass-through in the previous
collection tube (for total RNA purification) was mixed with an
equal volume of 70% ethanol prior to being transferred to another
new mini spin column (type W, blue ring). A series of
centrifugation steps for total RNA purification were performed
(Allspin™ Total DNA/RNA Purification kit; GeneAll Biotechnology
Co., Ltd., Seoul, Korea) according to the manufacturer's protocol.
The purified RNA bound to the membrane was then eluted with 50 µl
nuclease-free water. The mini column containing the membrane bound
DNA also underwent a series of centrifugation steps for total DNA
purification and was eluted in 100 µl nuclease-free water. DNA was
stored at −20°C until further use.
DNA analysis
Concentration and purity of the DNA was analyzed
using the Spectrophotometer NanoDrop ND-1000 (Thermo Fisher
Scientific, Inc., Waltham, MA, USA). The integrity of the genomic
DNA was analyzed using the Agilent Genomic DNA ScreenTape System
(Agilent Technologies, Inc., Santa Clara, CA, USA). The DNA
concentration ranged from 205.6 to 582.6 ng/µl while the mean DNA
integrity number was 8.28 with a standard deviation of 1.06.
RNA analysis
Concentration and purity of the RNA was analyzed
similar to that for DNA. The integrity of the RNA was analyzed
using the Agilent 2100 BioAnalyzer RNA 6000 Nano Chip platform
(Agilent Technologies, Inc.). The RNA concentration ranged from
87.6 to 894.4 ng/µl while the mean RNA integrity number was 7.15
with a standard deviation of 0.46.
aCGH
The DNA obtained from tissue samples were then
investigated for genetic aberrations by aCGH using the Cytosure
4×180K oligonucleotide array (Agilent Technologies, Inc.). For each
sample, 18 µl genomic DNA was used. The CytoSure Genomic DNA aCGH
labeling kit (Agilent Technologies, Inc.) was used to label the
reference and tumor DNA with Cy-3 and Cy-5 respectively. The
labeled targets were then purified according to the manufacturer's
protocol with a minimum labeling efficiency of 250 pmol/µg DNA for
Cy3 and 200 pmol/µg DNA for Cy-5. The purified Cy3 and Cy5-labelled
genomic DNA was then mixed with human Cot-1 DNA, blocking agent and
hybridization buffer, and then was denatured and incubated at 37°C
for 30 min. A total of 100 µl sample mix was then pipetted into the
hybridization chamber containing the microarray and placed in a
rotisserie hybridization oven at 65°C for 40 h. Subsequent to
hybridization, the slides were washed with 2 wash buffers, Wash
Buffer 1 for 5 min at room temperature and Wash Buffer 2 for 1 min
at 37°C. The slides were then scanned using the Agilent DNA scanner
to produce the raw image data. This data was further processed
using the Agilent Feature Extraction software (Agilent
Technologies, Inc.) and the results analyzed with the Cytosure
Interpret software v3.4.2 (Agilent Technologies, Inc.).
NGS-RNA quality control, library
construction and sequencing
RNA samples were analyzed using deep transcriptome
sequencing (Theragen Etex Bio Institute, Suwon, Gyeonggi, South
Korea). A total of 1 µg total RNA was mixed with oligo dT magnetic
beads to enrich for poly(A) mRNA. The purified mRNAs were disrupted
into short fragments and cDNAs were synthesized. The cDNAs were
subjected to end-repair, poly(A) addition and connected with
sequencing adapters using the TruSeq RNA Sample Preparation kit
(Illumina, Inc., San Diego, CA, USA). The suitable fragments were
purified using the BluePippin 2% agarose gel cassette (Sage
Science, Inc., MA, USA) and were selected as templates for
polymerase chain reaction amplification. The final library size and
quality were evaluated electrophoretically with an Agilent High
Sensitivity DNA kit (Agilent Technologies, Inc.) with fragment
sizes between 350–450 base pairs. The library was then sequenced
using an Illumina HiSeq 2500 sequencer (Illumina, Inc.).
Transcriptome data analysis
Filtering
The quality of reads were checked using the fast QC
tool (v.0.10.1) (21) for basic
sequence quality score, GC content, N content, length distribution
and duplication levels. Low quality reads were filtered according
to the following criteria: Reads containing >10% of skipped
bases (marked as ‘N's), reads containing >40% of bases whose
quality scores were <20 and reads whose mean quality scores for
each read was <20. The entire filtering process was performed
using in-house scripts.
Sequence alignment
Filtered reads were mapped to the human reference
genome (obtained from the online genome database Ensembl release
72) (22) using the aligner Tophat
(23).
Gene expression estimation
Gene expression levels were measured using Cufflinks
v2.1.1 (24,25) using the gene annotation database of
Ensembl release 72. Non-coding gene regions were removed using the
mask option. In order to improve the accuracy of measurements,
multi-read and fragment bias corrections were applied. All other
options were set to default values.
Results
aCGH
aCGH was performed on 3 oligodendroglioma samples, 1
of which was a low-grade oligodendroglioma (WHO Grade II; sample
1090) and 2 anaplastic oligodendroglioma (WHO Grade III; samples
193 and 1395). The purpose of this was to determine any known and
novel amplifications and/or deletions in any of the chromosomes in
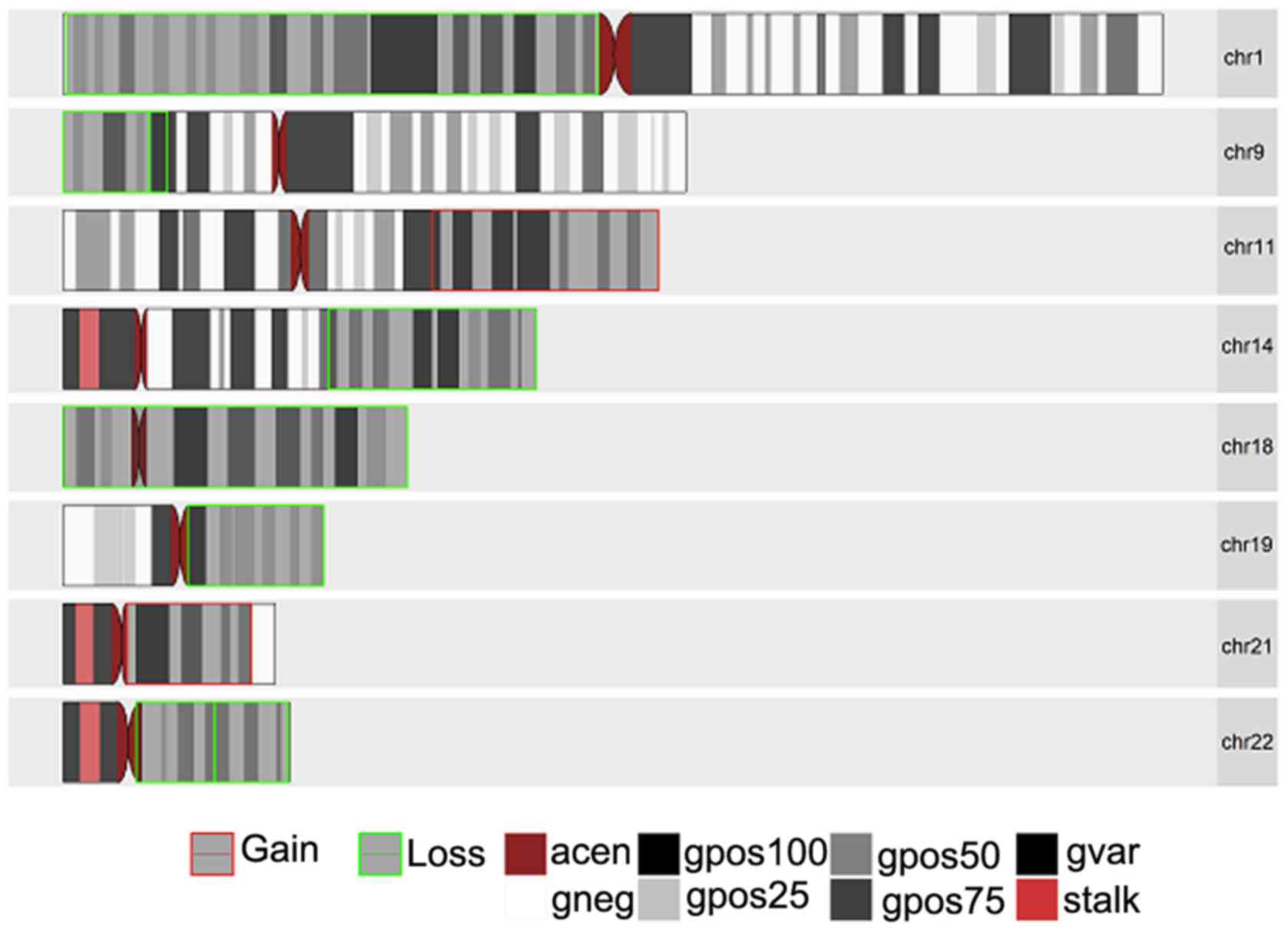
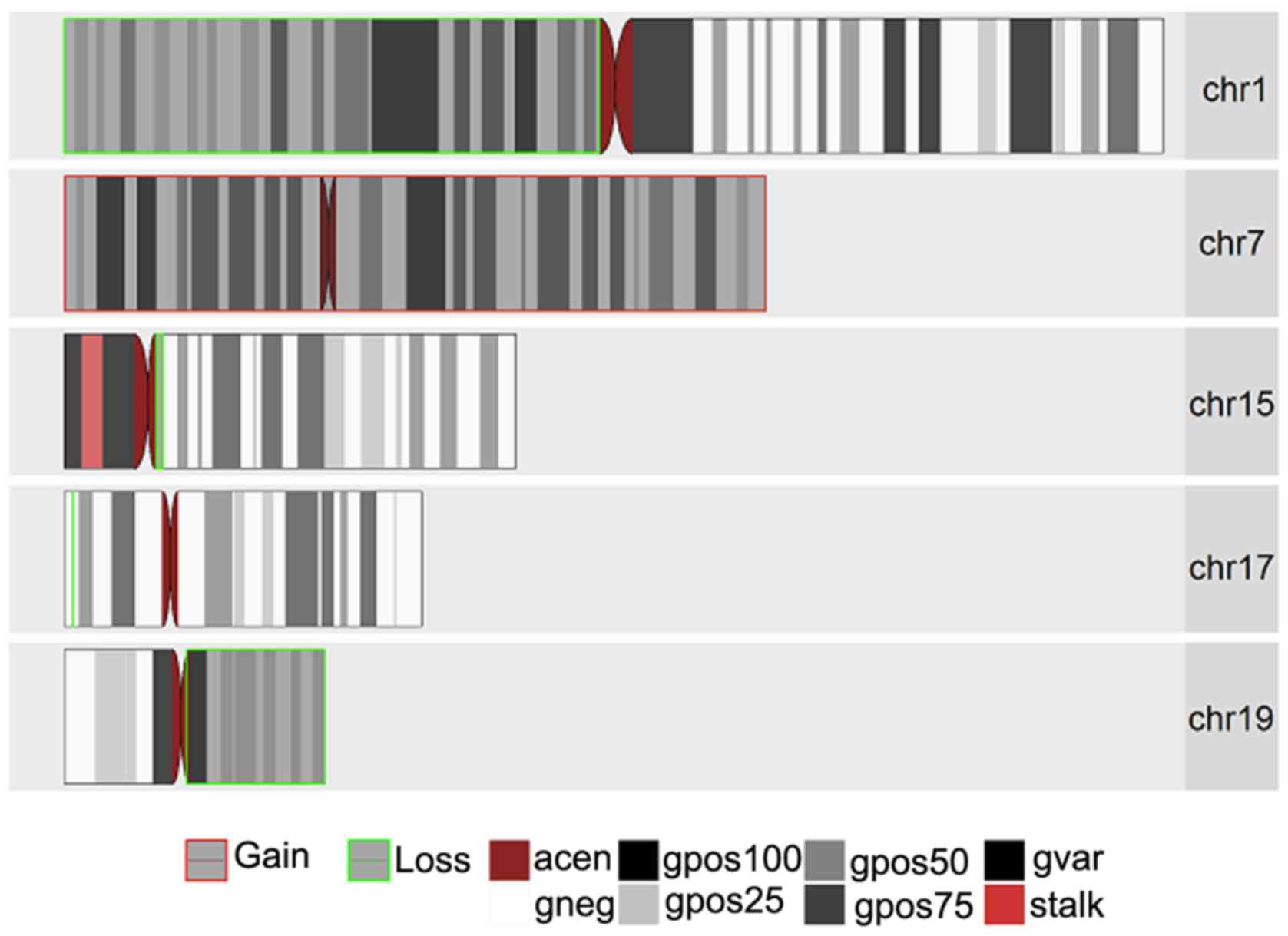
these samples. Figs. 1–3 present the chromosomal aberrations,
particularly the amplifications and deletions present in all 3
samples. Sample 1395, by far, had the greatest number of
chromosomal aberrations and sample 193 had the greatest number of
deletions out of the 3 samples.
The most common and notable losses were identified
on the 1p chromosomal arm, which was present in all 3 samples. This
was followed by a loss in chromosome 19q present only in the 2
anaplastic oligodendroglioma samples and a whole set of
amplification on chromosome 7 present in samples 1395 and 1090. In
addition, there was a partial loss in chromosome 14q (samples 193
and 1395) and deletions in chromosomes 15q (in samples 1395 and
1090). It was interesting to note that while there was a deletion
in chromosome 17p in samples 1395 and 1090, sample 1395 had an
extra mutation in the form of amplification on chromosomal arm 17q.
Another interesting result was observed on chromosome 11 for
samples 193 and 1395 with a partial amplification in the q arm for
sample 193 and a complete deletion of the p arm in sample 1395.
Less common losses were also observed in chromosomes
2p, 2q, 4p, 4q, 8p, 9p, 10q, 13q, 18 (whole chromosomal arm) and
22q. Excluding chromosomes 7,11q and 17q, losses were also
identified in chromososmes 12p, 12q, 20q and 21q.
Copy number variations (CNVs) in
anaplastic oligodendroglioma
Using aCGH for the genome-wide profiling of
oligodendroglioma not only revealed chromosomal alterations within
the tumor samples, but also allowed for the detection of genomic
CNVs. Based on the examination of sample 193, notable deletions in
the majority of the chromosomes associated with amplifications were
identified in the tumor sample. Chromosome 1p contained the largest
loss in its chromosomal arm of 120.63 Mb involving 2540 gene
overlaps. This was followed by chromosome 19q with a deletion of
1599 overlapping genes. Notable losses were also observed on
chromosomes 9p, 14q, 18, 19q and 22q with deletions of 303, 1172,
1032, 1599 and 1129 overlapping genes, respectively. There were an
additional 918 genes that were amplified on chromosome 11q and 484
genes amplified on chromosome 21q.
Two anaplastic oligodendroglioma samples (samples
193 and 1395) were compared to determine if there were any CNV
overlaps. The two samples had a common CNV at the same start and
stop positions (10478 and 121350692). Sample 1395 had a matching
region deleted that spanned 29663113 base pairs in chromosome 1, a
matching region that was amplified spanning 159022882 base pairs on
chromosome 7 and another matching deleted region spanning 1087 base
pairs on chromosome 17. For sample 193, there was a matching region
of 120627374 base pairs that was deleted in chromosome 1 and
another region of 30856201 base pairs that was deleted on
chromosome 19.
Gene ontology
A number of gene functions linked to the chromosomal
aberrations in the tumor cells were selected, particularly with
regards to immune regulation. This included deletions in genes
involved in the regulation of type I interferon-mediated signaling
pathway, T-cell activation, complement activation of the classical
pathway, circulating immunoglobulin complexes, immunoglobulin
receptor binding and antigen binding. The genes involved in the
amplification of certain chromosomes were not as common, however
they included functions in the regulation of interleukin-1 β
secretion, keratinization, cell-cell junction, intermediate
filament and endopeptidase activity. All overlapping genes involved
in chromosomal aberrations (amplifications/deletions) are listed in
Table II.
 | Table II.GO associated with chromosomal
aberrations. |
Table II.
GO associated with chromosomal
aberrations.
| A, List of
overlapping genes involved in loss/deleted regions |
|---|
|
|---|
| GO category | GO name | No. of genes | P-value | Term ID |
|---|
| BP | Regulation of type
I interferon-mediated signaling pathway | 18 |
1.68×10−04 | GO:0060338 |
| BP | T-cell activation
involved in immune response | 24 |
2.72×10−02 | GO:0002286 |
| BP | Defense response to
other organisms | 94 |
3.46×10−06 | GO:0098542 |
| BP | Positive regulation
of peptidyl-serine phosphorylation of signal transducer and
activator of transcription protein | 17 |
8.33×10−11 | GO:0033141 |
| BP | Response to
exogenous double stranded RNA | 19 |
5.93×10−05 | GO:0043330 |
| BP | Complement
activation, classical pathway | 67 |
3.01×10−29 | GO:0006958 |
| CC | Extracellular
region | 569 |
2.73×10−3 | GO:0005576 |
| CC | Blood
microparticle | 50 |
9.79×10−08 | GO:0072562 |
| CC | Immunoglobulin
complex, circulating | 37 |
9.21×10−22 | GO:0042571 |
| CC | External side of
plasma membrane | 52 |
9.21×10−3 | GO:0009897 |
| MF | Metal ion
binding | 489 |
7.15×10−4 | GO:0046872 |
| MF | Immunoglobulin
receptor binding | 37 |
1.76×10−20 | GO:0034987 |
| MF | Transcription
factor activity, sequence-specific DNA binding | 189 |
1.12×10−06 | GO:0003700 |
| MF | DNA binding | 343 |
5.49×10−09 | GO:0003677 |
| MF | Type I interferon
receptor binding | 16 |
3.25×10−12 | GO:0005132 |
| MF | Antigen
binding | 92 |
1.98×10−39 | GO:0003823 |
| MF | Serine-type
endopeptidase activity | 55 |
3.41×10−05 | GO:0004252 |
|
| B, List of
overlapping genes involved in gain/amplified regions |
|
| GO
category | GO name | No. of
genes | P-value | Term ID |
|
| BP | Regulation of
interleukin-1 β secretion | 9 |
4.61×10−4 | GO:0050706 |
| BP | Keratinization | 30 |
3.95×10−11 | GO:0031424 |
| BP | Multicellular
organismal catabolic process | 10 |
3.23×10−2 | GO:0044243 |
| CC | Cell-cell
junction | 26 |
2.65×10−2 | GO:0005911 |
| CC | Intermediate
filament | 33 |
2.45×10−15 | GO:0005882 |
| MF | Odorant
binding | 11 |
1.56×10−2 | GO:0005549 |
| MF | Endopeptidase
activity | 30 |
4.07×10−3 | GO:0004175 |
| MF | Interleukin-10
receptor activity | 4 |
1.50×10−2 | GO:0004920 |
NGS
High-throughput RNA sequencing was performed on all
6 oligodendroglioma samples. The NGS analysis of sample 193 only
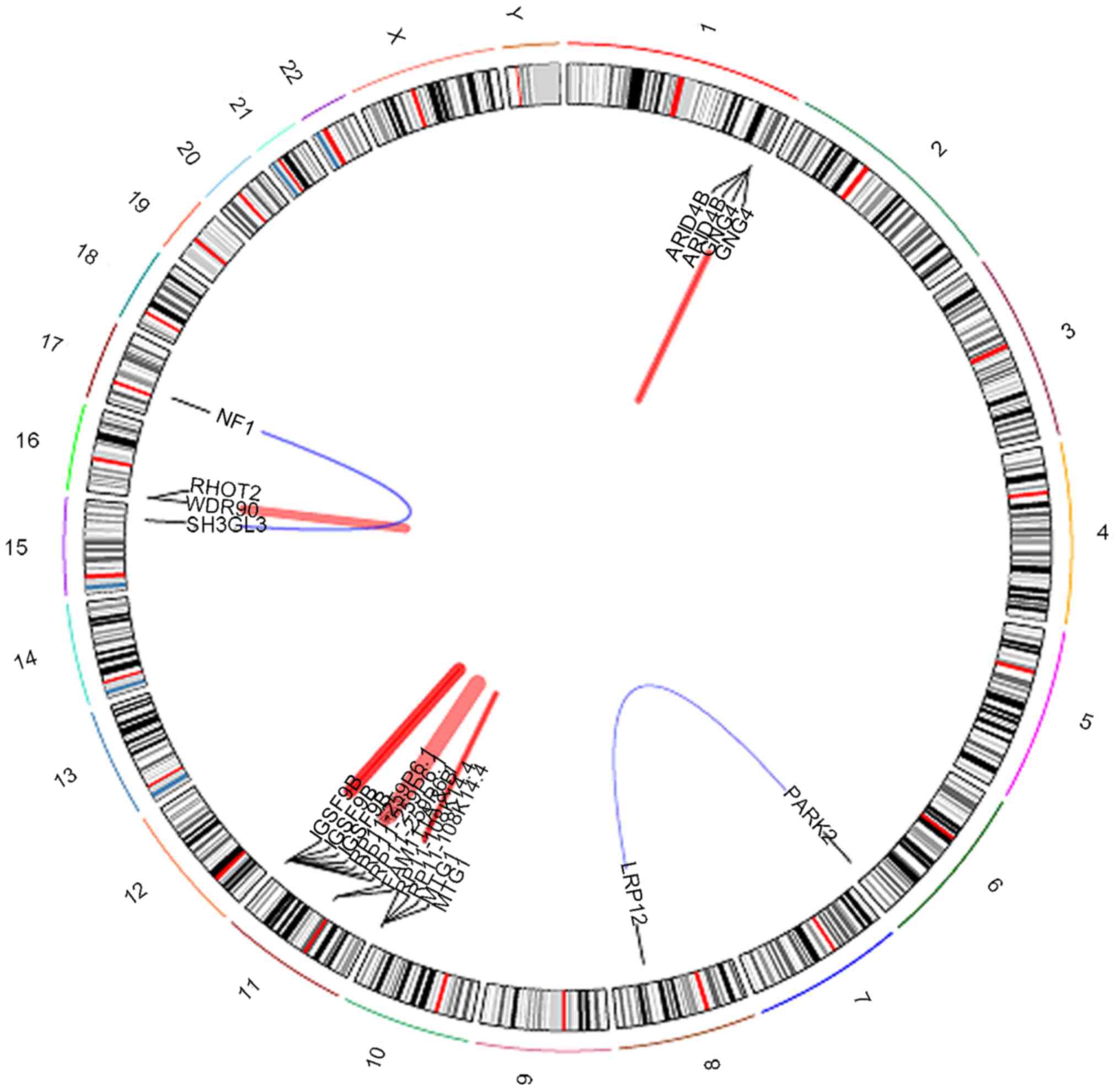
revealed a few notable gene fusions. Data revealed five novel
fusion genes (Fig. 4).
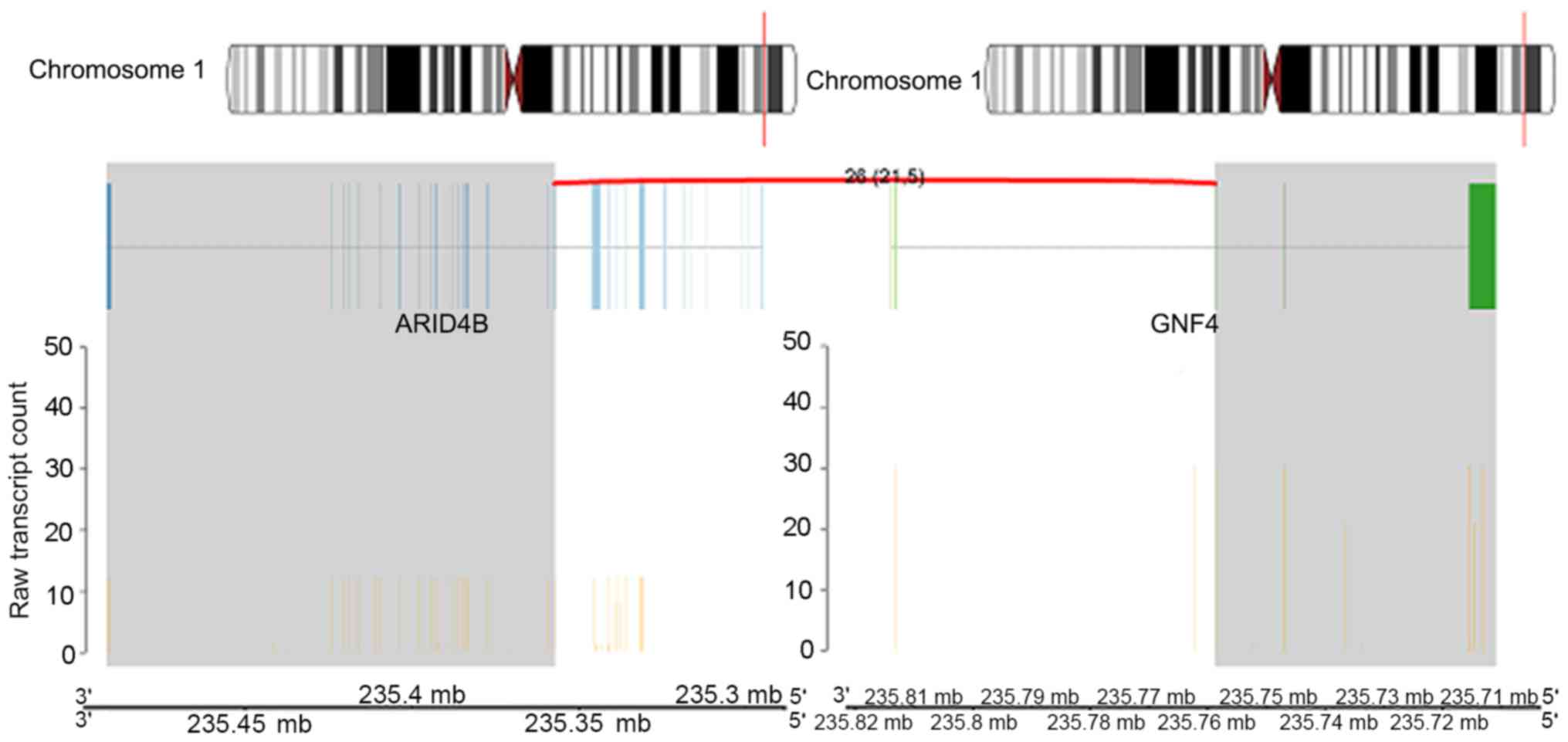
Intra-chromosomal rearrangements were identified on chromosome
1q42.3 between the AT-rich interaction domain 4B (ARID4B) and G
protein subunit γ 4 (GNG4) genes (Fig.
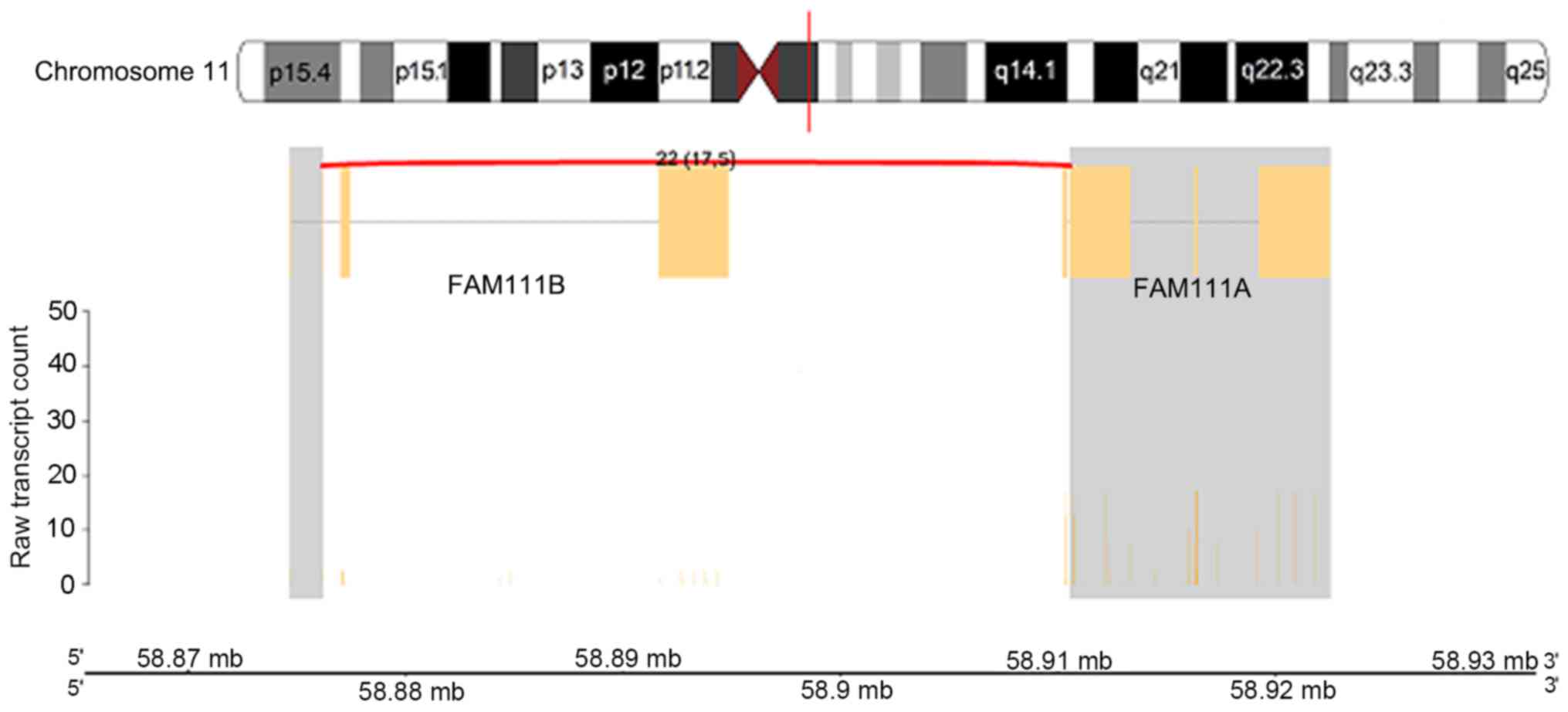
5), chromosome 11q12.1 between the family with sequence
similarity 111 member B (FAM111B) and FAM111A genes (Fig. 6) and on chromosome 16p13.3 between the
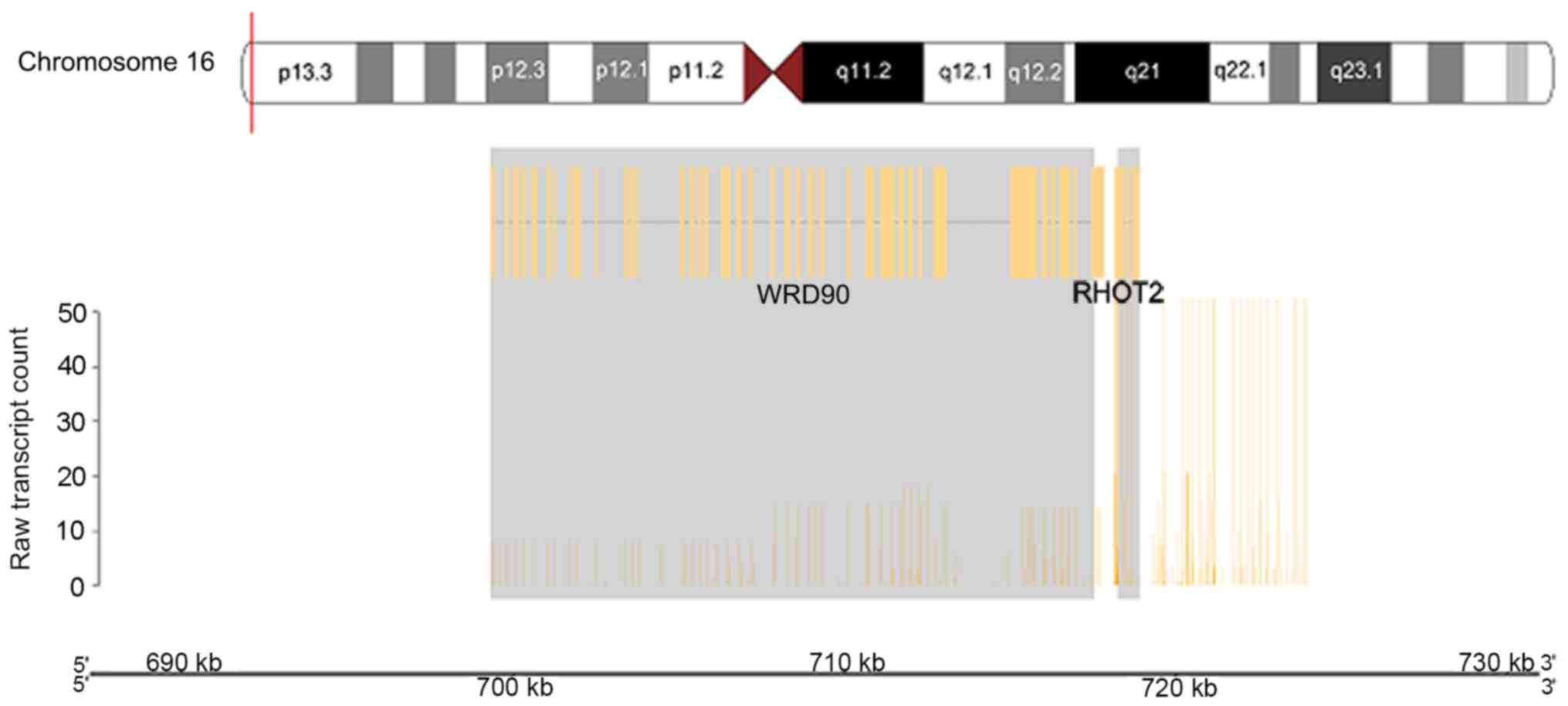
WD repeat domain 90 (WDR90) and ras homolog family member T2
(RHOT2) genes (Fig. 7).
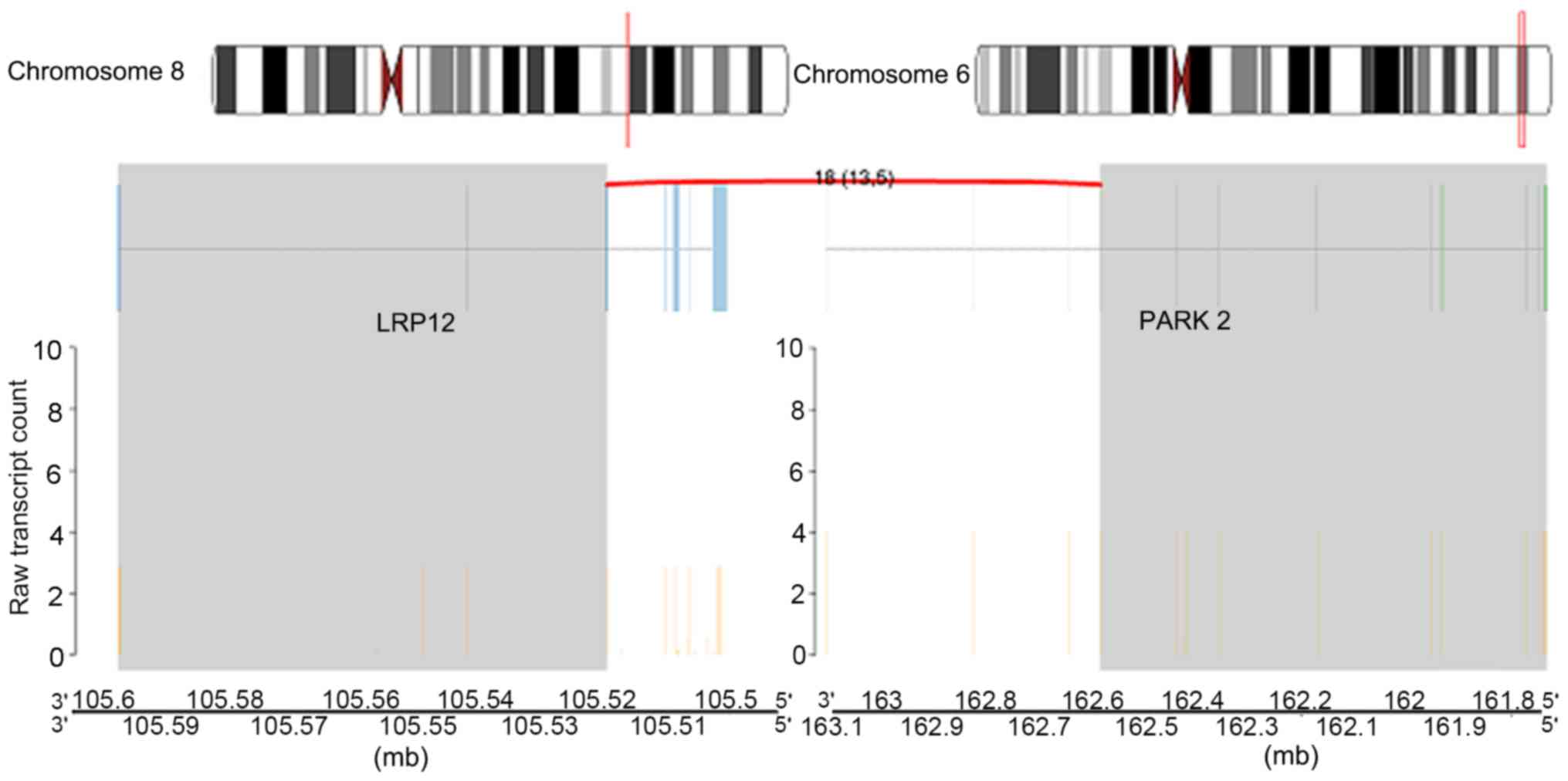
Inter-chromosomal rearrangements were identified between genes LDL
receptor related protein 12 (LRP12; on chromosome 8q22.3) and
parkin RBR E3 ubiquitin protein ligase (PARK2; on chromosome 6q26;
Fig. 8) and additionally between
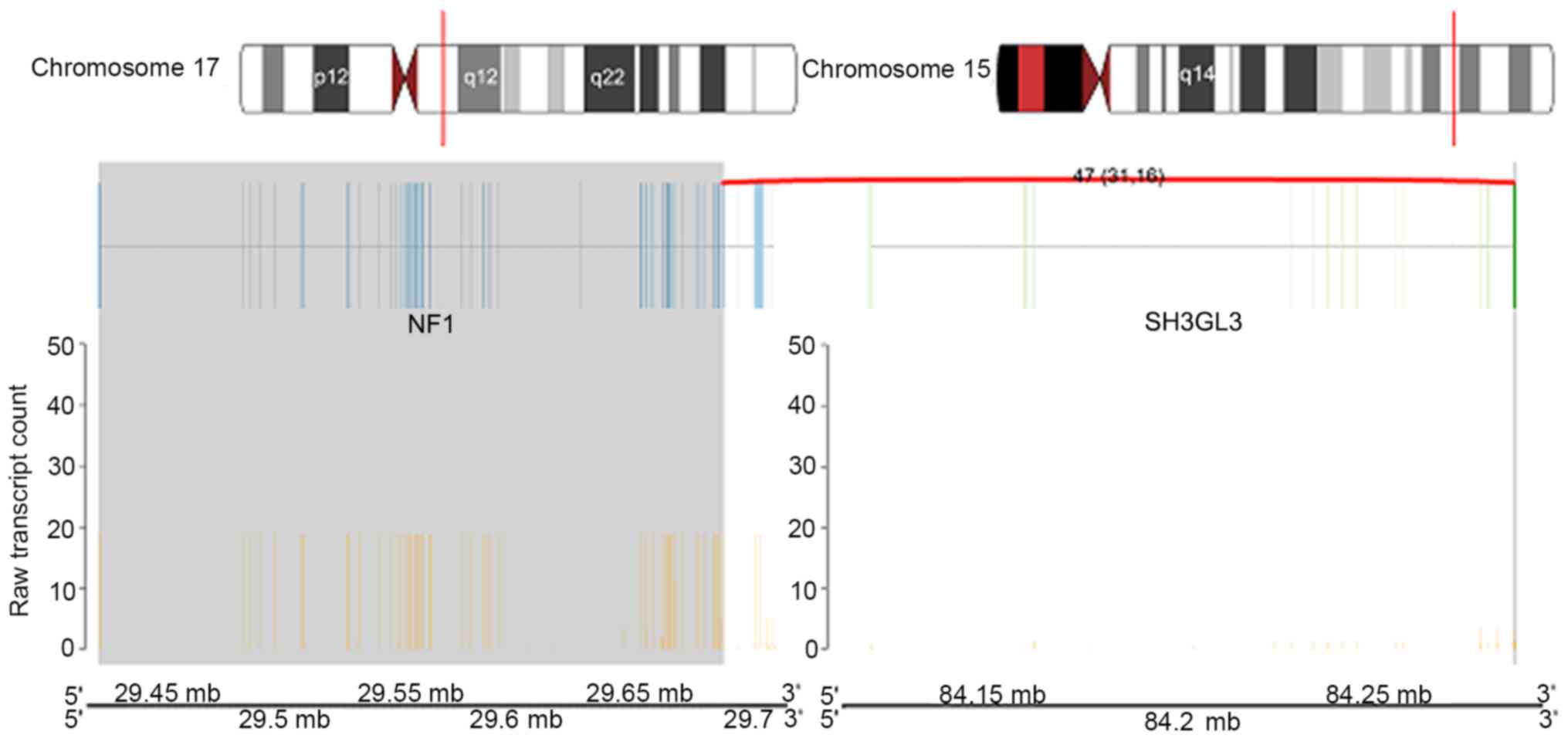
genes neurofibromin 1 (NF1) and SH3 domain containing GRB2 like 3,
endophilin A3 (SH3GL3) which mapped to chromosomes 17q11.2 and
15q25.2, respectively (Fig. 9).
The fusion events occurring in the majority of the
chromosomes in Sample 193 appeared to have different activity
levels. For the ARID4B_GNG4 fusion gene, the activity for GNG4 was
higher compared with ARID4B. This was also observed in the
FAM111B_FAM111A gene fusion where the activity of the FAM111A
isoform was higher compared with the FAM111B isoform. The most
notable difference in activity was observed in the fusion genes
WDR90_RHOT2 and NF1_SH3GL3, where RHOT2 and NF1 activity were
substantially higher compared with their respective pairings. On
the other hand, there was no notable difference in the activity for
the fusion gene pair LRP12_PARK2.
Discussion
In the present study, a comparative genomic analysis
of oligodendroglioma (WHO Grade II) and anaplastic
oligodendroglioma (WHO Grade III) samples was undertaken, using
aCGH and NGS. The aCGH results revealed that the most common
deletion present in oligodendroglioma samples were on chromosomes
1p followed by 19q. This is in keeping with previous studies that
have presented similar results (26,27). This
is not surprising as LOH 1p 19q is a predictor of radio- and
chemosensitivity in patients with improved survival (28,29).
Aberrations were identified in the majority of the
other chromosomes and included amplification/gain or deletion/loss.
This reiterates the fact that genetic alterations are common in
solid tumor types, including tumors of the central nervous system
(30–32).
Genetic alterations including CNVs are more commonly
observed in high-grade tumor types compared with low-grade tumor
types (33). Comparison between 2
anaplastic oligodendroglioma samples, namely samples 1395 and 193,
revealed that there was a large region of CNV overlap of >121
million base pairs. This suggests that high-grade tumor types
originating from a similar cell type have not only heterogeneity
but also substantial homogeneity.
Gene ontology analysis of the gene functions
associated with the chromosomal aberrations in the tumor cells
indicate that a large number of these genes are involved in the
innate and adaptive immune system. This included deletions in genes
involved in the regulation of the type I interferon-mediated
signaling pathway, T-cell activation, complement activation of the
classical pathway, circulating immunoglobulin complexes,
immunoglobulin receptor binding and antigen binding. This suggests
that the modulation of the immune system serves a critical role in
tumor response.
NGS analysis revealed 5 novel fusion genes in sample
193 (anaplastic oligodendroglioma). To the best of our knowledge,
this is the first study to have identified the presence of 5 novel
fusion genes in one anaplastic oligodendroglioma sample with 3
intra-chromosomal and 2 inter-chromosomal fusions. Fusion genes are
not uncommon in gliomas. The fibroblast growth factor receptor
3_transforming acidic coiled-coil containing protein 3 gene present
in patients with glioblastoma has been revealed to recur at a rate
of up to 8.3%. It is postulated to be involved in the extracellular
signal-regulated kinase (ERK) and signal transducer and activator
of transcription 3 (STAT3) signaling pathways, promoting tumor
growth and formation (34,35). Another fusion gene that is common is
KIAA1549_B-Raf proto-oncogene, serine threonine kinase (BRAF) which
occurs in 70% of patients diagnosed with pilocytic astrocytoma. A
mutation in the BRAF gene has been observed in a number of cancer
types, and is not exclusive to gliomas (36).
The 3 intra-chromosomal fusions identified were
ARID4B_GNG4, FAM111B_FAM111A and WDR90_RHOT2. ARID4B, also known as
retinoblastoma-binding protein-1-like protein-1 or breast
cancer-associated antigen 1, belongs to the ARID gene family and is
frequently expressed in human carcinomas and normal testis
(37). The gene family is involved in
regulating cell growth and mutations result in the loss of
inhibitory histone makers and DNA methylation (38). ARID4B in particular serves a dual role
in a tumor, where it contributes to the spread and invasive nature
of breast cancer cells; however, it also functions as a tumor
suppressor in leukemia (39,40). It has recently been reported to be
overexpressed in primary brain tumor types and therefore may be
able to function as an effective biomarker for and predictor of
tumor grade in meningioma and gliomas (41). GNG4, on the other hand, was recently
discovered to be hypermethylated and downregulated in GBM (42). It has been suggested that it may
function as a tumor suppressor gene in GBM through the inhibition
of the C-X-C motif chemokine ligand 12/C-X-C motif chemokine
receptor 4-dependent chemokine signaling pathway (42). Therefore, it is likely that a fusion
between the ARID4B and GNG4 genes has a role in the signaling
pathway that regulates DNA methylation.
FAM111B and FAM111A are members of the FAM111 family
of genes. Although the two genes contain a functional region called
a trypsin-like peptidase domain located close to each other, they
each have distinct functional properties (43). A mutated form of FAM111B protein is
thought to be involved in hereditary fibrosing poikiloderma,
characterised by mottled pigmentation, telangiectasia and atrophy
of the epidermis, tendon contractors and progressive pulmonary
fibrosis (43). Notably, it has also
been reported to be a cancer predisposition gene to pancreatic
cancer although a broader analysis and exploration is required to
confirm this (44). FAM111A is the
paralog of this gene and is associated with the diseases
Kenny-Caffey syndrome type 2 and osteocraniostenosis (45). It may also contribute to
carcinogenesis as it serves a role in cell cycle regulation and DNA
replication.
The activity of WDR90 was downregulated when fused
with RHOT2 in the anaplastic oligodendroglioma sample. WDR90 has
been thought to serve a role in signal transduction, transcription
regulation, cell cycle control, autophagy and apoptosis (46). However, RHOT2 was upregulated in the
tumor sample in the present study. The RHOT2 gene is part of the
Ras homolog gene family and encodes an enzyme called mitochondrial
Rho GTPase 2 (47). The enzyme is
thought to serve a role in mitochondrial trafficking and apoptosis
(47,48). RHOT2 has also been linked to cancer
progression, as it is upregulated in epithelial and hematological
tumor types (49). This may
corroborate with the hypothesis that an increase in RHOT2 may serve
a role in supporting tumor cell invasion in high-grade gliomas.
LRP12 encodes a protein named as the 12th member of
the low-density lipoprotein receptor related protein family.
Lipoproteins serve an important role in the development of the
brain. The low-density lipoprotein LRP12 has been postulated to be
a receptor for internalizing lipophilic molecules and may have a
potential function in signal transduction (50). Also known as suppressor of
tumorigenicity 7 protein, LRP12 has also been reported to be
differentially expressed in various cancer types including oral
squamous cell carcinoma (50),
lymphoma (51) and more recently
gangliogliomas and low-grade central nervous system tumor types
(52). PARK2 is a tumor suppressor
gene and when deleted is known to promote tumorigenicity. The
activation of PARK2 and its function in tumor growth is not fully
understood. However, the anti-apoptotic protein B-cell
lymphoma-extra large is thought to be involved to some extent in
the activation mechanism of PARK2 through regulation of the
permeability of the mitochondrial outer membrane (53). The fusion between these two genes may
imply the same manner by which high-grade gliomas including
anaplastic oligodendroglioma progress, although further
investigation is required in order to justify this.
The most notable fusion gene was NF1_SH3GL3. The NF1
gene is located on chromosome 17q11.2 and encodes the cytoplasmic
and multi-domain regions of neurofibromin, a protein that functions
as a tumor suppressor and negatively regulates Ras through the
mitogen-activated protein kinase (MAPK)- and mechanistic target of
rapamycin-pathway (54). A mutation
or inactivation of the gene will likely increase Ras activity
(55). Ras activation will induce a
series of cascades involved in regulating transcription,
translation, cell cycle progression, metabolic processes, immune
inflammatory responses and survival.
NF1 mutations are mostly identified in low-grade
gliomas, particularly with regards to the optic pathway (56). However the mutation may also be
observed in higher-grade gliomas commonly during adult life
(57). In the majority of cases,
there are mutations that occur in additional genes together with
NF1, including TP53 and cyclin dependent kinase inhibitor 2A
(58,59). This is clearly observed in the
anaplastic oligodendroglioma sample 193 that has multiple gene
amplifications and deletions, along with gene fusions in different
chromosomes.
Currently, there are 5 known NF1 fusion genes based
on The Cancer Genome Atlas RNA-Seq analysis data that are involved
in certain cancer types including oesophageal carcinoma, lung
adenocarcinoma and ovarian cancer (60). Based on the Ensembl genome browser,
there are 23 alternative transcripts that have been discovered from
gene splicing. However, the full significance of the NF1 alteration
has yet to be elucidated for this particular glioma subtype and
requires further study. One previous report revealed a linkage
between NF1 gains in a subset of diffuse gliomas, whilst NF1 loss
is restricted to only a small subset of GBM (60). In the case of anaplastic
oligodendroglioma, there has only been one case report that
suggested an association between a somatic mutation in NF1 and the
tumor (61).
The SH3GL3 gene is a member of a family of genes
highly expressed in the central nervous system (62). More specifically, the gene becomes
enriched in myelinating and newly formed oligodendrocytes. The
carboxyl and amino-terminal of the Src-homology-3 domain (SH3)
binds to proline-rich ligands and is involved in signal
transduction pathways for protein-protein interaction. The disease
most frequently associated with the SH3GL3 gene is Huntington's
disease (HD) (63). The SH3 domain
binds to the proline-rich region in the first exon of the HD gene,
which contributes to the production of high levels of insoluble
polyglutamine-containing aggregates in the brain. These aggregates
interfere with cellular functions, resulting in cellular death and
neuronal degradation that are atypical in HD (63). SH3GL3 mutations have been implicated
in a number of cancer types including colorectal cancer, multiple
myeloma and breast cancer (64–66).
Recently, it has been suggested that SH3GL3 is one of three novel
genes expressed in infiltrating glioma cells and is likely a key
component involved in the invasive nature of malignant gliomas
(67).
The growth factor receptor bound protein (GRB2), via
its SH3 domain, serves a key role in the Ras/MAPK pathway. GRB2
facilitates Ras activation via the co-localisation of SOS Ras/Rac
guanine nucleotide exchange factor 1 and Ras, which in turn
activates MAPK/e mitogen-activated protein kinase (MEK). This then
activates MAPK/ERK phosphorylation through Raf activation,
promoting cell proliferation and survival (68). A mutation or deletion/amplification in
part of the SH3 domain or GRB2 adaptor protein would affect the
Ras/Raf/MEK/ERK cascade, affecting cellular responses including
cell cycle progression and potentially contributing to the spread
of solid tumor types through local invasion and metastasis, for
example in oligodendroglioma. Its signaling has been reported to be
linked with several human diseases including breast cancer and
chronic myelogenous leukaemia (36).
Another paralogous gene, SH3GL2, has already been
reported to be downregulated in glioblastoma cells (69). Loss of SH3GL2 may increase the
activity of STAT3/matrix metalloproteinase-2 signaling and promote
the migration and invasion of high-grade glioma cells. It may be
fused to a part of the SH3 domain of GRB2, similar to the
interaction between NF2 and GRB2 signaling in neurofibromatosis
type 2 (70). However, it still
remains to be seen how the interaction between NF1 and SH3GL3 may
define a particular glioma cell type, for example anaplastic
oligodendroglioma.
In the present study, five novel fusion transcripts
were identified, the most notable of which was the identification
of the NF1_SH3GL3 fusion gene in an anaplastic oligodendroglioma
sample. To the best of our knowledge, this is the first report of
its type to identify the involvement of these five fusion genes in
the pathogenesis of oligodendroglioma. The functional consequences
of these fusions and their underlying mechanisms have yet to be
explored and require further investigation and validation.
Acknowledgements
The authors would like to thank Mr Ranganth
Gudimella (Sengenics, Kuala Lumpur, Malaysia) for help with the
bioinformatics and database analysis, Dr Zubaidah Zakaria (Cancer
Research Center, Institute for Medical Research) for helpful
comments and suggestions, the Brain Tumor Foundation of Canada for
providing some of the tissue samples, and the Director General of
Health Malaysia for permission to publish this study.
Funding
The present study was supported by the Ministry of
Health Malaysia (grant no. NMRR-14-599-19912) awarded to Dr Stephen
Navendran Ponnampalam.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contribution
SAH performed the majority of the experimental work.
SNP provided substantial contributions to the conception and design
of the study, coordinated the project and revised the manuscript
critically for important intellectual content. MNA performed some
of the experimental work.
Ethical approval
The present study received ethical approval from the
Medical Research Ethics Council of the Ministry of Health Malaysia
on 6th August 2014 [approval no. (6)
KKM/NIHSEC/P14-679] and was renewed annually until October
2016.
Patient consent for publication
All subjects involved in the present study provided
written informed consent to participate in this study.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Stewart BW and Wild CP: World cancer
report. 2014, International agency for research on cancer (IARC)
Publications; Lyon, France: 2014, http://www.iarc.fr/en/publications/books/wcr/index.php20–01.
2018
|
|
2
|
Ferlay J, Soerjomataram I, Ervik M,
Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D and
Bray F: GLOBOCAN 2012 v1.0, Cancer incidence and mortality
worldwide. IARC CancerBase no.11. IARC Press; Lyon: 2013
|
|
3
|
US Mortality Data: 2006. National Centre
for Health Statistics. Centres for Disease Control and Prevention.
2009.
|
|
4
|
Louis DN, Ohgaki H, Wiestler OD, Cavenee
WK, Burger PC, Jouvet A, Scheithauer BW and Kleihaus P: The 2007
WHO classification of tumors of the central nervous system. Acta
Neuropathol. 114:97–109. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhang X, Zhang W, Cao WD, Cheng G and
Zhang YQ: Glioblastoma multiforme: Molecular characterization and
current treatment strategy (Reviw). Exp Ther Med. 3:9–14. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Li J, Miao N, Liu M, Ciu W, Liu X, Shi X,
Qing S, Ma Y, Zhang W and Biekemituofu H: Clinical significance of
chromosome 1p/19q loss of heterozygosity and Sox17 expression in
oligodendrogliomas. Int J Clin Exp Pathol. 7:8609–8615.
2014.PubMed/NCBI
|
|
7
|
Kakkar A, Suri V, Jha P, Srivasta A,
Sharma V, Pathak P, Sharma MC, Sharma MS, Kale SS, Chosdol K, et
al: Loss of heterozygosity on chromosome 10q in glioblastomas, and
its association with other genetic alterations and survival in
Indian patients. Neurol India. 59:254–261. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hartmann C, Hentschel B, Tatagiba M,
Schramm J, Schnell O, Seidel C, Stein R, Reifenberger G, Pietsch T,
von Deimling A, et al: Molecular markers in low-grade gliomas:
Predictive or prognostic? Clin Cancer Res. 17:4588–4599. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Burger PC, Minn AY, Smith JS, Borell TJ,
Jedlicka AE, Huntley BK, Goldthwaite PT, Jenkins RB and Feuerstein
BG: Losses of chromosomal arms 1p and 19q in the diagnosis of
oligodendroglioma. A study of paraffin-embedded sections. Mod
Pathol. 14:842–853. 2001.
|
|
10
|
Haynes HR, Camelo-Piragua S and Kurian KM:
Prognostic and predictive biomarkers in adult and pediatric
gliomas: Toward personalized treatment. Front Oncol. 4:472014.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Gadji M, Fortin D, Tsanaclis AN and Drouin
R: Is the 1p/19q deletion a diagnostic marker of
oligodendrogliomas? Cancer Genet Cytogenet. 194:12–22. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Hartmann C, Meyer J, Balss J, Capper D,
Mueller W, Christians A, Felsberg J, Wolter M, Mawrin C, Wick W, et
al: Type and frequency of IDH1 and IDH2 mutations are related to
astrocytic and oligodendroglial differentiation and age: A study of
1,010 diffuse gliomas. Acta Neuropathol. 118:469–474. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Nayak A, Ralte AM, Sharma MC, Singh VP,
Mahapatra AK, Mehta VS and Sarkar C: p53 protein alterations in
adult astrocytic tumors and oligodendrgliomas. Neurol India.
52:228–232. 2004.PubMed/NCBI
|
|
14
|
Blakely J and Grossman S: Anaplastic
oligodendroglioma. Curr Treat Options Neurol. 10:295–307. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Mohapatra G, Betensky RA, Miller ER, Carey
B, Gaumont LD, Engler DA and Loius DN: Glioma test array for use
with formalin-fixed, paraffin-embedded tissue: Array comparative
genomic hybridization correlates with loss of heterozygosity and
fluorescence in situ hybridization. J Mol Diagn. 8:268–276. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Meyerson M, Gabriel S and Getz G: Advances
in understanding cancer genomes through second-generation
sequencing. Nat Rev Genet. 11:685–696. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Robinson K: Application of
second-generation sequencing to cancer genomics. Brief Bioinform.
11:524–534. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Patil V, Pal J and Somasundaram K:
Elucidating the cancer-specific genetic alteration spectrum of
glioblastoma derived cell lines from whole exome and RNA
sequencing. Oncotarget. 6:43452–43471. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhang Y, Chen K, Sloan SA, Bennet ML,
Scholze AR, O'Keefe S, Phatnani HP, Guarnieri P, Caneda C,
Ruderisch N, et al: An RNA-sequencing transcriptome and splicing
database of glia, neurons, and vascular cells of the cerebral
cortex. J Neurosci. 34:11929–11947. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhao Z, Meng F, Wang W, Wang Z, Zhang C
and Jiang T: Comprehensive RNA-seq transcriptomic profiling in the
malignant progression of gliomas. Sci Data. 4:1700242017.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Andrews S: FastQC: A quality control tool
for high throughput sequence data. http://www.bioinformatics.babraham.ac.uk/projects/fastqc15–06.
2017
|
|
22
|
Flicek P, Ahmed I, Amode MR, Barrell D,
Beal K, Brent S, Carvalho-Silva D, Clapham P, Coates G, Fairley S,
et al: Ensembl 2013. Nucleic Acids Res. 41:(Database Issue).
D48–D55. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Trapnell C, Pachter L and Salzberg SL:
Tophat: Discovering splice junctions with RNA-Seq. Bioinformatics.
25:1105–1111. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Trapnell C, Roberts A, Goff L, Pertea G,
Kim D, Kelley DR, Pimentel H, Salzburg SL, Rinn JL and Pachter L:
Differential gene and transcript expression analysis of RNA-seq
experiments with TopHat and Cufflinks. Nat Protoc. 7:562–578. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Trapnell C, Williams BA, Pertea G,
Mortazavi A, Kwan G, van Baren MJ, Salzburg SL, Wold BJ and Pachter
L: Transcript assembly and quantification by RNA-Seq reveals
unannotated transcripts and isoform switching during cell
differentiation. Nat Biotechnol. 28:511–515. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Bello MJ, Vaquero J, de Campos JM, Kusak
ME, Sarasa JL, Saez-Castresana J, Pestana A and Rey JA: Molecular
analysis of chromosome 1 abnormalities in human gliomas reveals
frequent loss of 1p in oligodendroglial tumors. Int J Cancer.
57:172–175. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Bello MJ, Leone PE, Vaquero J, de Campos
JM, Kusak ME, Sarasa JL, Pestaña A and Rey JA: Allelic loss at 1p
and 19q frequently occurs in association and may represent early
oncogenic events in oligodendroglial tumors. Int J Cancer.
64:207–210. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Cairncross JG, Ueki K, Zlatescu MC, Lisle
DK, Finkelstein DM, Hammond RR, Silver JS, Stark PC, Macdonald DR,
Ino Y, et al: Specific genetic predictors of chemotherapeutic
response and survival in patients with anaplastic
oligodendrogliomas. J Natl Cancer Inst. 90:1473–1479. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Baumann GS, Ino Y, Ueki K, Zlatescu MC,
Fisher BJ, Macdonald DR, Stitt L, Louis DN and Cairncross JG:
Allelic loss of chromosome 1p and radiotherapy plus chemotherapy in
patients with oligodendrogliomas. Int J Radiat Oncol Biol Phys.
48:825–830. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Hagel C, Krog B, Laas R and Stavrou DK:
Prognostic relevance of TP53 mutations, p53 protein, Ki-67 index
and conventional histologic grading in oligodendrogliomas. J Exp
Clin Cancer Res. 18:305–309. 1999.PubMed/NCBI
|
|
31
|
Korshunov A and Golanov A: The prognostic
significance of vascular endothelial growth factor (VEGF C-1)
immunoexpression in oligodendroglioma. An analysis of 91 cases. J
Neurooncol. 48:13–19. 2000. View Article : Google Scholar
|
|
32
|
Di Rocco F, Carroll RS, Zhang J and Black
PM: Platelet-derived growth factor and its receptor expression in
human oligodendrogliomas. Neurosurg. 42:341–346. 1998. View Article : Google Scholar
|
|
33
|
Iwabuchi H, Sakamoto M, Sakunaga H, Ma YY,
Carcangiu ML, Pinkel D, Yang-Feng TL and Gray JW: Genetic analysis
of benign, low-grade, and high-grade ovarian tumors. Cancer Res.
55:6172–6180. 1995.PubMed/NCBI
|
|
34
|
Parker BC, Annala MJ, Cogdell DE, Granberg
KJ, Sun Y, Ji P, Li X, Gumin J, Zheng H, Hu L, et al: The
tumorigenic FGFR3-TACC3 gene fusion escapes miR-99a regulation in
glioblastoma. J Clin Invest. 123:855–865. 2013.PubMed/NCBI
|
|
35
|
Singh D, Chan JM, Zoppoli P, Niola F,
Sullivan R, Castano A, Liu EM, Reichel J, Porrati P, Pellegatta S,
et al: Transforming fusions of FGFR and TACC genes in human
glioblastoma. Science. 337:1231–1235. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Jones DT, Kocialkowski S, Liu L, Pearson
DM, Bäcklund LM, Ichimura K and Collins VP: Tendem duplication
producing a novel oncogenic BRAF fusion gene defines the majority
of pilocytic astrocytomas. Cancer Res. 68:8673–8677. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Cao J, Gao T, Stanbridge EJ and Irie R:
RBP1L1, a retinoblastomabinding protein-related gene encoding an
antigenic epitope abundantly expressed in human carcinomas and
normal testis. J Natl Cancer Inst. 93:1159–1165. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Neidhart M: Chapter 14-DNA Methylation in
Growth Retardation. In DNA Methylation and Complex Human Disease.
Academic Press; Oxford: pp. 241–259. 2016, View Article : Google Scholar
|
|
39
|
Goldberger N, Walker RC, Kim CH, Winter S
and Hunter KW: Inherited variation in miR-290 expression suppresses
breast cancer progression by targeting the metastasis
susceptibility gene Arid4b. Cancer Res. 73:2671–2681. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Wu MY, Eldin KW and Beaudet AL:
Identification of chromatin remodeling genes Arid4a and Arid4b as
leukemia suppressor genes. J Natl Cancer Inst. 100:1247–1259. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Tsai WC, Hueng DY, Nieh S and Gao HW:
ARID4B is a good biomarker to predict tumour behaviour and decide
WHO grades in gliomas and meningiomas. J Clin Pathol. 70:162–167.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Pal J, Patil V, Mondal B, Shukla S, Hegde
AS, Arivazhagan A, Santosh V and Somasundaram K: Epigenetically
silenced GNG4 inhibits SDF1α/CXCR4 signaling in mesenchymal
glioblastoma. Genes Cancer. 7:136–147. 2016.PubMed/NCBI
|
|
43
|
Mercier S, Küry S, Shaboodien G, Hounier
DT, Khumalo NP, Bou-Hanna C, Bodak N, Cormier-Daire V, David A,
Faivre L, et al: Mutations in FAM111B cause hereditary fibrosing
poikiloderma with tendon contracture, myopathy, and pulmonary
fibrosis. Am J Hum Genet. 93:1100–1107. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Goussot R, Prasad M, Stoetzel C, Lenormand
C, Dollfus H and Lipsker D: Expanding phenotype of hereditary
fibrosing poikiloderma with tendon contractures, myopathy, and
pulmonary fibrosis caused by FAM111B mutations: Report of an
additional family raising the question of cancer predisposition and
a short review of early-onset poikiloderma. JAAD Case Rep.
3:143–150. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Unger S, Górna MW, Le Béchec A, Do
Vale-Pereira S, Bedeschi MF, Geiberger S, Grigelioniene G,
Horemuzova E, Lalatta F, Lausch E, et al: FAM111A mutations result
in hypoparathyroidism and impaired skeletal development. Am J Hum
Genet. 92:990–995. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Stirnimann CU, Petsalaki E, Russell RB and
Müller CW: WD40 proteins propel cellular networks. Trends Biochem
Sci. 35:565–574. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Fransson A, Ruusala A and Aspenström P:
Atypical Rho GTPases have roles in mitochondrial homeostasis and
apoptosis. J Biol Chem. 278:6495–6502. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Fransson S, Ruusala A and Aspenström P:
The atypical Rho GTPases Miro-1 and Miro-2 have essential roles in
mitochondrial trafficking. Biochem Biophys Res Commun. 344:500–510.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Caino MC, Seo JH, Aguinaldo A, Wai E,
Bryant KG, Kossenkov AV, Hayden JE, Vaira V, Morotti A, Ferrero S,
et al: A neuronal network of mitochondrial dynamics regulates
metastasis. Nat Commun. 7:137302016. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Garnis C, Coe BP, Zhang L, Rosin MP and
Lam WL: Overexpression of LRP12, a gene contained within an 8q22
amplicon identified by high-resolution array CGH analysis of oral
squamous cell carcinomas. Oncogene. 23:2582–2586. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Bethge N, Honne H, Andresen K, Hilden V,
Trøen G, Liestøl K, Holte H, Delabie J, Lind GE and Smeland EB: A
gene panel, including LRP12, is frequently hypermethylated in major
types of B-cell lymphoma. PLoS One. 9:e1042492014. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Robens BK, Gembé E, Fassunke J, Becker AJ,
Schoch S and Grote A: Abundance of LRP12 C-rs9694676 allelic
promoter variant in epilepsy-associated gangliogliomas. Life Sci.
155:70–75. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Gong Y, Schumacher SE, Wu WH, Tang F,
Beroukhim R and Chan TA: Pan-cancer analysis links PARK2 to
BCL-XL-dependent control of apoptosis. Neoplasia. 19:75–83. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Andersen LB, Ballester R, Marchuk DA,
Chang E, Gutmann DH, Saulino AM, Camonis J, Wigle M and Collins FS:
A conserved alternative splice in the von Recklinghausen
neurofibromatosis (NF1) gene produces two neurofibromin isoforms,
both of which have GTPase-activating protein activity. Mol Cell
Biol. 13:487–495. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Banerjee S, Crouse NR, Emnett RJ, Gianino
SM and Gutmann DH: Neurofibromatosis-1 regulates mTOR-mediated
astrocyte growth and glioma formation in a TSC/Rheb-independent
manner. Proc Natl Acad Sci USA. 108:15996–16001. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Szudek J, Birch P, Riccardi VM, Evans DG
and Friedman JM: Associations of clinical features in
neurofibromatosis 1 (NF1). Genet Epidemiol. 19:429–439. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Gutmann DH, Rasmussen SA, Wolkenstein P,
MacCollin MM, Guha A, Inskip PD, North KN, Poyhonen M, Birch PH and
Friedman JM: Gliomas presenting after age 10 in individuals with
neurofibromatosis type 1 (NF1). Neurology. 59:759–761. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Nielsen GP, Stemmer-Rachamimov AO, Ino Y,
Moller MB, Rosenberg AE and Louis DN: Malignant transformation of
neurofibromas in neurofibromatosis 1 is associated with CDKN2A/p16
inactivation. Am J Pathol. 155:1879–1884. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Legius E, Dierick H, Wu R, Hall BK,
Marynen P, Cassiman JJ and Glover TW: TP53 mutations are frequent
in malignant NF1 tumors. Genes Chromosomes Cancer. 10:250–255.
1994. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Vizcaíno MA, Shah S, Eberhart CG and
Rodriguez FJ: Clinico-pathologic implications of NF1 gene
alterations in diffuse gliomas. Human Pathology. 46:1323–1330.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Bruzek AK, Zureick AH, McKeever PE, Garton
HJL, Robertson PL, Mody R and Koschmann CJ: Molecular
characterization reveals NF1 deletions and FGFR1-activating
mutations in a pediatric spinal oligodendroglioma. Pediatr Blood
Cancer. 64:e263462017. View Article : Google Scholar
|
|
62
|
Giachino C, Lantelme E, Lanzetti L,
Saccone S, Bella Valle G and Migone N: A Novel SH3-containing human
gene family preferentially expressed in the central nervous system.
Genomics. 41:427–434. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Sittler A, Walter S, Wedemeyer N,
Hasenbank R, Scherzinger E, Eickhoff H, Bates GP, Lehrach H and
Wanker EE: SH3GL3 associates with the huntingtin exon 1 protein and
promotes the formation of polygln-containing protein aggregates.
Mol Cell. 2:427–436. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Huebner K, Kastury K, Druck T, Salcini AE,
Lanfrancone L, Pelicci G, Lowenstein E, Li W, Park SH, Cannizzaro
L, et al: Chromosome locations of genes encoding human signal
transduction adapter proteins, Nck (NCK), Shc (SHC1), and Grb2
(GRB2). Genomics. 22:281–287. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Fang WJ, Zheng Y, Wu LM, Ke QH, Shen H,
Yuan Y and Zheng SS: Genome-wide analysis of aberrant DNA
methylation for identification of potential biomarkers in
colorectal cancer patients. Asian Pac J Cancer Prev. 13:1917–1921.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Chen R, Zhao H, Wu D, Zhao C, Zhao W and
Zhou X: The role of SH3GL3 in myeloma cell migration/invasion,
stemness and chemo-resistance. Oncotarget. 7:73101–73113.
2016.PubMed/NCBI
|
|
67
|
Delic S, Lottmann N, Jetschke K,
Reifenberger G and Riemenschneider MJ: Identification and
functional validation of CDH11, PCSK6 and SH3GL3 as novel glioma
invasion-associated candidate genes. Neuropathol Appl Neurobiol.
38:201–212. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Aphrothiti JH, Gopa I and David B: Solit:
2-Intracellular Signaling, In Abeloff's Clinical Oncology (Fifth
edition). John EN, James OA, James HD, Michael BK and Joel ET:
Tepper, Content Repository Only; Philadelphia: pp. 22–39. 2014
|
|
69
|
Zhu Y, Zhang X, Wang L, Ji Z, Xie M, Zhou
X, Liu Z, Shi H and Yu R: Loss of SH3GL2 promotes the migration and
invasion behaviours of glioblastoma cells through activating the
STAT3/MMP2 signalling. J Cell Mol Med. 21:2685–2694. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Wiederhold T, Lee MF, James M, Neujahr R,
Smith N, Murthy A, Hartwig J, Gusella JF and Ramesh V: Magicin, a
novel cytoskeletal protein associates with the NF2 tumor suppressor
merlin and Grb2. Oncogene. 23:8815–8825. 2004. View Article : Google Scholar : PubMed/NCBI
|