Introduction
Oral cancer is considered to be part of the head and
neck group of cancers. Oral cancer may arise from any part the oral
cavity or oropharynx (1). Oral
squamous cell carcinoma (OSCC) is the predominant histological
type, accounting for ~95% of oral cancer cases in the USA in 2008
(2). Tobacco smoking, alcohol
consumption, poor oral hygiene and human papilloma virus (HPV)
infections are the main causes of OSCC (3,4).
Conventional oral examination has been the mainstay of oral cancer
screening for decades. This examination is able to detect tumors
located in the oral cavity from the early stages, but fails to
identify all premalignant oral lesions (5).
Surgery is the primary therapy for oral cancer,
especially for patients in advanced stages. Chemotherapy and
radiation therapy are often combined with surgical resection to
treat patients with advanced oral cancer (6). Nevertheless, the 5-year survival rate is
only ~50% (7). Therefore, the
mechanism underlying OSCC development must be elucidated, in order
to optimize treatment and improve patient survival.
MicroRNAs (miRNAs) are a type of non-protein coding
RNA, conserved in the genomes of animals and plants (8). miRNAs regulate numerous genes
post-transcriptionally by binding the 3′-untranslated regions
(UTRs) of targeted genes (9). A
previous study revealed numerous types of miRNA dysregulation in
cancer (10). In OSCC, multiple
miRNAs have been identified to be differentially expressed, which
indicates that miRNAs may be involved in the pathogenesis of OSCC
(11). Previous studies have
demonstrated that miR-545 serves inhibitory functions in pancreatic
ductal adenocarcinoma (12), lung
cancer (13) and hepatocellular
carcinoma (14) and the target genes
of miR-545 is retinoic acid-inducible gene-I (RIG-I) (12,14) and
cyclin D1 and CDK4 (13).
In the present study, the role of miR-545 in OSCC
was assessed in vitro and in vivo, aiming to identify
the underlying molecular mechanism of the pathogenesis of OSCC. We
hypothesize that miR-545 inhibits OSCC by targeting RIG-I.
Materials and methods
Tissue samples
A total of 20 OSCC tissue samples and their matched
adjacent normal tissues were acquired from the Department of
Stomatology, Shunde Hospital of Guangzhou Medical University
(Foshan, China) from September 2012 to July 2014. Matched adjacent
normal tissues were used as controls. The patient information is
listed as follows: Median age, 57 years; age range: 41–76 years;
sex ratio (M/F): 13/7. The senior pathologists of Shunde Hospital
of Guangzhou Medical University confirmed the pathological
diagnosis of all OSCC patients. All tissues were frozen immediately
and preserved in a −80°C freezer for further analysis, including
the detection of miR-545 and retinoic acid-inducible gene (RIG)-I
mRNA. Written informed consent was obtained from all patients and
the present study was approved by the Ethics Committees of
Guangzhou Medical University (Guangzhou, China).
Cell culture
Four OSCC cell lines (HSC2, HSC4, SAS, KON) were
acquired from the cell bank of the Chinese Academy of Medical
Sciences (Shanghai, China). One normal oral keratinocyte cell line
(HOK) was purchased from Applied Biological Material (Vancouver,
Canada). OSCC lines were cultured in Dulbecco's modified Eagle's
medium and Ham's F-12 medium (Sigma-Aldrich; Merck KGaA, Darmstadt,
Germany) with 10% fetal bovine serum (Gibco; Thermo Fisher
Scientific, Inc., Waltham, MA, USA). HOK cells were cultured in
Prigrow series medium (cat. no. TM4074; Applied Biological
Material) with 10% FBS.
Detection of miR-545 in OSCC tissue
samples and cell lines
The levels of miR-545 in 20 OSCC tissue samples and
7 cell lines was detected using reverse transcription-quantitative
polymerase chain reaction (RT-qPCR). The total RNA was extracted
from each of the 20 specimens using TRIzol® reagent,
according to the manufacturer's protocol (Life Technologies; Thermo
Fisher Scientific, Inc.). RNA (100 ng) was reverted by ImProm-II ™
Reverse Transcription system (Promega Corporation, Madison, WI,
USA). Normalization was performed using the 2−ΔΔCq
method (15) using SYBR-Green reagent
(Sangon Biotech Co, Ltd., Shanghai, China). The primers for miR-545
were: Primer-Sense, TCAGTAAATGTTTATTAGATGA; Primer-Anti-Sense,
GTGCAGGGTCCGAGGTATTC. U6 snRNA was used as the reference gene. The
primer of U6 snRNA was listed as follows: Forward,
5′-CTCGCTTCGGCAGCACA-3′; and reverse, 5′-AACGCTTCACGAATTTGCGT-3′.
The amplification conditions were: an initial denaturation step at
95°C for 10 min, followed by 40 cycles at 95°C for 15 sec and 60°C
for 60 sec.
The RIG-I mRNA level in the OSCC tissues was assayed
using SYBR-Green reagent (Sangon Biotech Co, Ltd.). The primers for
RIG-I (human) were as follows: Forward, 5′-GGACGTGGCAAAACAAATCAG-3′
and reverse, 5′-GCAATGTCAATGCCTTCATCA-3′. The amplification
conditions were: 94°C for 5 min; 30 cycles of 94°C for 45 sec, 55°C
for 45 sec and 72°C for 1 min; and 72°C for 10 min.
Overexpression and downregulation of
miR-545 in OSCC cells
In the OSCC cell lines, miR-545 was overexpressed by
miR-545 mimics and decreased by miR-545 antisense oligonucleotides
(ASO), as described in a previous study (11). miR-545 mimics, miR-545 ASO and control
miRNA were purchased from Sangon Biotech Co, Ltd. miRNAs were
transfected into cells using the Lipofectamine 2000 reagent
(Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol. In short, cells were seeded to 70–90%
confluent at transfection, the miR-545 (30 nM) and Lipofectamine
2000 reagent complex were added into cells according to the
manufacturer's protocol. Cells were incubated at 37°C in a
CO2 incubator for 24 h prior to testing for transgene
expression.
Cell proliferation assay
Cellular growth was analyzed using an MTT assay, as
previously described (16–20). Briefly, cells were placed into 96-well
plates at a density of 5×103 cells/well. The MTT reagent
was added into the medium at a final concentration of 0.1 mg/ml,
and 100 µl of dimethyl sulfoxide was added. The optical density was
measured on a microplate reader with a 570 nm filter.
Cell migration assay
Transwell systems were used to assess cell migration
(21). The Transwell chambers (8.0 µm
pore size; Sigma-Aldrich; Merck KGaA) were placed in 24-well
plates. The miR-545 mimics- or ASO-transfected cells were deprived
of FBS for 12 h, and subsequently added to the upper chambers. DMEM
Medium containing 10% FBS was placed in the lower chambers. The
cells were incubated in a humidified incubator at 37°C for 24 h.
Cells in the upper chambers were removed with cotton swabs. The
cells attached to the lower surface were fixed in 70% ethanol for
10 min at room temperature. The remaining ethanol was removed from
the top of the membrane using a cotton-tipped applicator. Cells
were then stained with 0.2% crystal violet into for 10 min at room
temperature. The number of cells that had attached to the lower
surface was counted in five randomly selected fields under a
microscope (The Eclipse Ti2, Tokyo, Japan) (light, magnification,
×200).
Prediction of the putative targets of
miR-545
The Targetscan software (http://www.targetscan.org/, accessed September 2017)
was used to predict the putative targets of miR-545.
Dual luciferase reporter assays
Cells were seeded at 1×105 per well and
were serum-starved for 6 h pre-transfection. Since RIG-I is coded
by the DExD/H-Box Helicase 58 (DDX58) gene and there are two
miR-545 binding sites on DDX58 (22), the 3′-UTR of RIG-I and mutated
controls were cloned and inserted into the reporter plasmid (cat.
no. E1761; 500 ng) and the pGL3-control (cat. no. E1741; 100 ng;
Promega Corporation, Madison, WI, USA). miR-545 mimics (50 nM) were
then transfected into the HSC4 cells containing the wild-type or
mutant 3′-UTR plasmids, using Lipofectamine 2000 (Invitrogen;
Thermo Fisher Scientific, Inc.). Cells were harvested, and
luciferase activity was measured for each specimen after 24 h using
the Dual-Luciferase Reporter assay system (cat. no. E1910; Promega
Corporation). Mutants of RIG-I 3′-UTR were generated using the
Site-Directed Mutagenesis kit (Thermo Fisher Scientific, Inc.).
Western blotting analysis
Cells were frozen and lysed in lysis buffer (150 mM
NaCl, 50 mM Tris-HCI, 1% Triton X-100 and 0.1% SDS) with a Protease
Inhibitor Cocktail (cat. no. S8820; Sigma-Aldrich; Merck KGaA,
Darmstadt, Germany) and a Phosphatase Inhibitor (cat. no. P0044;
Sigma-Aldrich). For RIG-I analysis, a RIG-I antibody (ab132505) was
used at a dilution of 1:1,000 and incubated at 4°C, overnight),
followed by detection with a peroxidase-linked antibody (ab6759;
both Abcam, Cambridge, UK) to rabbit antibody IgG of 1:2,000
dilution incubated at room temperature for 2 h. Proteins were
detected using an Enhance Chemiluminesence Western Blotting
Detection Reagent (GE Healthcare, Chicago, IL, USA). Images were
analyzed using Image J (National Institutes of Health, Bethesda,
MD, USA).
Statistical analysis
All experiments were repeated three times. Data are
presented as the mean ± the standard deviation. A two-tailed
Student's t-test was used to analyze the mean value between two
groups. One way Analysis of variance was used to test the mean
value among three groups or more with post hoc contrasts by
Student-Newman-Keuls test. The correlation between miR-545 and
RIG-I levels were examined by Pearson correlation coefficient
analysis. P<0.05 was considered to indicate a statistically
significant difference. All calculations were performed using SPSS
software (version 16.0; SPSS, Inc., Chicago, IL, USA).
Results
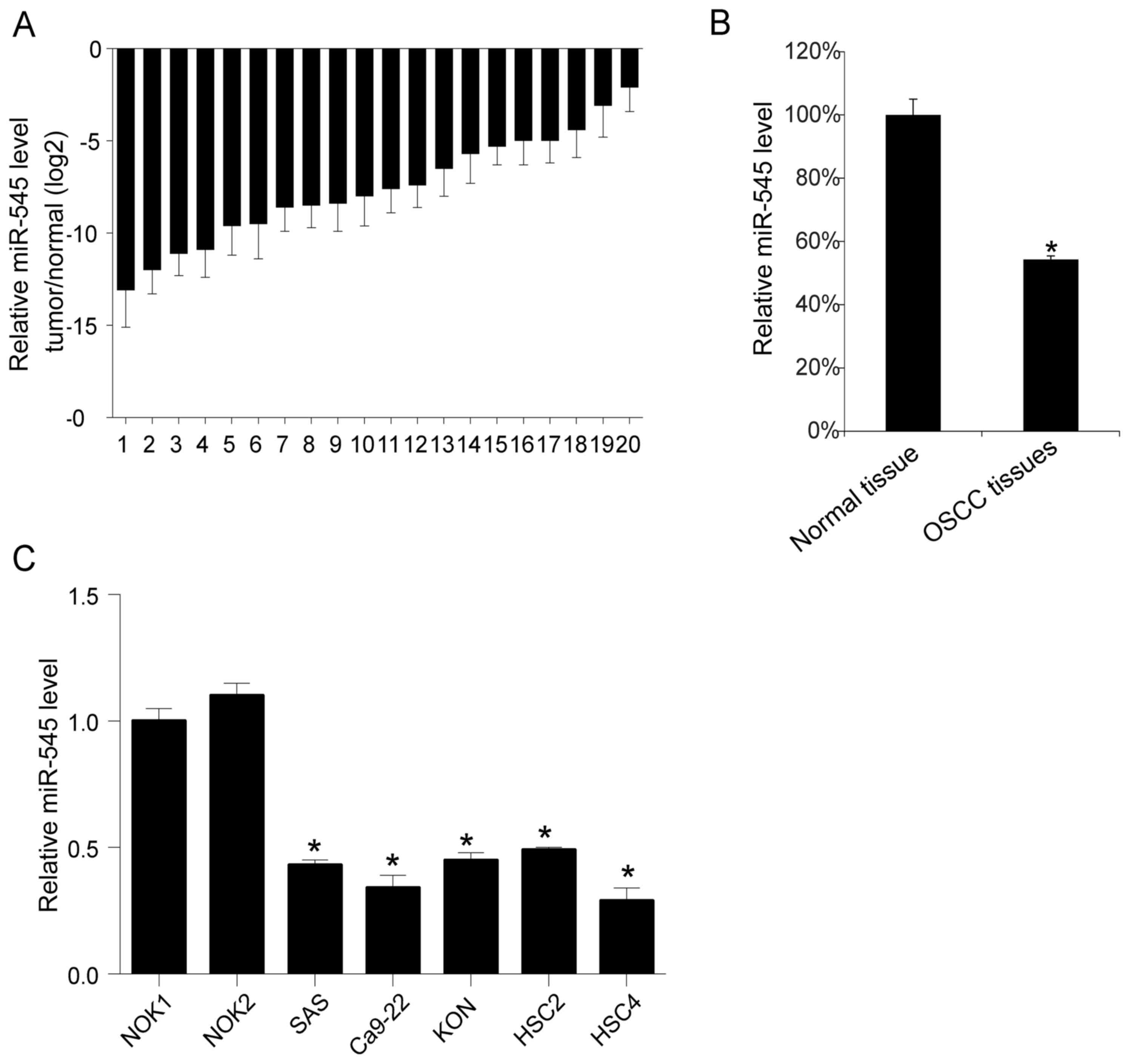
miR-545 levels in OSCC tissues
To understand the function of miR-545, the miR-545
levels in 20 OSCC tissue samples and their matching adjacent normal
tissues were evaluated using RT-qPCR. Overall, the analysis
revealed a decreased level of miR-545 in each tumor tissue compared
with the matched normal tissues (Fig.
1A). The mean level of miR-545 in the 20 OSCC samples was
calculated and compared to the mean level of miR-545 in the normal
tissues. As presented in Fig. 1B, the
mean level of miR-545 obtained for the 20 OSCC tissues was
decreased compared with the mean in normal tissues (P<0.05).
Similarly, a subsequent comparison revealed that miR-545 was more
abundant in normal oral keratinocyte cell lines (HOK) than in four
OSCC cell lines (HSC2, HSC4, SAS, KON; Fig. 1C) (the mean value in NOK1 or NOK2 were
compared with the four OSCC cell line separately, P<0.05).
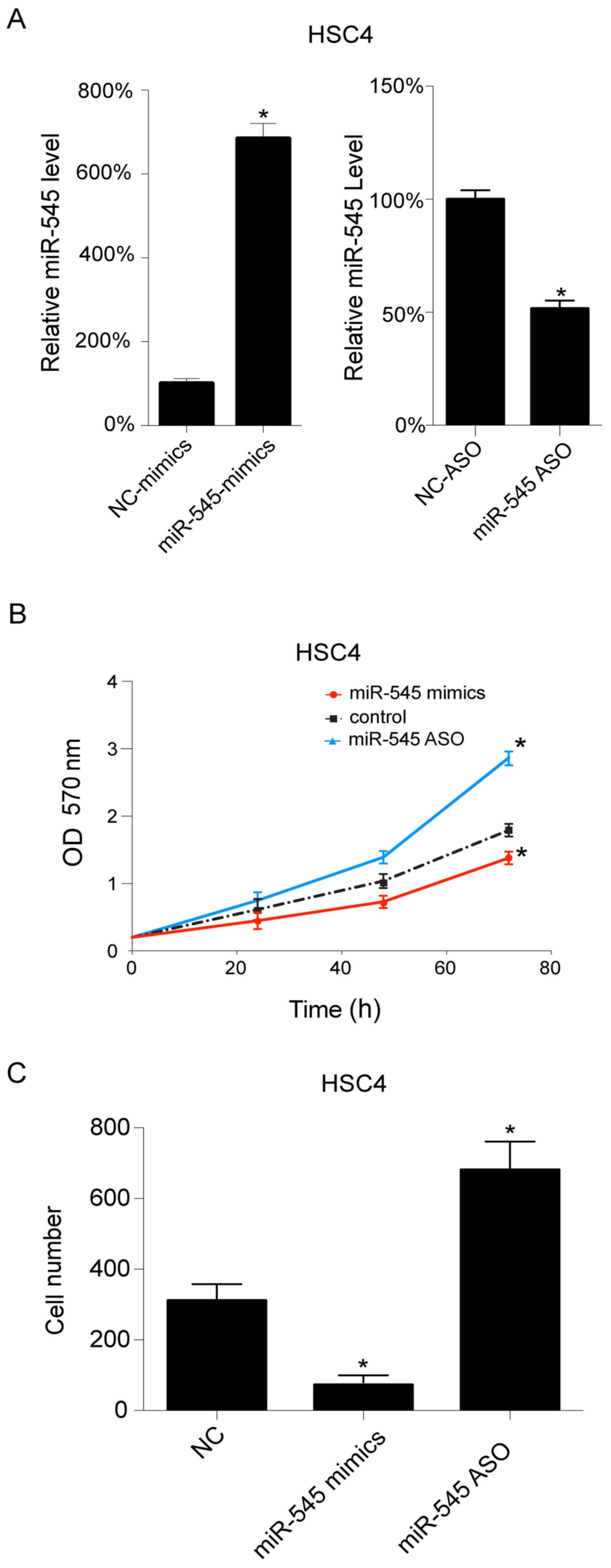
The in vitro role of miR-545 in OSCC
cells
As HSC4 cells exhibited relatively lower miR-545
levels when compared with the other examined OSCC cell lines, the
HSC4 cells were selected for further investigation. The miR-545
levels were regulated using miR-545 mimics and miR-545 ASO
transfections, which upregulated and downregulated miR-545,
respectively. miR-545 mimics transfection increased the level of
miR-545 in HSC4 cells, whereas miR-545 ASO transfection resulted in
a decrease (Fig. 2A) (P<0.05).
Additionally, cell growth was assessed following transfection. As
presented in Fig. 2B, the
upregulation of miR-545 inhibited cell growth, whereas its
downregulation promoted cell growth in HSC4 cells. Subsequently,
cell migration was investigated following transfection, and it was
observed that miR-545 mimics decrease the number of migrating
cells, whereas miR-545 ASO transfection results in an increase in
cell migration in HSC4 cells (Fig.
2C) (P<0.05).
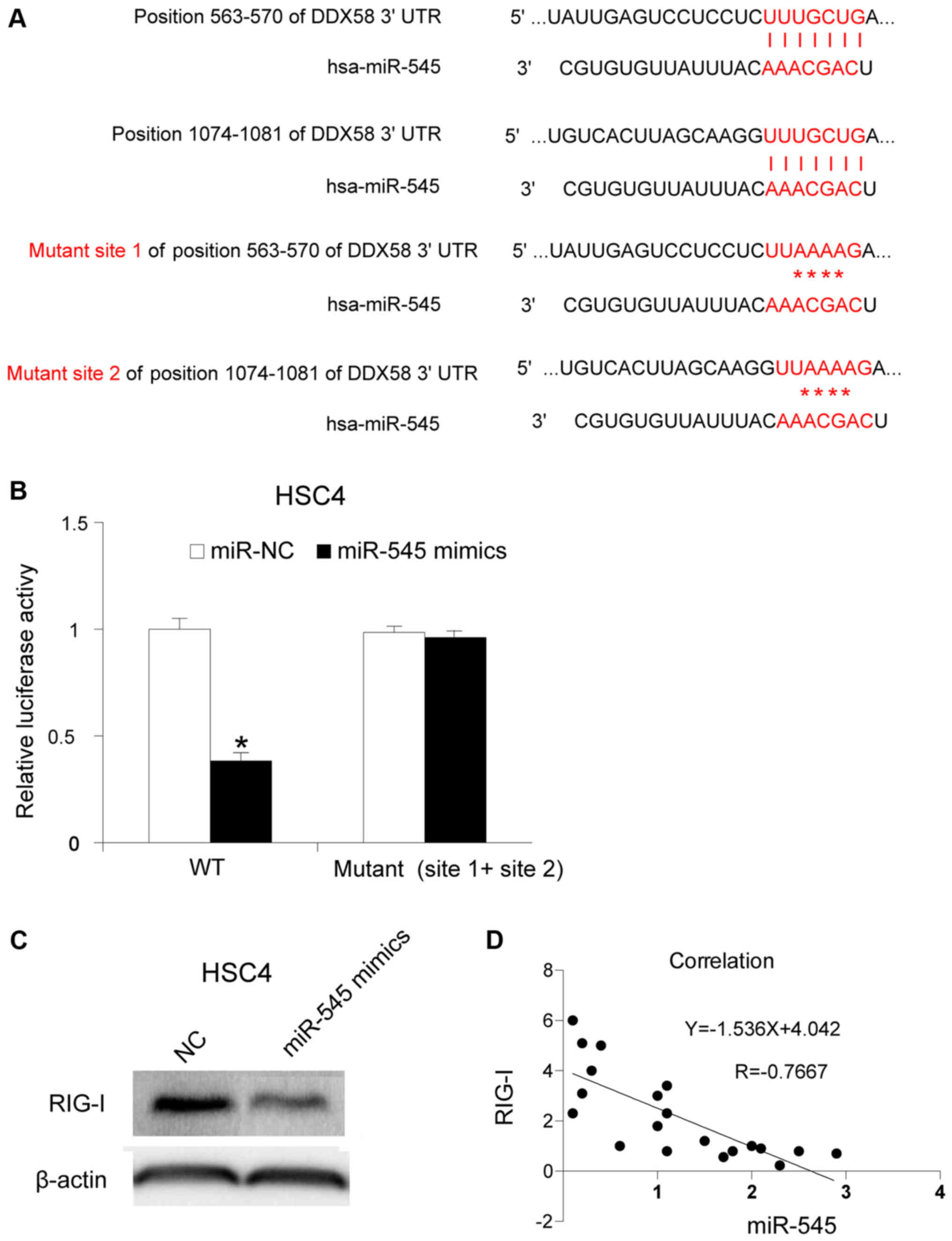
RIG-I is a target gene of miR-545
To understand the molecular mechanism of miR-545 in
OSCC, its potential target gene was investigated. It has been
reported that the 3′-UTR of RIG-I may be targeted by miR-545 in
pancreatic ductal adenocarcinoma (12). RIG-I is localized in the cytosol,
where it recognizes the 5′-triphosphate RNA (3p-RNA) generated by
viral RNA polymerases (22,23). In the present study, the association
between miR-545 and RIG-I in OSCC was investigated. The binding
sites were mutated as described in a previous study (Fig. 3A) (12).
The mutated sites were cloned into a luciferase reporter plasmid.
miR-545 mimics and the reporter plasmid were co-transfected into
HSC4 cells. The activity of luciferase was assessed 12 h after
transfection. The upregulation of miR-545 inhibited luciferase
activity (P<0.05), whereas the mutation of binding site 1 and
site 2 partly restored it, indicating that miR-545 targets RIG-I in
HSC4 cells (Fig. 3B). Subsequently,
the RIG-I protein levels following miR-545 mimics transfection were
determined, and it was identified that miR-545 transfection
inhibited the RIG-I protein levels in HSC4 cells (Fig. 3C). Furthermore, the RIG-I mRNA
expression levels were assessed in 20 OSCC tissues, and it was
identified that RIG-I mRNA expression was associated with miR-545
expression. The results obtained revealed a negative association
between RIG-I mRNA expression and miR-545 expression in the 20 OSCC
samples (Fig. 3D) (P<0.05).
Discussion
In the present study, the function of miR-545 in
OSCC was investigated. The present data revealed that miR-545
levels in OSCC tissues were lower than levels in matched adjacent
normal tissues. Overexpression of miR-545 inhibited HSC4
proliferation and migration, and vice versa. The potential target
gene of miR-545 was identified to be RIG-I.
The antitumor role of miR-545 have been confirmed in
other types of cancer, including lung cancer (13), pancreatic ductal adenocarcinoma
(12), and epithelial ovarian cancer
(23). To the best of our knowledge,
there are no reports pertaining to the oncogenic function of
miR-545. Thus, we hypothesize that miR-545 exhibited an antitumor
function in various types of cancer.
The present data demonstrated the inhibitory role of
miR-545 in OSCC. Furthermore, a negative association between RIG-I
mRNA expression and miR-545 expression was observed in the OSCC
tissues. To our knowledge, this is the first study to highlight the
function of miR-545 and RIG-I in OSCC. Notably, RIG-I is a part of
the key pathway of human papilloma virus (HPV) infection. Non-self
dsDNA of HPV may serve as template for transcription and induce
type I interferon and nuclear factor-κ -B through the RIG-I pathway
(24–26). Additionally, HPV infection was an
independent factor associated with OSCC after adjusting for age,
smoking and alcohol use (27). Thus,
it is hypothesized that HPV may exploit the regulation mechanism of
miR-545 and RIG-I in the pathogenesis of OSCC. The present study
identified that miR-545 was able to target RIG-I in OSCC cells.
Besides RIG-I, miR-545 has been demonstrated to target cyclin D1
and CDK1 in lung cancer (13),
implying that miR-545 has multiple targets in cancer. In
conclusion, the present data suggest an inhibitory role of miR-545
in OSCC.
Acknowledgements
The authors would like to thank Mr. Li Dayin
(Department of Oral and Maxillofacial Surgery, Shunde Hospital of
Guangzhou Medical University) for technical assistance.
Funding
No funding was received.
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
GY collected patient data and performed cell
experiments. HW and YD performed PCR, western blotting and other
molecular experiments. FH contributed to study design and
manuscript writing.
Ethics approval and consent to
participate
Written informed consent was obtained from all
patients, and the Ethics Committees of Guangzhou Medical University
approved the present study.
Patient consent for publication
All patients have provided their consent for the use
of their information and samples for scientific research and
publication.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Rivera C: Essentials of oral cancer. Int J
Clin Exp Pathol. 8:11884–11894. 2015.PubMed/NCBI
|
|
2
|
Choi S and Myers JN: Molecular
pathogenesis of oral squamous cell carcinoma: Implications for
therapy. J Dent Res. 87:14–32. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Gupta PC: Mouth cancer in India: A new
epidemic? Indian Med Assoc. 97:370–373. 1999.
|
|
4
|
Mehrotra R, Singh M, Kumar D, Pandey AN,
Gupta RK and Sinha US: Age specific incidence rate and pathological
spectrum of oral cancer in Allahabad. Indian J Med Sci. 57:400–404.
2003.PubMed/NCBI
|
|
5
|
Lingen MW, Kalmar JR, Karrison T and
Speight PM: Critical evaluation of diagnostic aids for the
detection of oral cancer. Oral Oncol. 44:10–22. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
O'neill V and Twelves C: Oral cancer
treatment: Developments in chemotherapy and beyond. Br J Cancer.
87:933–937. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ries LAG, Melbert D, Krapcho M, Stinchcomb
DG, Howlader N, Horner MJ, Mariotto A, Miller BA, Feuer EJ and
Altekruse SF: SEER Cancer Statistics Review, 1975–2005 (Based on
November 2007 SEER data submission). National Cancer Institute;
Bethesda, MD: 2008
|
|
8
|
Du T and Zamore PD: microPrimer: The
biogenesis and function of microRNA. Development. 132:4645–4652.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Farazi TA, Hoell JI, Morozov P and Tuschl
T: MicroRNAs in human cancer. Adv Exp Med Biol. 1–774. 1–20. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Manikandan M, Rao AKDM, Arunkumar G,
Manickavasagam M, Rajkumar KS, Rajaraman R and Munirajan AK: Oral
squamous cell carcinoma: microRNA expression profiling and
integrative analyses for elucidation of tumourigenesis mechanism.
Mol Cancer. 15:282016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Song B, Ji W, Guo S, Liu A, Jing W, Shao
C, Li G and Jin G: miR-545 inhibited pancreatic ductal
adenocarcinoma growth by targeting RIG-I. FEBS Lett. 588:4375–4381.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Du B, Wang Z, Zhang X, Feng S, Wang G, He
J and Zhang B: MicroRNA-545 suppresses cell proliferation by
targeting cyclin D1 and CDK4 in lung cancer cells. PLoS One.
9:e880222014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Liu Z, Dou C, Yao B, Xu M, Ding L, Wang Y,
Jia Y, Li Q, Zhang H, Tu K, et al: Ftx non coding RNA-derived
miR-545 promotes cell proliferation by targeting RIG-I in
hepatocellular carcinoma. Oncotarget. 7:25350–25365.
2016.PubMed/NCBI
|
|
15
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Mosmann T: Rapid colorimetric assay for
cellular growth and survival: Application to proliferation and
cytotoxicity assays. J Immuno Methods. 65:55–63. 1983. View Article : Google Scholar
|
|
17
|
Roehm NW, Rodgers GH, Hatfield SM and
Glasebrook AL: An improved colorimetric assay for cell
proliferation and viability utilizing the tetrazolium salt XTT. J
Immunol Methods. 142:257–265. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Gerlier D and Thomasset N: Use of MTT
colorimetric assay to measure cell activation. J Immunol Methods.
94:57–63. 1986. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Berridge MV, Tan AS, McCoy KD and Wang R:
The biochemical and cellular basis of cell proliferation assays
that use tetrazolium salts. Biochemica. 4:14–19. 1996.
|
|
20
|
Weichert H, Blechschmidt I, Schröder S and
Ambrosius H: The MTT-assay as a rapid test for cell proliferation
and cell killing: Application to human peripheral blood lymphocytes
(PBL). Allergie Immunol (Leipz). 37:139–144. 1991.
|
|
21
|
Rüster B, Grace B, Seitz O, Seifried E and
Henschler R: Induction and detection of human mesenchymal stem cell
migration in the 48-well reusable transwell assay. Stem Cells Dev.
14:231–235. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Hornung V, Ellegast J, Kim S, Brzózka K,
Jung A, Kato H, Poeck H, Akira S, Conzelmann KK, Schlee M, et al:
5′-Triphosphate RNA is the ligand for RIG-I. Science. 314:994–997.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Jia X, Liu X, Li M, Zeng Y, Feng Z, Su X,
Huang Y, Chen M and Yang X: Potential tumor suppressing role of
microRNA-545 in epithelial ovarian cancer. Oncol Lett.
15:6386–6392. 2018.PubMed/NCBI
|
|
24
|
Pichlmair A, Schulz O, Tan CP, Näslund TI,
Liljeström P, Weber F and Reis e Sousa C: RIG-I-mediated antiviral
responses to single-stranded RNA bearing 5′-phosphates. Science.
314:997–1001. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Chiu YH, Macmillan JB and Chen ZJ: RNA
polymerase III detects cytosolic DNA and induces type I interferons
through the RIG-I pathway. Cell. 138:576–591. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ablasser A, Bauernfeind F, Hartmann G,
Latz E, Fitzgerald KA and Hornung V: RIG-I-dependent sensing of
poly(dA:dT) through the induction of an RNA polymerase
III-transcribed RNA intermediate. Nat Immunol. 10:1065–1072. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Gan LL, Zhang H, Guo JH and Fan MW:
Prevalence of human papillomavirus infection in oral squamous cell
carcinoma: A case-control study in Wuhan, China. Asian Pac J Cancer
Prev. 15:5861–5865. 2014. View Article : Google Scholar : PubMed/NCBI
|