Introduction
Renal cell carcinoma (RCC), accounting for
approximately 3% of human malignant tumors, is a common type of
adult kidney tumor (1). Globally,
over 260,000 patients are diagnosed with kidney tumor annually and
over 116,000 patients succumb to the disease (2). The 5-year survival rate for RCC patients
in advanced stages remains poor due to metastasis or recurrence
(3). Generally, the standard
therapies for RCC involve nephron sparing surgery and radical
nephrectomy (4,5). However, these types of therapies are not
expected to have curative effects as they only slightly extend
progression-free survival (6).
Therefore, further understanding of the molecular mechanisms
underlying RCC metastasis and progression is of great importance to
identify novel prognostic and diagnostic biomarkers for RCC.
miRNAs are a class of conserved non-coding RNAs,
serving important roles in regulating gene expression levels via
directly targeting the mRNA 3′-UTR to induce translation inhibition
or mRNA degradation (7,8). miRNAs have been confirmed to be involved
in different kinds of cellular processes, such as apoptosis,
metabolism, and differentiation (9–11). The
study of miRNAs has provided a new direction in our understanding
of the tumorigenic mechanism of RCC. Additionally, aberrant
expression levels of various miRNAs have been observed in RCC. For
instance, Song et al reported that miR-384 could repress RCC
cell invasion and growth via regulating astrocyte elevated gene 1
(12). Wang et al reported
that migration and invasion abilities of RCC were suppressed by
miR-182 through the regulation of IGF1R (13). Hu et al revealed that miR-138
repressed RCC cell invasion and proliferation by mediating
SOX9 (14). Thus, miRNAs may
be underlying diagnostic markers for RCC.
Epithelial-mesenchymal transition (EMT) is a
pathological and physiological process where epithelial cells lose
their polarity and cell-cell adhesion signature, acquiring the
mesenchymal characteristics (15,16).
During this progress, the expression levels of epithelial markers,
such as E-cadherin, increase, while the expression levels of
mesenchymal markers, such as vimentin, decrease (17). EMT plays a critical role in many
aspects of tumor behavior, including metastasis (18). Thus, further investigation is
essential to better understand the mechanisms underlying EMT
progression in RCC.
ATAD2 (ATPase family AAA domain-containing
protein 2), also known as AAA+ nuclear coregulator
cancer associated (ANCCA), is a member of the AAA+
ATPase family (19). ATAD2
contains both an ATPase domain and a bromodomain. Additionally,
ATAD2 maps to chromosome 8q24, which is the most commonly
amplified region in multiple tumors (20,21). The
special structures of ATAD2 indicated that it was implicated
in the regulation of genome, including cell differentiation,
apoptosis, division and proliferation (22,23).
Recently, dysregulation of ATAD2 has been found in various
tumors, playing significant roles in tumor progression. For
example, ATAD2 overexpression was associated with prognosis
and progression in colorectal cancer (24); ATAD2 overexpression in gastric
cancer served as a poor prognostic biomarker (25). Thus, studies have revealed that
ATAD2 manifests oncogenic functions in various malignancies.
However, the significance of ATAD2 in RCC remains
uncertain.
Materials and methods
RCC clinical tissue specimens
Fifty-two pairs of RCC tissue specimens and matched
non-cancerous tissues were collected from RCC patients who
underwent radical nephrectomy in Beijing Ditan Hospital Capital
Medical University (Beijing, China) between September 2015 and June
2017. None of the patients had received chemotherapy or
radiotherapy prior to the surgery. All the tissues were immediately
frozen in liquid nitrogen after resection and stored at −80°C for
further assays.
Written consent was obtained from the RCC patients
involved in the current study. The study was approved by the Ethics
Committees of Beijing Ditan Hospital Capital Medical
University.
Cell lines and cell culture
Five human RCC cell lines (Caki-1, 786-O, A498, ACHN
and Caki-2) and the immortalized normal human renal proximal tubule
epithelial cell line HK-2 were obtained from the American Type
Culture Collection (Manassas, VA, USA). All the RCC cells were
maintained in RPMI-1640 medium (Gibco; Thermo Fisher Scientific,
Inc., Waltham, MA, USA) while HK-2 was cultured in keratinocyte
serum-free medium (Gibco; Thermo Fisher Scientific, Inc.). Both
culture media contained 10% FBS (Gibco; Thermo Fisher Scientific,
Inc.), 100 U/ml penicillin and 100 µg/ml streptomycin (Corning
Incorporated, Corning, NY, USA). All the cells were kept in a
humidified incubator containing 5% CO2 at 37°C.
Cell transfection
The miR-372 mimics, inhibitors, ATAD2
overexpression plasmids and ATAD2 siRNA were produced by
GenePharmaCo. (Shanghai, China) and transfected into RCC cells
using Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) in line with the manufacturer's protocol. The
cells were harvested 48 h post-transfection for further assays.
RT-qPCR
TRIzol (Invitrogen; Thermo Fisher Scientific, Inc.)
was applied to isolate the total RNAs from RCC tissues and cultured
cells following the manufacturers' protocol. Reverse transcription
for cDNA was performed using the TaqMan MicroRNA Reverse
Transcription kit (Applied Biosystems; Thermo Fisher Scientific,
Inc.). Then, cDNA was amplified using a SYBR-Green mix kit and the
ABI 7900 Real-Time PCR system (Applied Biosystems). Primer
sequences are shown in Table I. GAPDH
was used as endogenous controls for ATAD2 and EMT-related
genes. U6 served as the internal control for miR-372. The relative
expression levels were determined using the 2−∆∆Cq
method (26). RT-qPCR was performed
with the following thermocycling conditions: Initial denaturation
at 95°C for 10 min, followed by 40 cycles of 95°C for 15 sec,
annealing at 50°C for 30 sec, and extension at 72°C for 30 sec.
 | Table I.Primer sequences for RT-qPCR. |
Table I.
Primer sequences for RT-qPCR.
| Primer | Sequence |
|---|
| miR-372 | F:
5′-AGCCTAAAGTGCTGCGACATT-3′ |
|
| R:
5′-GTGCAGGGTCCGAGGT-3′ |
| U6 | F:
5′-CTCGCTTCGGCAGCACA-3′ |
|
| R:
5′-AACGCTTCACGAATTTGCGT-3′ |
| ATAD2 | F:
5′-GGAATCCCAAACCACTGGACA-3′ |
|
| R:
5′-GGTAGCGTCGTCGTAAAGCACA-3′ |
| GAPDH | F:
5′-GAAGGTGAAGGTCGGAG-3′ |
|
| R:
5′-GAAGATGGTGATGGGAT-3′ |
| E-cadherin | F:
5′-CACCTGGAGAGAGGCCATGT-3′ |
|
| R:
5′-TGGGAAACATGAGCAGCTCT-3′ |
| Vimentin | F:
5′-AGCTGCAGGCTCAGATTCAGGA-3′ |
|
| R:
5′-CGGTTGGCAGCCTCAGAGAGGT-3′ |
Transwell assays
Transwell chamber (8.0 µm pore size, Corning
Incorporated) was applied to determine the RCC cell invasion and
migration abilities with or without the Matrigel (Corning
Incorporated) being precoated. For invasion assay, 1×104
RCC cells in serum-free RPMI-1640 medium were placed into top
chambers of the inserts which had been precoated with Matrigel.
Then, culture medium containing 10% FBS, as a chemoattractant, was
added into bottom chambers. After incubation at 37°C for 48 h in a
5% CO2 atmosphere, the cells that remained on the top
side of the inserts were removed using a cotton swab while the
cells that invaded the bottom side were fixed with 0.1%
paraformaldehyde for 15 min and stained with 0.1% crystal violet
for 15 min at room temperature. Then, the stained cells were
counted under a microscope (IX53; Olympus Corporation, Tokyo,
Japan) from five randomly selected visual fields. The difference
between the migration and invasion assays was that the insert was
not plated with Matrigel for the migration assays.
Western blotting
Cultured cells were harvested and lysed with RIPA
buffer (Beyotime, Shanghai, China) containing the protease
inhibitors (Roche, Basel, Switzerland) on ice for 15 min. The BCA
method (BCA protein assay kit; Beyotime) was used to determine the
concentration of the proteins. Proteins (40 µg) were then separated
by 10% SDS-PAGE and transferred onto PVDF membranes (Millipore,
Bedford, MA, USA). The membranes were blocked with 5% non-fat milk
in TBST at room temperature for 2 h, followed by incubation
overnight at 4°C with appropriate primary antibodies which were as
follows: antibodies against ATAD2 (1:1000, ab118664, Abcam,
Cambridge, MA, USA), Vimentin (1:1000, ab92547, Abcam), E-cadherin
(1:1000, ab15148, Abcam) and GAPDH (1:1000, ab9485, Abcam). After
incubation with the corresponding horseradish peroxidase-conjugated
secondary antibodies (1:2000, ab6721, Abcam) for 2 h at room
temperature, the protein bands were visualized using a
chemiluminescent imaging system (Pierce, Rockford, IL, USA).
Densitometry analysis of the protein blots was conducted using
Image-Pro Plus 6.0 software (Media Cybernetics, Inc., Rockville,
MD, USA). GAPDH was used as an internal control.
Luciferase reporter assays
The wild-type (WT) or mutant (MUT)
ATAD2−3′UTR which contained the miR-372 binding sites was
inserted into the luciferase genes in the pGL3 vectors (Promega,
Madison, WI, USA). RCC cells were co-transfected with pGL3-WT
ATAD2−3′-UTR or pGL3-MUT ATAD2−3′-UTR and miR-372
mimics. The luciferase activities were detected with the
dual-luciferase reporter assay kit (Promega) 48 h after the
transfections.
Statistical analysis
Statistical analyses were performed using SPSS
software version 17.0 (SPSS Inc., Chicago, IL, USA). Student's
t-test, ANOVA and Scheffe post-hoc analysis were applied, where
appropriate. A paired Student's t-test was performed to analyze the
paired data. For analysis on clinical correlation between miR-372
expression and RCC patients' clinicopathological features,
Pearson's Chi-square test was used. Spearman's correlation method
was applied to estimate the correlations between mRNA and miRNA.
Kaplan-Meier method was applied to determine the survival rates,
and the log-rank test was performed to compare the difference
between the survival curves. P<0.05 was considered to be
statistically significant difference.
Results
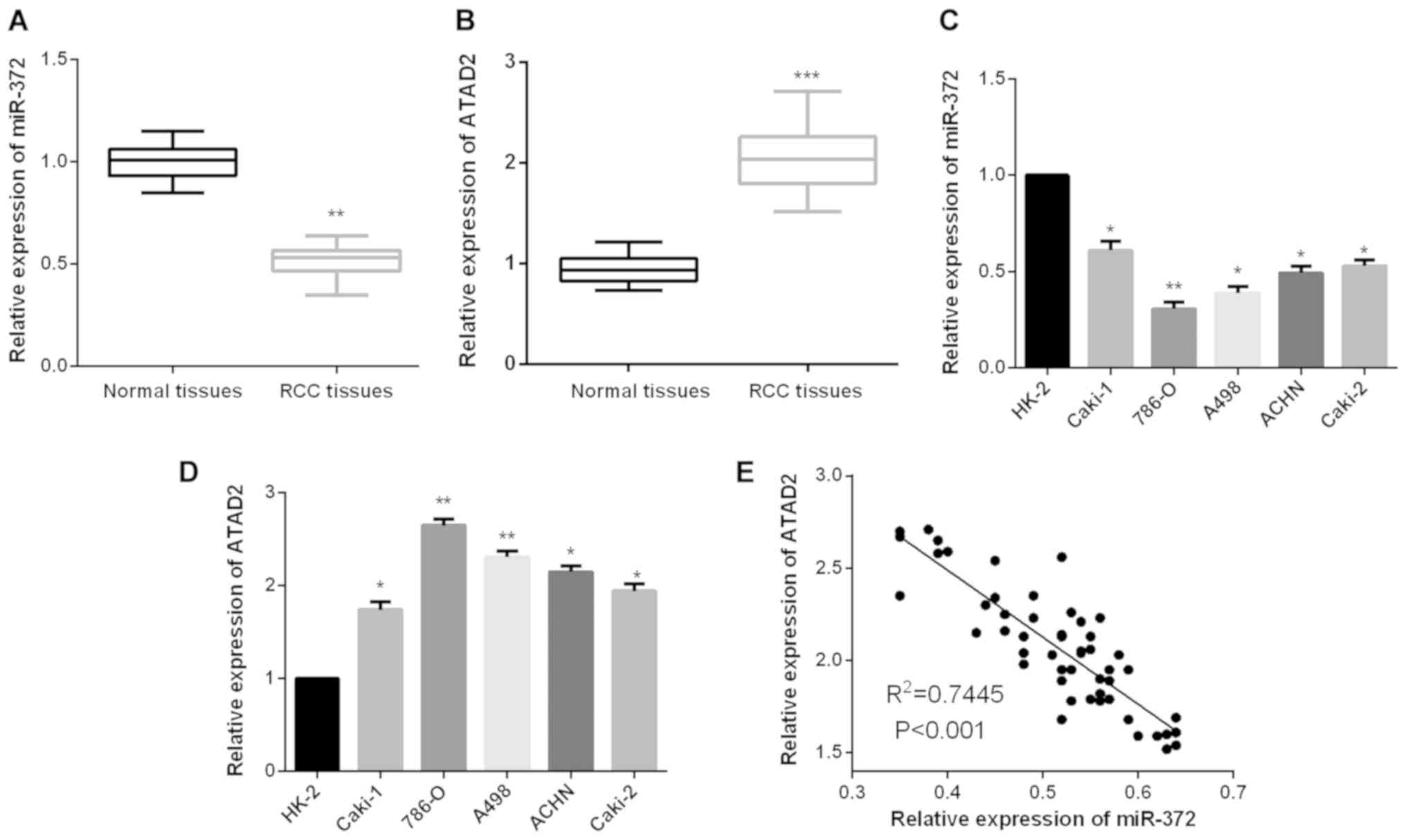
Aberrant expression levels of miR-372
and ATAD2 in RCC
To validate whether miR-372 functionally modulates
ATAD2 expression levels in RCC, firstly, we measured miR-372
and ATAD2 expression levels in RCC tissue samples. Results
revealed that RCC tissues presented a lower miR-372 expression
level in comparison with the non-tumor tissues (Fig. 1A), whereas, the expression levels of
ATAD2 in RCC tissues were prominently upregulated when
compared to the paired adjacent normal tissues (Fig. 1B). Similarly, significantly decreased
miR-372 expression levels were detected in RCC cells as compared to
the normal renal cell HK-2 (Fig. 1C).
We further evaluated ATAD2 expression levels in RCC cells.
As shown in Fig. 1D, ATAD2
expression levels were found to be markedly increased in RCC cells
compared to HK-2. Additionally, the correlation between miR-372 and
ATAD2 expression levels in RCC tissues was analyzed. The
finding of Spearman's correlation rank test demonstrated a negative
correlation between miR-372 and ATAD2 expression levels
(Fig. 1E).
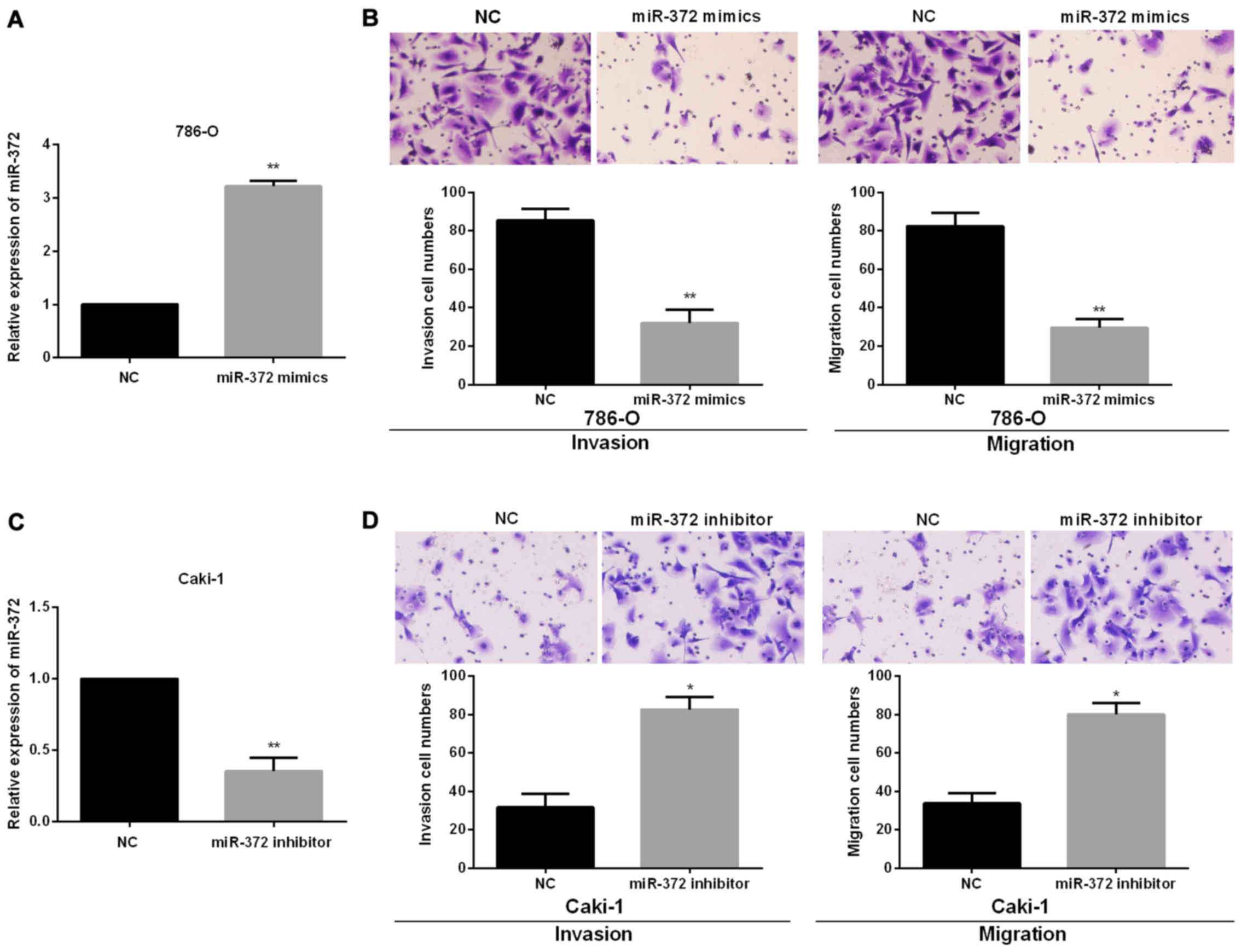
miR-372 suppresses RCC cell migration
and invasion
To investigate the functional roles of miR-372 in
RCC progression, we performed miR-372 inhibited or overexpressed
assays in Caki-1 and 786-O cells, which had the relatively highest
or lowest endogenous miR-372 expression levels, by transfecting the
miR-372 mimics or inhibitor into 786-O or Caki-1 cells. Successful
overexpression or knockdown of miR-372 in 786-O or Caki-1 cells was
confirmed using RT-qPCR (Fig. 2A and
2C). Then, Transwell assays were conducted to confirm the
functions of miR-372 in RCC cell migration and invasion. The
findings showed that miR-372 overexpression significantly reduced
786-O cell migration and invasion (Fig.
2B). By contrast, there was a significant increase in cell
invasion and migration of Caki-1 treated with miR-372 inhibitor
(Fig. 2D).
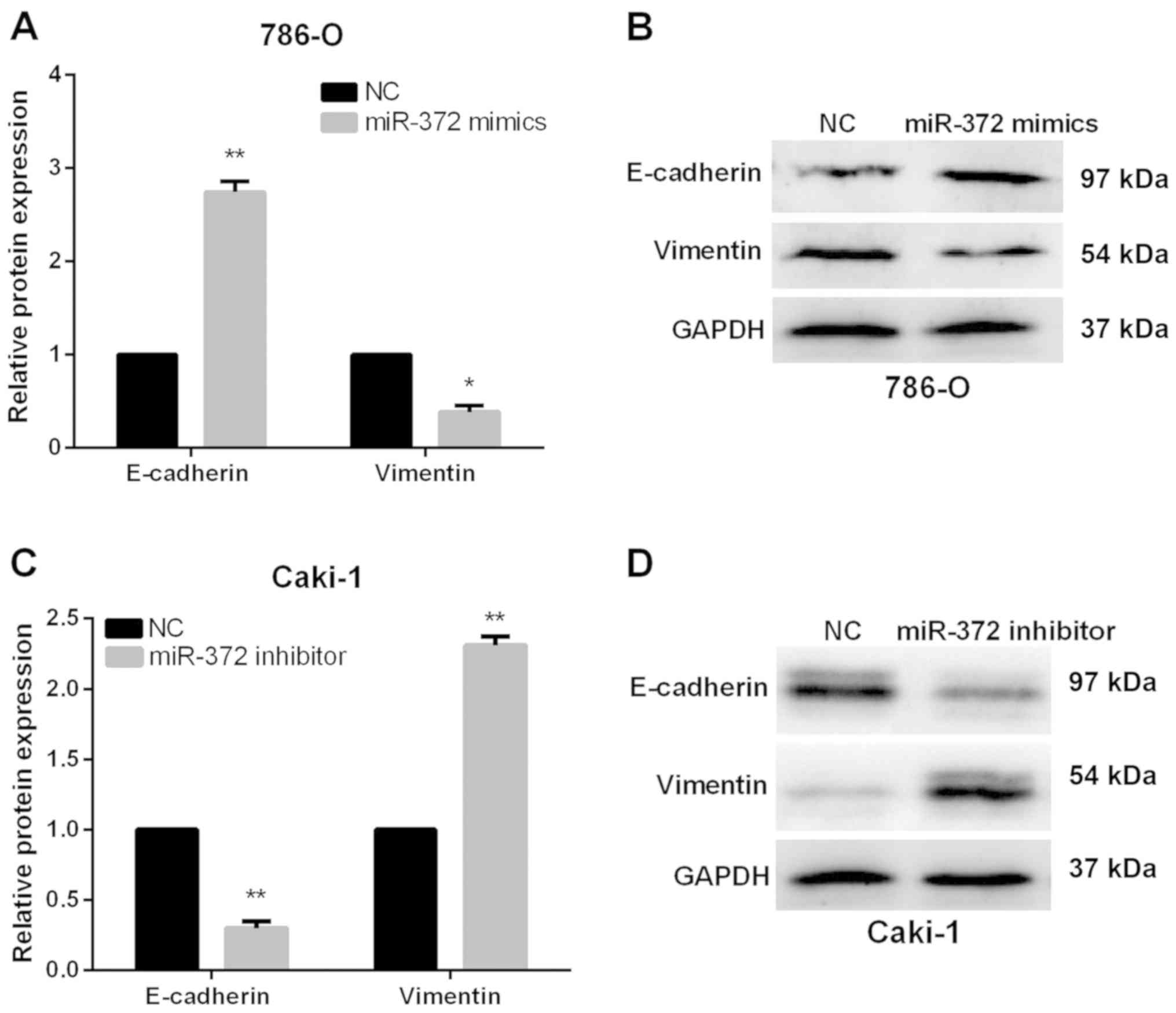
miR-372 suppresses RCC cell EMT
Next, we monitored alterations of EMT markers,
including Vimentin and E-Cadherin, to investigate the functions of
miR-372 in EMT of RCC cells. As expected, results of RT-qPCR and
western blot analysis both demonstrated that E-Cadherin expression
levels were obviously increased, whereas the Vimentin expression
levels were markedly reduced in miR-372 overexpressed 786-O cells
(Fig. 3A and B). By contrast,
Vimentin was upregulated in miR-372-suppressed Caki-1 cells, while
E-Cadherin was prominently downregulated (Fig. 3C and D). Our data revealed that
miR-372 overexpression could suppress the RCC cell EMT
progression.
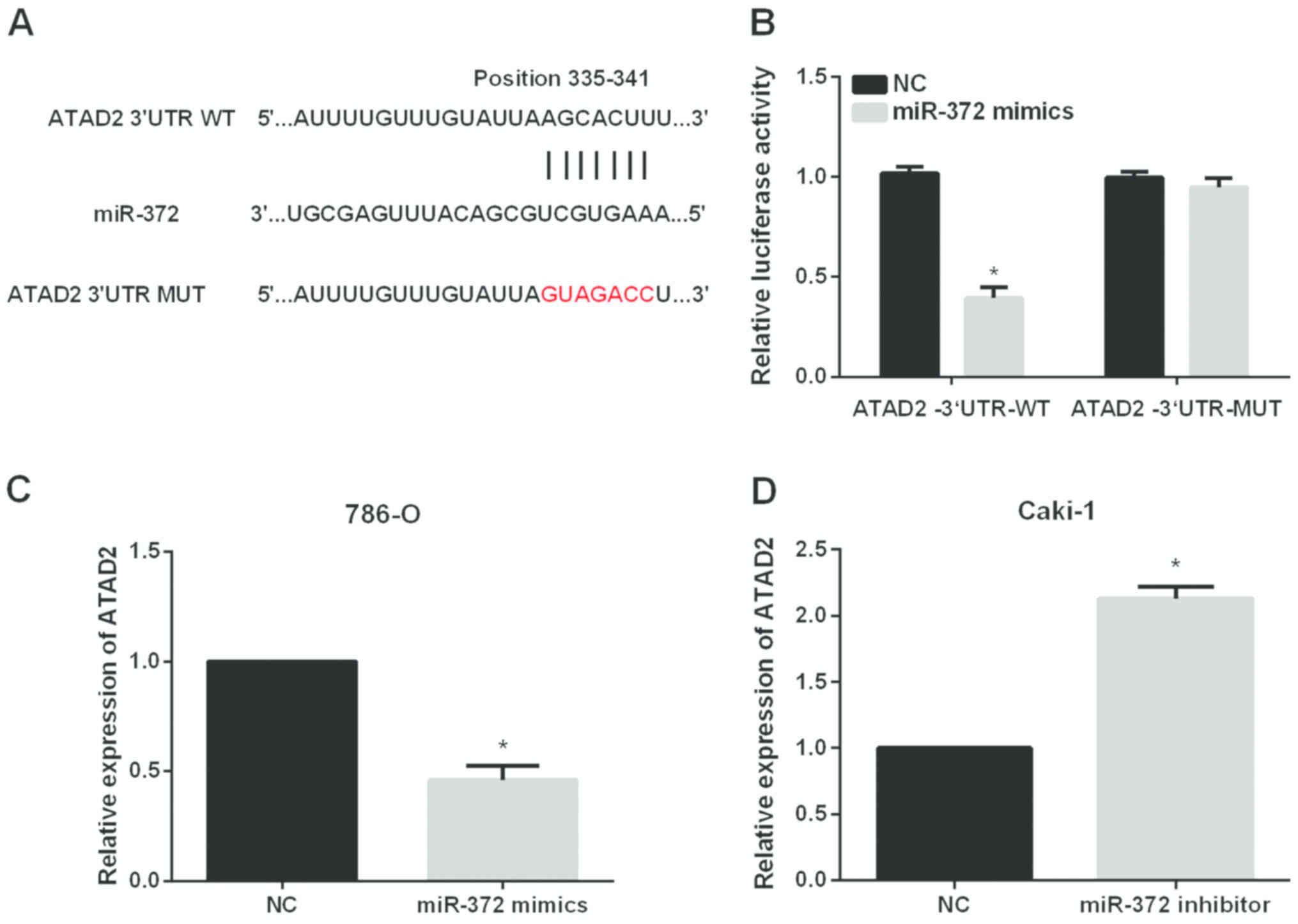
ATAD2 is a direct target of miR-372 in
RCC cell lines
The significant impact of miR-372 on the metastasis
of RCC cells prompted us to investigate the mechanisms underlying
its functional effects. The data from Targetscan database revealed
that ATAD2 had complementary binding sites for miR-372
(Fig. 4A). Then, dual-luciferase
reporter assays were performed to validate the association between
ATAD2 and miR-372. The results showed that miR-372
significantly reduced the luciferase activities of
ATAD2−3′UTR-WT without having obvious effects on that of
ATAD2−3′UTR-Mut (Fig. 4B). We
also investigated whether miR-372 could regulate the ATAD2
expression levels in RCC cells. The results of RT-qPCR indicated
that miR-372 overexpression significantly reduced ATAD2
expression levels in 786-O cells (Fig.
4C) while inhibition of miR-372 significantly elevated ATAD2
expression levels in Caki-1 cells (Fig.
4D).
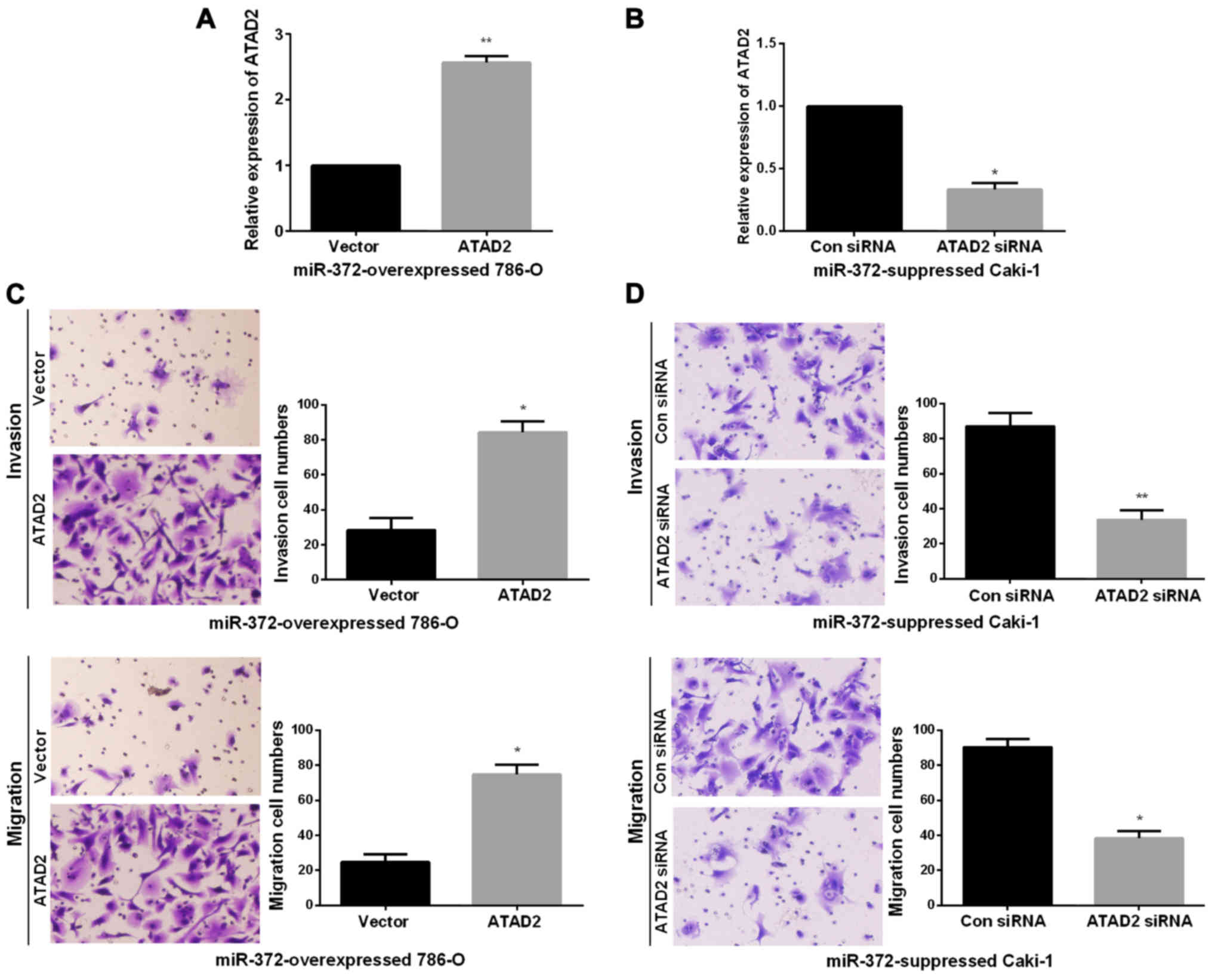
ATAD2 mediated the biological
functions of miR-372 in RCC cell invasion and migration
Next, we performed rescue experiments to verify that
miR-372 inhibited RCC metastasis ability by directly reducing
ATAD2. First, we obtained miR-372 overexpressed 786-O cells
or miR-372 suppressed Caki-1 cells by transfecting miR-372 mimics
or inhibitor. Then, to verify that the biological functions of
miR-372 in RCC was modulated by ATAD2, we transfected ATAD2
overexpressing vector or ATAD2 siRNA into miR-372
overexpressed 786-O cells or miR-372 suppressed Caki-1 cells,
inducing an increase or decrease of ATAD2 expression
(Fig. 5A and B). Next, Transwell
assays were performed to determine the migration and invasion
capacities. Findings demonstrated that ATAD2 upregulation
markedly enhanced the invasion and migration abilities of miR-372
overexpressed 786-O cells (Fig. 5C).
On the contrary, knockdown of ATAD2 dramatically reduced the
invasion and migration abilities of miR-372 suppressed Caki-1 cells
(Fig. 5D). miR-372 inhibited RCC cell
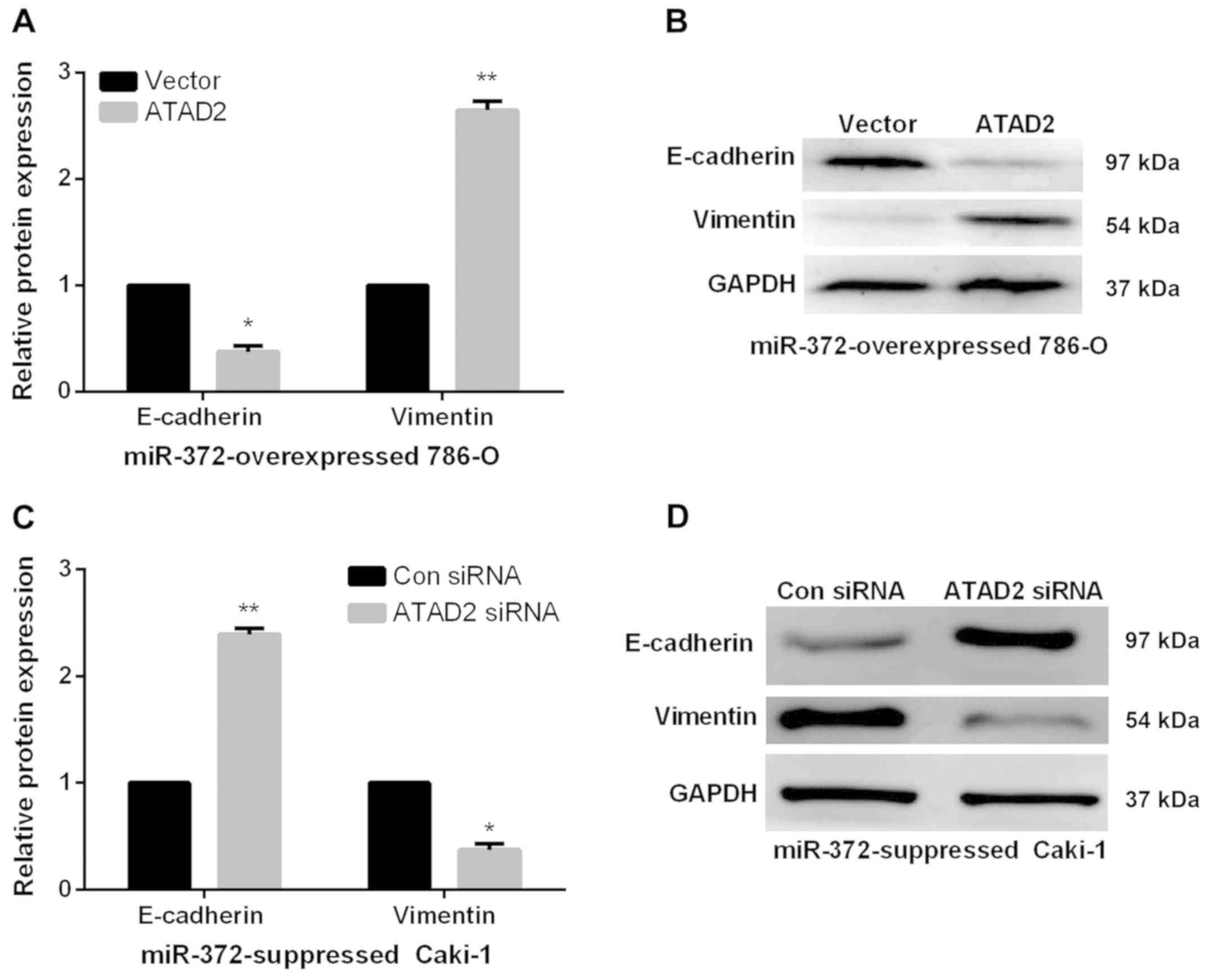
EMT by regulating ATAD2. We further investigated the
functions of ATAD2 in mediating the functional roles of
miR-372 in RCC EMT progress by qRT-PCR and western blots. As
expected, in miR-372 overexpressed 786-O cells, E-cadherin
expression levels were significantly reduced and vimentin
expression levels were prominently enhanced by ATAD2
overexpression (Fig. 6A and 6B).
However, ATAD2 silence significantly elevated the E-cadherin
expression levels and decreased the vimentin expression levels in
miR-372 suppressed Caki-1 cells (Fig. 6C
and D). The data indicated that miR-372 suppressed RCC cell EMT
process by regulation of ATAD2.
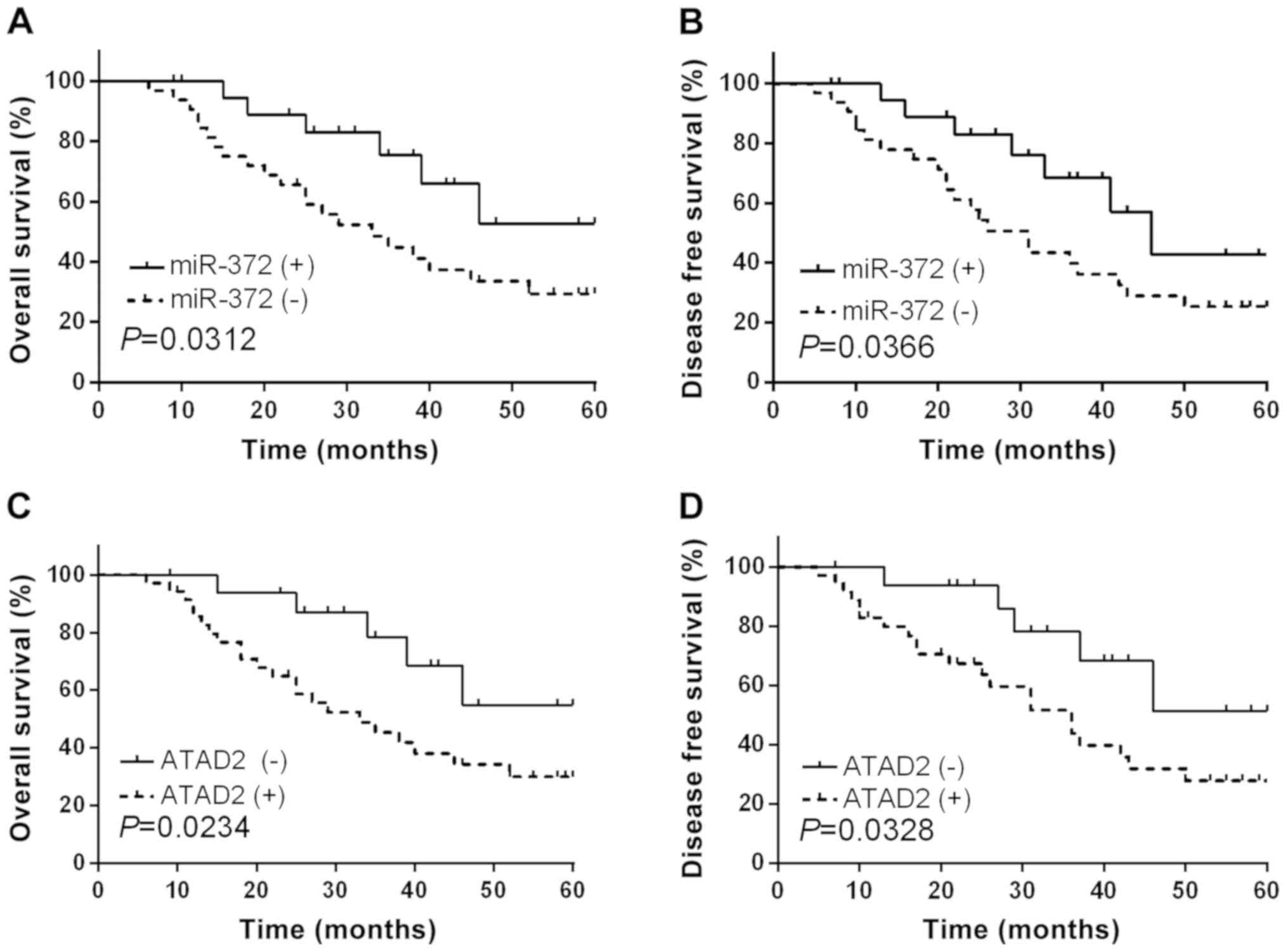
Aberrant expression levels of miR-372
and ATAD2 were correlated with clinicopathological features and
prognosis of RCC patients
As we had confirmed the functional effects of
miR-372 and ATAD2 in RCC, subsequently, we further
investigated the clinical significance of miR-372 and ATAD2
in RCC. The RCC patients involved in current study were divided
into two groups based on the mean expression level of miR-372.
Kaplan-Meier abalysis demonstrated that RCC patients with
relatively low miR-372 expression levels had significantly
decreased OS (Fig. 7A, P=0.0312) and
DFS (Fig. 7B, P=0.0366). In addition,
ATAD2 high-expressing RCC patients presented shorter OS and
DFS compared to ATAD2 low-expressing cases (Fig. 7C and D). Correlation between miR-372
expression and clinical features analysis implied that miR-372
expression levels were relevant to lymph node metastasis, TNM stage
and tumor differentiation (P=0.0015, 0.0075 and 0.0103,
respectively, Table II).
 | Table II.Correlation of miR-372 expression
with the clinicopathological characteristics of the RCC
patients. |
Table II.
Correlation of miR-372 expression
with the clinicopathological characteristics of the RCC
patients.
|
|
|
miR-372a
expression |
|
|
|---|
|
|
|
|
|
|
|---|
| Clinicopathological
features | Cases (n=52) | High (n=21) | Low (n=31) | P-value | χ2
value |
|---|
| Age (years) |
|
|
| 0.2016 | 0.3718 |
|
>60 | 27 | 11 | 16 |
|
|
|
≤60 | 25 | 10 | 15 |
|
|
| Sex |
|
|
| 0.3124 | 0.0132 |
|
Male | 26 | 9 | 17 |
|
|
|
Female | 26 | 12 | 14 |
|
|
| Tumor size
(cm) |
|
|
| 0.0614 | 2.1431 |
|
≥5.0 | 24 | 6 | 18 |
|
|
|
<5.0 | 28 | 15 | 13 |
|
|
| TNM stage |
|
|
| 0.0075b | 7.3920 |
|
I–II | 26 | 17 | 9 |
|
|
|
III | 26 | 4 | 22 |
|
|
| Histological
type |
|
|
| 0.0765 | 1.0125 |
| Clear
cell | 28 | 12 | 16 |
|
|
|
Papillary | 24 | 9 | 15 |
|
|
| Tumor
differentiation |
|
|
| 0.0103b | 3.7846 |
| Well
and Moderate | 25 | 16 | 9 |
|
|
|
Poor | 27 | 5 | 22 |
|
|
| Lymph node
metastasis |
|
|
| 0.0015b | 9.5623 |
|
Present | 26 | 4 | 22 |
|
|
|
Absent | 26 | 17 | 9 |
|
|
Discussion
RCC is the most common neoplasm of the adult kidney,
and the morbidity and mortality rates are increasing at a rate of
2–3% per decade (27). However, the
curative effects of radiotherapy and traditional chemotherapy in
advanced RCC therapy are limited (28). Additionally, although surgery is often
curative for localized disease, majority of these patients
experience metastasis or relapses, which is associated with poor
prognosis. Therefore, it is of crucial significance to understand
the mechanisms underlying RCC metastasis and recurrence to explore
novel anti-RCC therapeutic targets. Previous studies revealed that
miRNAs may provide new directions for earlier diagnosis as well as
better prognosis of RCC (29).
Recent studies have proposed the antitumor effects
of miR-372 in various malignancies. Aberrant miR-372 expression has
been identified as novel prognostic as well as diagnostic marker
for osteosarcoma (30) and colorectal
cancer (31). Additionally, studies
by Wu et al indicated that declined miR-372 expression in
hepatocellular carcinoma was relevant to tumor metastasis and poor
prognosis (32); Kong et al
reported that miR-372 repressed prostate cancer cell invasion and
migration via regulating p65 (33); Liu et al found that miR-372
inhibited the progression of endometrial carcinoma via the
regulation of RhoC (34).
Consistent with these findings, we found that miR-372 was obviously
downregulated in RCC and its decreased expression levels were
related to poor prognosis and worse clinicopathological parameters
of RCC patients. Furthermore, miR-372 overexpression suppressed RCC
cell EMT and metastasis. The data suggested that miR-372 played an
inhibitory role in RCC metastasis and progression.
Previous studies have identified and validated an
array of possible direct targets of miR-372, such as TXNIP,
ULK1 and CDK2 (35–37),
suggesting that miR-372 is an antitumor miRNA which plays a pivotal
role in the initiation and progression of cancer. To extend that
observation, we assessed the relationship between miR-372 and
ATAD2 in RCC. ATAD2 has been identified as a novel
candidate oncogene and possibly a therapeutic target for several
types of human cancers (38). For
instance, Hwang et al found that ATAD2 functioned as
a poor prognostic biomarker in hepatocellular carcinoma (39); Wan et al reported that
ATAD2 overexpression in ovarian carcinoma indicated poor
prognosis (40); Shang et al
reported that overexpression of ATAD2/ANCCA in endometrial
carcinoma was related to poor prognosis and tumor progression
(41). These studies suggested that
ATAD2 might have a potential as a therapeutic target for
malignancies. Although the correlation between clinicopathological
features and ATAD2 expression levels has been studied in
multiple tumors, little is known about ATAD2 in RCC. Herein,
the ectopic upregulation of ATAD2 in RCC was presented,
which demonstrated a poor prognosis of RCC patients. Moreover,
ATAD2 was identified as a direct target of miR-372 in RCC
cells. Importantly, the inhibitory functions of miR-372 in RCC cell
metastasis and EMT were confirmed to be regulated by
ATAD2.
In conclusion, the findings in current research
demonstrated a significant decrease of miR-372 expression levels
and a notable increase of ATAD2 expression levels in RCC.
Additionally, miR-372 overexpression had similar effects on
repressing RCC cell invasion, migration and EMT as ATAD2
silence. Moreover, ATAD2 was confirmed to be a direct target
of miR-372 and the antitumor effects of miR-372 on RCC progression
were partially mediated by ATAD2. The findings suggested
that miR-372/ATAD2 axis may be attractive biomarkers and
therapeutic targets for RCC patients.
Acknowledgements
Not applicable.
Funding
The study was supported by the Key Youth Project for
Training Excellent Talents of Beijing (3101-03-36-10) and The
Fundamental clinical cooperation project of Capital Medical
University (17JL37).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
SJ and XS contributed to the conception of the
study; HZ contributed significantly to data analysis and manuscript
preparation; ZH and YZ performed the data analyses and wrote the
manuscript; QL performed the statistical analysis with constructive
discussions. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
The study was approved by the Ethics Committee of
Beijing Ditan Hospital Capital Medical University (Beijing, China).
Signed informed consents were obtained from the patients or the
guardians.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
Statistics, 2017. CA Cancer J Clin. 67:7–30. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ridge CA, Pua BB and Madoff DC:
Epidemiology and staging of renal cell carcinoma. Semin Intervent
Radiol. 31:3–8. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Gupta K, Miller JD, Li JZ, Russell MW and
Charbonneau C: Epidemiologic and socioeconomic burden of metastatic
renal cell carcinoma (mRCC): A literature review. Cancer Treat Rev.
34:193–205. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Jansi Prema KS, Devanathan KS and Kurien
AA: Renal cell carcinoma with t(6,11): A case report and review of
literature. Indian J Pathol Microbiol. 60:574–576. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Gill DM, Hahn AW, Hale P and Maughan BL:
Overview of current and future first-line systemic therapy for
metastatic clear cell renal cell carcinoma. Curr Treat Options
Oncol. 19:62018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Margulis V, Master VA, Cost NG, Leibovich
BC, Joniau S, Kuczyk M, Mulders PF, Kirkali Z, Wirth MP, Hirao Y,
et al: International consultation on urologic diseases and the
European Association of Urology international consultation on
locally advanced renal cell carcinoma. Eur Urol. 60:673–683. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lin S and Gregory RI: MicroRNA biogenesis
pathways in cancer. Nat Rev Cancer. 15:321–333. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Cirilo PDR, de Sousa Andrade LN, Corrêa
BRS, Qiao M, Furuya TK, Chammas R and Penalva LOF: MicroRNA-195
acts as an anti-proliferative miRNA in human melanoma cells by
targeting Prohibitin 1. BMC Cancer. 17:7502017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Gang L, Qun L, Liu WD, Li YS, Xu YZ and
Yuan DT: MicroRNA-34a promotes cell cycle arrest and apoptosis and
suppresses cell adhesion by targeting DUSP1 in osteosarcoma. Am J
Transl Res. 9:5388–5399. 2017.PubMed/NCBI
|
|
11
|
Xu Y, Du J, Zhang P, Zhao X, Li Q, Jiang
A, Jiang D, Tang G, Jiang Y, Wang J, et al: MicroRNA-125a-5p
mediates 3T3-L1 preadipocyte proliferation and differentiation.
Molecules. 23:E3172018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Song H, Rao Y, Zhang G and Kong X:
MicroRNA-384 inhibits the growth and invasion of renal cell
carcinoma cells by targeting astrocyte elevated gene 1. Oncol Res.
Aug 25–2017.(Epub ahead of print).
|
|
13
|
Wang X, Li H, Cui L, Feng J and Fan Q:
MicroRNA-182 suppresses clear cell renal cell carcinoma migration
and invasion by targeting IGF1R. Neoplasma. 63:717–725. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hu B, Wang J and Jin X: MicroRNA-138
suppresses cell proliferation and invasion of renal cell carcinoma
by directly targeting SOX9. Oncol Lett. 14:7583–7588.
2017.PubMed/NCBI
|
|
15
|
Chilosi M, Caliò A, Rossi A, Gilioli E,
Pedica F, Montagna L, Pedron S, Confalonieri M, Doglioni C, Ziesche
R, et al: Epithelial to mesenchymal transition-related proteins
ZEB1, β-catenin, and β-tubulin-III in idiopathic pulmonary
fibrosis. Mod Pathol. 30:26–38. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Cruz-Solbes AS and Youker K: Epithelial to
mesenchymal transition (EMT) and endothelial to mesenchymal
transition (EndMT): Role and implications in kidney fibrosis.
Results Probl Cell Differ. 60:345–372. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Fedele M, Cerchia L and Chiappetta G: The
epithelial-to-mesenchymal transition in breast cancer: Focus on
basal-like carcinomas. Cancers (Basel). 9:E1342017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Saito N, Mine N, Kufe DW, Von Hoff DD and
Kawabe T: CBP501 inhibits EGF-dependent cell migration, invasion
and epithelial-to-mesenchymal transition of non-small cell lung
cancer cells by blocking KRas to calmodulin binding. Oncotarget.
8:74006–74018. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zou JX, Revenko AS, Li LB, Gemo AT and
Chen HW: ANCCA, an estrogen-regulated AAA+ ATPase
coactivator for ERalpha, is required for coregulator occupancy and
chromatin modification. Proc Natl Acad Sci USA. 104:18067–18072.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Beroukhim R, Mermel CH, Porter D, Wei G,
Raychaudhuri S, Donovan J, Barretina J, Boehm JS, Dobson J,
Urashima M, et al: The landscape of somatic copy-number alteration
across human cancers. Nature. 463:899–905. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Raeder MB, Birkeland E, Trovik J, Krakstad
C, Shehata S, Schumacher S, Zack TI, Krohn A, Werner HM, Moody SE,
et al: Integrated genomic analysis of the 8q24 amplification in
endometrial cancers identifies ATAD2 as essential to MYC-dependent
cancers. PLoS One. 8:e548732013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zheng L, Li T, Zhang Y, Guo Y, Yao J, Dou
L and Guo K: Oncogene ATAD2 promotes cell proliferation, invasion
and migration in cervical cancer. Oncol Rep. 33:2337–2344. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Cattaneo M, Morozumi Y, Perazza D,
Boussouar F, Jamshidikia M, Rousseaux S, Verdel A and Khochbin S:
Lessons from yeast on emerging roles of the ATAD2 protein family in
gene regulation and genome organization. Mol Cells. 37:851–856.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hou M, Huang R, Song Y, Feng D, Jiang Y
and Liu M: ATAD2 overexpression is associated with progression and
prognosis in colorectal cancer. Jpn J Clin Oncol. 46:222–227. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhang M, Zhang C, Du W, Yang X and Chen Z:
ATAD2 is overexpressed in gastric cancer and serves as an
independent poor prognostic biomarker. Clin Transl Oncol.
18:776–781. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Hakimi AA, Furberg H, Zabor EC, Jacobsen
A, Schultz N, Ciriello G, Mikklineni N, Fiegoli B, Kim PH, Voss MH,
et al: An epidemiologic and genomic investigation into the obesity
paradox in renal cell carcinoma. J Natl Cancer Inst. 105:1862–1870.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Massari F, Di Nunno V, Ciccarese C, Graham
J, Porta C, Comito F, Cubelli M, Iacovelli R and Heng DYC: Adjuvant
therapy in renal cell carcinoma. Cancer Treat Rev. 60:152–157.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhu J, Ma X, Zhang Y, Ni D, Ai Q, Li H and
Zhang X: Establishment of a miRNA-mRNA regulatory network in
metastatic renal cell carcinoma and screening of potential
therapeutic targets. Tumour Biol. 37:15649–15663. 2016. View Article : Google Scholar
|
|
30
|
Xu SY, Xu PF and Gao TT: MiR-372-3p
inhibits the growth and metastasis of osteosarcoma cells by
targeting FXYD6. Eur Rev Med Pharmacol Sci. 22:62–69.
2018.PubMed/NCBI
|
|
31
|
Yu J, Jin L, Jiang L, Gao L, Zhou J, Hu Y,
Li W, Zhi Q and Zhu X: Serum miR-372 is a diagnostic and prognostic
biomarker in patients with early colorectal cancer. Anticancer
Agents Med Chem. 16:424–431. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wu G, Wang Y, Lu X, He H, Liu H, Meng X,
Xia S, Zheng K and Liu B: Low miR-372 expression correlates with
poor prognosis and tumor metastasis in hepatocellular carcinoma.
BMC Cancer. 15:1822015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Kong X, Qian X, Duan L, Liu H, Zhu Y and
Qi J: MicroRNA-372 suppresses migration and invasion by targeting
p65 in human prostate cancer cells. DNA Cell Biol. 35:828–835.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Liu BL, Sun KX, Zong ZH, Chen S and Zhao
Y: MicroRNA-372 inhibits endometrial carcinoma development by
targeting the expression of the Ras homolog gene family member C
(RhoC). Oncotarget. 7:6649–6664. 2016.PubMed/NCBI
|
|
35
|
Ragusa M, Statello L, Maugeri M, Majorana
A, Barbagallo D, Salito L, Sammito M, Santonocito M, Angelica R,
Cavallaro A, et al: Specific alterations of the microRNA
transcriptome and global network structure in colorectal cancer
after treatment with MAPK/ERK inhibitors. J Mol Med (Berl).
90:1421–1438. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Chen H, Zhang Z, Lu Y, Song K, Liu X, Xia
F and Sun W: Downregulation of ULK1 by microRNA-372 inhibits the
survival of human pancreatic adenocarcinoma cells. Cancer Sci.
108:1811–1819. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Tian RQ, Wang XH, Hou LJ, Jia WH, Yang Q,
Li YX, Liu M, Li X and Tang H: MicroRNA-372 is down-regulated and
targets cyclin-dependent kinase 2 (CDK2) and cyclin A1 in human
cervical cancer, which may contribute to tumorigenesis. J Biol
Chem. 286:25556–25563. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Ciró M, Prosperini E, Quarto M, Grazini U,
Walfridsson J, McBlane F, Nucifero P, Pacchiana G, Capra M,
Christensen J, et al: ATAD2 is a novel cofactor for MYC,
overexpressed and amplified in aggressive tumors. Cancer Res.
69:8491–8498. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Hwang HW, Ha SY, Bang H and Park CK: ATAD2
as a poor prognostic marker for hepatocellular carcinoma after
curative resection. Cancer Res Treat. 47:853–861. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Wan WN, Zhang YX, Wang XM, Liu YJ, Zhang
YQ, Que YH and Zhao WJ: ATAD2 is highly expressed in ovarian
carcinomas and indicates poor prognosis. Asian Pac J Cancer Prev.
15:2777–2783. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Shang P, Meng F, Liu Y and Chen X:
Overexpression of ANCCA/ATAD2 in endometrial carcinoma and its
correlation with tumor progression and poor prognosis. Tumour Biol.
36:4479–4485. 2015. View Article : Google Scholar : PubMed/NCBI
|