Introduction
Annually, ~780,000 patients are diagnosed worldwide
with liver cancer; this type of cancer accounted for ~740,000 cases
of mortality in 2012. Between 70 and 90% of primary liver cancer
cases are hepatocellular carcinoma (1–3). Viral
infection with hepatitis B or C, and alcohol consumption, are among
the most common factors that promote the occurrence and development
of liver cancer. However, despite investigation of the underlying
causes and pathogenesis of hepatocellular carcinoma, survival
remains poor (4,5).
MicroRNAs (miRNAs/miRs) are a type of noncoding RNA
that contain 19–22 nucleotide pairs. miRNAs inhibit
post-transcriptional gene expression by binding to the
3′-untranslated region (3′-UTR) of the target gene (6). miRNAs are associated with tumor cell
proliferation, differentiation, apoptosis, migration and invasion
(7), and have been implicated in
hepatocellular carcinoma growth (8).
miR-20b belongs to the miR-106a-363 gene cluster, and together with
the miR-17-92 and miR-106b-25 clusters, it forms the miR-17 gene
(9), which is active in the
oncogenesis of certain types of human tumor (10). Increased miR-20b expression has
previously been associated with decreased survival rate in gastric
cancer (11), promotion of
proliferation and migration of prostate cancer cells (12), and proliferation and DNA synthesis of
breast cancer cells (13). miR-20b
expression is altered in hepatocellular carcinoma, although its
activity has not yet been described. In the present study, it was
suggested that miR-20b promoted the proliferation of the mouse
hepatocellular carcinoma H22 cell line by directly targeting the
phosphatase and tensin homolog (PTEN) gene and negatively
regulating its expression. PTEN was at least partially
involved in the promotion of H22 cell proliferation by miR-20b.
Materials and methods
Cell culture and transfection
Mouse H22 hepatocellular carcinoma cells were
purchased from Nanjing Keygen Biotech Co., Ltd. (Nanjing, China).
Cells were cultured in RPMI-1640 complete medium (Gibco; Thermo
Fisher Scientific, Inc., Waltham, MA, USA) containing 10% fetal
bovine serum (HyClone; GE Healthcare Life Sciences, Logan, UT, USA)
at 37°C in a humidified incubator containing 5% CO2.
Cells were passaged once every 3–4 days. miR-20b inhibitor and a
scrambled RNA control were designed and synthesized by Shanghai
GenePharma Co., Ltd. (Shanghai, China). Cells (106
cells/ml) were transfected with the miR-20b inhibitor and control
(20 nmol/l) at 37°C for 24 h, using Lipofectamine® 2000
(Invitrogen; Thermo Fisher Scientific, Inc.) and Opti-MEM (Gibco;
Thermo Fisher Scientific, Inc.) reagents according to the
manufacturer's protocols. The sequences were as follows: miR-20b
inhibitor sequence, 5′-CUACCUGCACUAUGAGCACUUUG-3′; miR-20b
inhibitor control sequence, 5′-CAGUACUUUUGUGUAGUACAA-3′; miR-20b
mimics sequence forward, 5′-CAAAGUGCUCAUAGUGCAGGUAG-3′ and reverse,
5′-ACCUGCACUAUGAGCCACUUUGUU-3′; miR-20b mimics control sequence
forward, 5′-UUCUCCGAACGUGUCACGUTT-3′ and reverse,
5′-ACGUGACACGUUCGGAGAATT-3′.
Cell proliferation assay
Cell proliferation was assessed using the Cell
Counting Kit (CCK)-8 assay (Beyotime Institute of Biotechnology,
Haimen, China). Briefly, H22 cells were seeded in 96-well plates at
3×103 cells/well, and were incubated with 10 µl CCK-8
reagent for 2 h at 37°C. Absorbance was measured at 24, 48, 72 and
96 h at 450 nm using a microplate reader.
Construction of dual luciferase
recombinant plasmids
Total RNA was isolated from H22 cells with
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.) and was reverse transcribed to first strand cDNA using
TransScript First-Strand cDNA Synthesis SuperMix kit (TransGen
Biotech Co., Ltd., Beijing, China), according to the manufacturer's
protocol. The miR-20b binding site in the PTEN 3′-UTR region
sequence was predicted using the miRanda algorithm. The 3′-UTR
sequence of the PTEN gene was then amplified by polymerase
chain reaction (PCR) using TransStart FastPfu DNA Polymerase
(TransGen Biotech Co., Ltd., Beijing, China). The reaction
conditions were: 95°C predenaturation, 1 min; 95°C denaturation, 20
sec; 57°C annealing, 20 sec; 72°C extension, 30 sec; 40 cycles. The
length of the amplified product was ~318 bp. The primer PCR
amplification sequences were as follows: Forward
5′-CCGCTCGAGCCCTCCTTGCTATCCT-3′ and reverse
5′-GAATGCGGCCGCTCCCGATGAAACCTC-3′. The miRNA reporter vector
plasmid (pmiR-RB-Report system; Applied Biosystems; Thermo Fisher
Scientific, Inc.) and the amplified 3′-UTR PTEN gene
sequence were double digested with XhoI and NotI. The
digested products were separated using electrophoresis on a 1.5%
agarose gel, and the gels were cut to recover the purified product.
The ligation reaction of the purified product was performed at 16°C
using the TaKaRa DNA Ligation kit Ver. 2.0 (Takara Bio, Inc., Otsu,
Japan), according to the manufacturer's protocol, and the ligation
product was transformed into competent DH5α cell lines (Beyotime
Institute of Biotechnology, Haimen, China) for expansion culture
for 16 h at 37°C. The pmiR-RB-Reporter-PTEN 3′-UTR dual luciferase
recombinant plasmid was extracted by concentration from bacterial
culture supernatant using DNA extraction kit (Axygen Biosciences,
Union City, CA, USA). Recombinant plasmids were digested by
XhoI and NotI restriction enzymes and sent to Sangon
Biotech Co., Ltd. (Shanghai, China) for sequencing.
Luciferase activity assay
The recombinant plasmid or empty plasmid was
co-transfected with miR-20b mimics or miR-20b scramble into HeLa
cells. The detection of luciferase activity was divided into four
groups: (i) pmiR-RB-Report empty plasmid+miR-20b scramble; (ii)
pmiR-RB-Report empty plasmid+miR-20b mimics; (iii)
pmiR-RB-Report-PTEN 3′-UTR recombinant plasmid+miR-20b scramble;
and (iv) pmiR-RB-Report-PTEN 3′-UTR recombinant plasmid+miR-20b
mimics. Luciferase activity was assessed by the luciferase activity
assay kit (Promega Corporation, Madison, WI, USA), according to the
manufacturer's protocol. HeLa cells were washed with PBS and lysed
with freshly prepared lysis buffer (Promega Corporation, Madison,
WI, USA). The luciferase activity was evaluated in 96-well black
plates and the results were expressed as the ratio of
Renilla and firefly luciferase fluorescence.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was extracted from H22 cells with
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.) according to the manufacturer's protocol. RNA was then
reverse transcribed to cDNA (Easy Script First-Strand cDNA
Synthesis SuperMix kit; TransGen Biotech Co., Ltd.) and amplified
by PCR (TransStart Tip Green qPCR SuperMix kit; TransGen Biotech
Co., Ltd.), according to the manufacturer's protocol (Beijing
TransGen Biotech Co., Ltd., Beijing, China). The PCR conditions
were 94°C for 30 sec; 94°C for 5 sec, 60°C for 30 sec, for a total
of 40 cycles. The 2−ΔΔCq method (14) was used to determine the relative
expression of each gene; miR-20b expression was normalized against
U6 small nuclear RNA as an internal reference. Primer sequences
were as follows: miR-20b RT primer,
GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACCTACCT; forward,
5′-ATGCCAAAGTGCTCATAGTG-3′; reverse, 5′-GTGCAGGGTCCGAGGT-3′; U6 RT
primer, 5′-AACGCTTCACGAATTTGCGT-3′; forward,
5′-CTCGCTTCGGCAGCACA-3′; and reverse,
5′-AACGCTTCACGAATTTGCGT-3′.
RNA interference
Transfection of interfering RNA fragments was
performed with Lipofectamine® 2000 reagent (Invitrogen;
Thermo Fisher Scientific, Inc.), according to the manufacturer's
protocol. Cells (106 cells/ml) were transfected 48 h at
37°C with 30 nM PTEN small interfering (si)RNA or scramble
siRNA, which was synthesized by Shanghai GenePharma Co., Ltd.
(Shanghai, China). PTEN siRNA sense, 5′-GGUGAAACUAUACUUUACATT-3′
and antisense, 5′-UGUAAAGUAUAGUUUCACCTT-3′; scramble siRNA sense,
5′-UUCUCCGAACGUGUCACGUTT-3′ and antisense,
5′-ACGUGACACGUUCGGAGAATT-3′.
Western blotting
Western blot analyses were performed as previously
described (15). Briefly, cells were
harvested and lysed using NP-40 lysis buffer (Beyotime Institute of
Biotechnology) and the protein concentration of the cell lysates
was quantified using the bicinchoninic acid assay. Equal aliquots
of sample protein were separated by 10% SDS-PAGE and transferred to
polyvinylidene difluoride membranes. The membranes were blocked at
room temperature with 5% non-fat dry milk for 2 h, and were
subsequently incubated with anti-GAPDH antibody (cat. no. AF0006;
Beyotime Institute of Biotechnology) and anti-PTEN antibody (cat.
no. AF1426; Beyotime Institute of Biotechnology) at a 1:1,000
dilution, overnight at 4°C. Subsequently, the membranes were
incubated with a horseradish peroxidase-conjugated secondary
antibody (cat. no. A0208; Beyotime Institute of Biotechnology) for
2 h at room temperature and bands were visualized by enhanced
chemiluminescence using Immobilon™ Western Chemiluminescent HRP
Substrate (EMD Millipore, Billerica, MA, USA).
Statistical analysis
Data are expressed as the mean ± standard error of
the mean from at least three experiments. Statistical analysis was
performed with SPSS 16.0 software (SPSS Inc., Chicago, IL, USA).
Analysis of variance followed by the least significant difference
test was used for comparison between groups. P<0.05 was
considered to indicate a statistically significant difference.
Results
miR-20b promotes the proliferation of
H22 cells in vitro
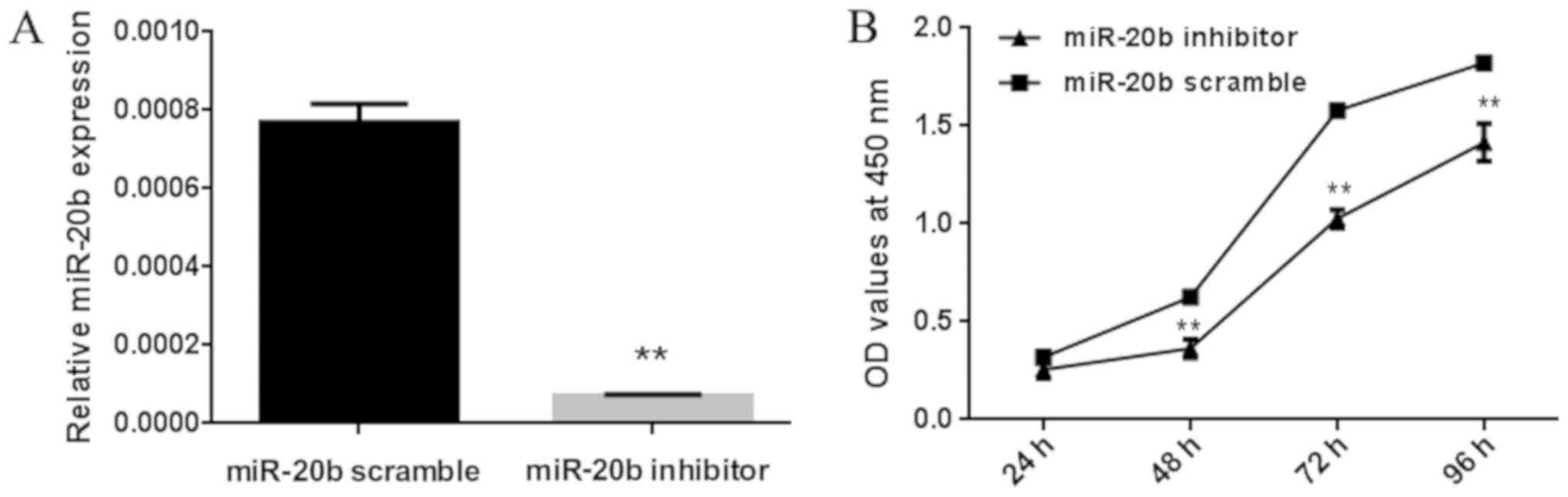
The effects of miR-20b on the proliferation of H22
cells were investigated using the CCK-8 assay, by comparing
proliferation between cells that were transfected with miR-20b
inhibitor or miR-20b scramble control. Compared with the
transfected control group, transfection with the miR-20b inhibitor
significantly reduced miR-20b expression levels in H22 cells
(Fig. 1A). CCK-8 assay results at 48,
72 and 96 h revealed a significant inhibition in the proliferation
of miR-20b-transfected H22 cells compared with that in the control
cells (Fig. 1B).
miR-20b negatively regulates PTEN
expression in H22 cells
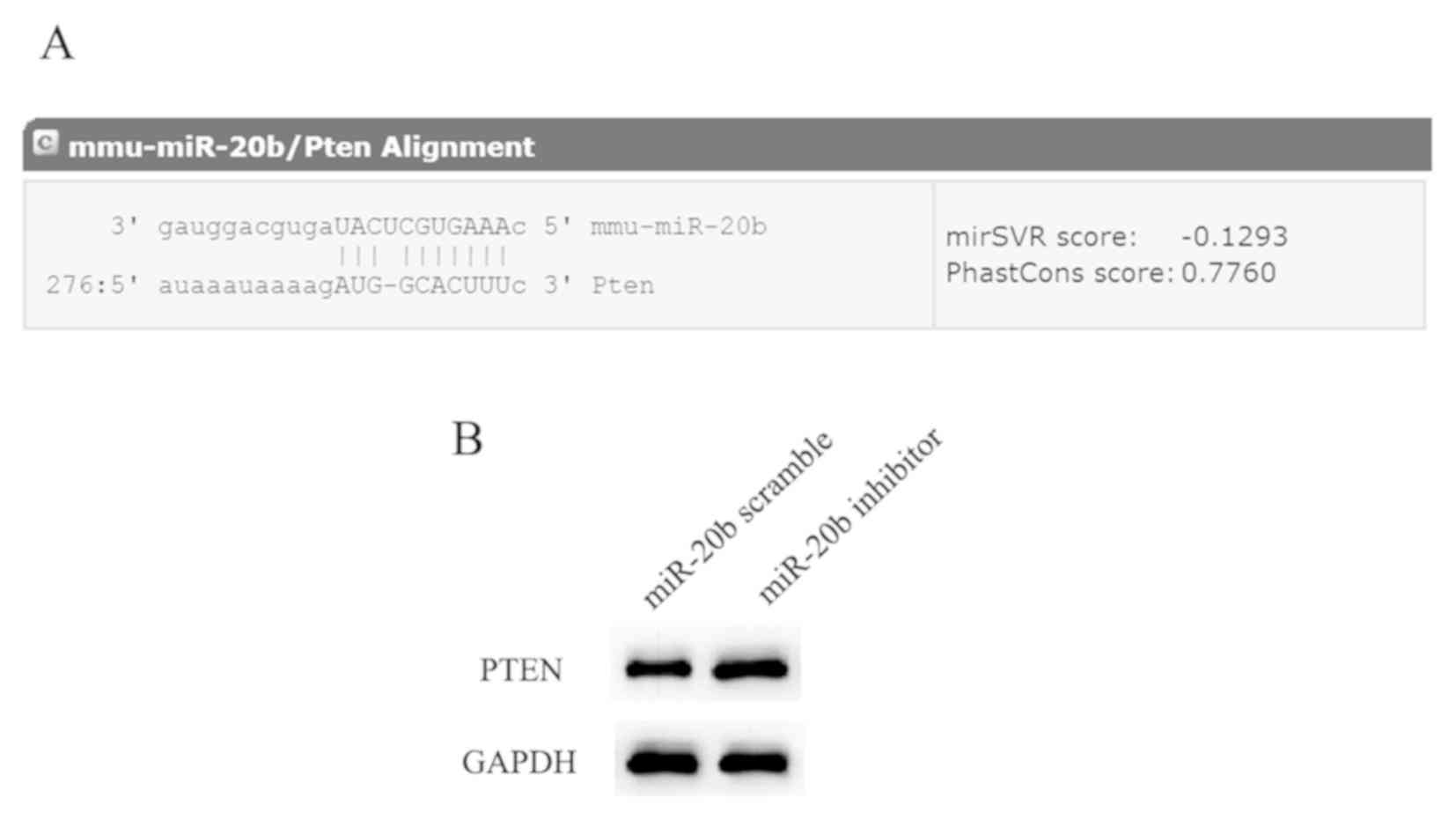
Investigation of the mechanism of action of miR-20b
was guided by the miRanda prediction of PTEN as a potential
target gene of miR-20b (microRNA.org)
(Fig. 2A). PTEN is a tumor
suppressor gene which inhibits the proliferation of various types
of tumor, including bladder cancer, breast cancer and colon
carcinoma (16). In the present
study, downregulation of miR-20b in H22 cells significantly
upregulated PTEN expression (Fig.
2B). These findings suggested that the expression of
PTEN was negatively regulated by miR-20b.
PTEN is a direct target gene of
miR-20b
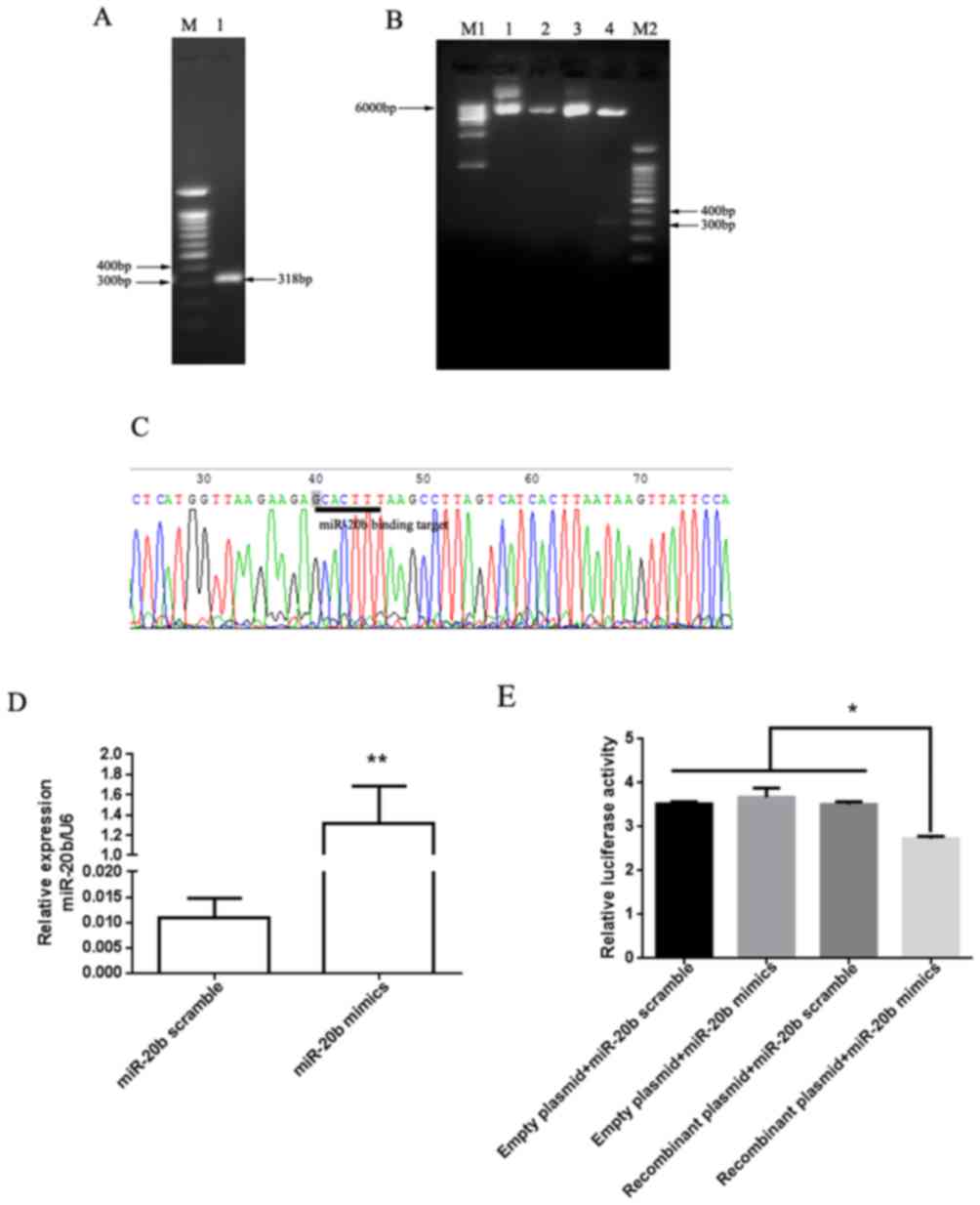
The direct targeting of PTEN by miR-20b was
confirmed with a dual luciferase reporter vector assay. The
PTEN 3′-UTR fragment containing the miR-20b binding site
(Fig. 3A) was cloned and inserted
into the pmiR-RB-Reporter plasmid vector system. The
pmiR-RB-Reporter-PTEN 3′-UTR dual luciferase recombinant plasmid
vectors were identified by double enzyme digestion electrophoresis
(Fig. 3B) and DNA sequencing
(Fig. 3C). The miR-20b mimics or
control, and recombinant or empty plasmids were co-transfected into
H22 cells. Post-transfection of H22 cells with miR-20b mimics, the
expression levels of miR-20b were increased by ~120-fold compared
with the control group (Fig. 3D). The
results of the dual luciferase reporter assay revealed that miR-20b
significantly reduced recombinant plasmid luciferase activity
(Fig. 3E). These results indicated
that miR-20b may directly target PTEN in H22 cells.
Downregulation of PTEN partially
reverses the anti-proliferative effects of miR-20b inhibitor in H22
cells
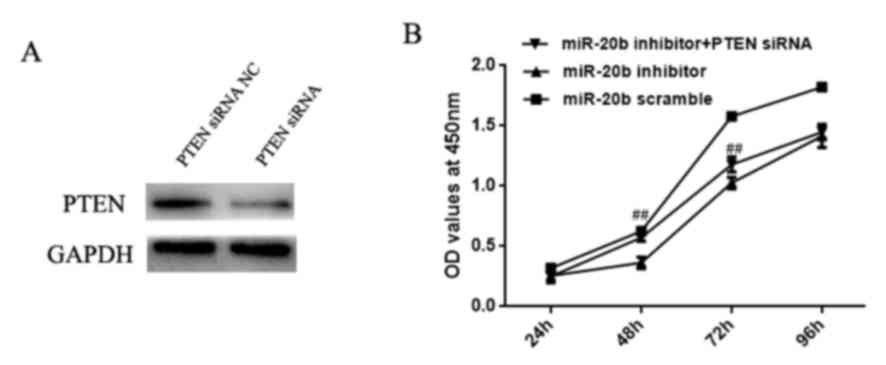
To confirm that the effect of miR-20b on H22 cell
proliferation may be mediated directly by targeting PTEN,
H22 cells were transfected with PTEN siRNA and miR-20b
inhibitor. Initially, total protein was extracted from the
siRNA-transfected H22 cells and PTEN protein expression was
assessed by western blotting. As illustrated in Fig. 4A, PTEN expression was
downregulated in response to PTEN siRNA. The CCK-8 results
(Fig. 4B) revealed that the miR-20b
inhibitor significantly reduced H22 cell growth, whereas the
PTEN siRNA partially reversed this effect.
Discussion
In the present study, downregulation of miR-20b
inhibited H22 cell growth. It is well known that miRNAs participate
in oncogenesis by regulating the expression of their target genes
(17). miR-20b is a member of the
miR-106a-363 and miR-7 gene families (9), which are active in various types of
human tumor (10). For example,
miR-20b activity is increased in liver, gastric and breast tumors
(18–20), and it is also a plasmatic marker for
non-small cell lung cancer (21). In
addition, anti-angiomiR-miR-20b has been described as a potential
therapeutic target for refractory large B-cell lymphoma (22). In this study, miR-20b was revealed to
promote H22 cell growth by targeting PTEN.
PTEN is a tumor suppressor gene located on
chromosome 10 at 10q23.31 (23). It
codes for a lipid phosphatase that dephosphorylates the second
messenger inositol triphosphate and negatively regulates the
phosphoinositide 3-kinase pathway (24,25).
PTEN regulates tumor cell migration, cell cycle progression
and apoptosis (26). The loss of
PTEN expression has been demonstrated to promote
transforming growth factor-β-induced cell invasiveness, and
PTEN deletion is associated with progression and liver
metastasis in colon cancer (27). In
addition, PTEN is regulated by miRNAs in ovarian, colon and
breast cancer, glioma and other malignant tumors (28–31).
Notably, miR-20b regulates proliferation and migration of prostate
cancer cells by directly targeting PTEN (12). Chu et al (32) demonstrated that the expression of PTEN
is inhibited by miR-205, which is why miR-205 promotes
proliferation and invasion of ovarian cancer cells. miR-106a
promotes proliferation of prostate cancer cells by regulating the
expression of PTEN (33). PTEN
expression is significantly lower in primary hepatocellular
carcinoma than in normal liver tissue, and its expression is
correlated with tumor stage, invasion and metastasis (34). In the present study, miR-20b
negatively regulated PTEN expression, whereas PTEN
siRNA partially reversed the anti-proliferative effect of the
miR-20b inhibitor on H22 cells. Therefore, it was suggested that
miR-20b promoted H22 cell growth by directly targeting and
downregulating PTEN expression, and PTEN expression
may be involved in the effects of miR-20b on H22 cell growth. The
importance of the present study is that it may further reveal the
growth mechanism of hepatocellular carcinoma, and also provide a
theoretical basis for clinical tumor therapy with miR-20b as the
target.
Acknowledgements
Not applicable.
Funding
This project was supported by the National Science
Foundation of China (grant no. 81273273), the Anhui Provincial
Natural Science Foundation (grant no. 1708085MH218), the Key
Program of Anhui Province for Outstanding Talents in University
(grant nos. gxbjZD2016071 and 2014H012), and the Scientific
Research Innovation Team Project of Anhui Colleges and Universities
(grant no. 2016-40).
Availability of data and materials
The datasets used and analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
CWS and HZL conceived and designed the experiments.
JH, MMM, YLL, and HLW performed the experiments. CWS, HM, SJG, QF,
ZQQ and HZL analyzed and interpreted the data, and wrote the
manuscript. HZL revised the paper. All authors reviewed and
approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
de Martel C, Maucort-Boulch D, Plummer M
and Franceschi S: World-wide relative contribution of hepatitis B
and C viruses in hepatocellular carcinoma. Hepatology.
62:1190–1200. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
He RQ, Wu PR, Xiang XL, Yang X, Liang HW,
Qiu XH, Yang LH, Peng ZG and Chen G: Downregulated miR-23b-3p
expression acts as a predictor of hepatocellular carcinoma
progression: A study based on public data and RT-qPCR verification.
Int J Mol Med. 41:2813–2831. 2018.PubMed/NCBI
|
|
4
|
M'Bengue AK, Doumbia M, Denoman SR,
Ouattara DN, Adoubi I and Pineau P: A major shift of viral and
nutritional risk factors affects the hepatocellular carcinoma risk
among Ivorian patients: A preliminary report. Infect Agent Cancer.
10:182015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhang E, Liu Q, Wang Y, Wang H, He L, Jin
X and Li N: MicroRNA miR-147b promotes tumor growth via targeting
UBE2N in hepatocellular carcinoma. Oncotarge. 8:114072–114080.
2017. View Article : Google Scholar
|
|
6
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lu J, Getz G, Miska EA, Alvarez-Saavedra
E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA,
et al: MicroRNA expression profiles classify human cancers. Nature.
435:834–838. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Liu Z, Wang Y, Dou C, Sun L, Li Q, Wang L,
Xu Q, Yang W, Liu Q and Tu K: MicroRNA-1468 promotes tumor
progression by activating PPAR-γ-mediated AKT signaling in human
hepatocellular carcinoma. J Exp Clin Cancer Res. 37:492018.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Tanzer A and Stadler PF: Molecular
evolution of a microRNA cluster. J Mol Biol. 339:327–335. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hayashita Y, Osada H, Tatematsu Y, Yamada
H, Yanagisawa K, Tomida S, Yatabe Y, Kawahara K, Sekido Y and
Takahashi T: A polycistronic microRNA cluster, miR-17-92, is
overexpressed in human lung cancers and enhances cell
proliferation. Cancer Res. 65:9628–9632. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Katada T, Ishiguro H, Kuwabara Y, Kimura
M, Mitui A, Mori Y, Ogawa R, Harata K and Fujii Y: microRNA
expression profile in undifferentiated gastric cancer. Int J Oncol.
34:537–542. 2009.PubMed/NCBI
|
|
12
|
Guo J, Xiao Z, Yu X and Cao R: miR-20b
promotes cellular proliferation and migration by directly
regulating phosphatase and tensin homolog in prostate cancer. Oncol
Lett. 14:6895–6900. 2017.PubMed/NCBI
|
|
13
|
Zhou W, Shi G, Zhang Q, Wu Q, Li B and
Zhang Z: MicroRNA-20b promotes cell growth of breast cancer cells
partly via targeting phosphatase and tensin homologue (PTEN). Cell
Biosci. 4:622014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Song C, Ma H, Yao C, Tao X and Gan H:
Alveolar macrophage-derived vascular endothelial growth factor
contributes to allergic airway inflammation in a mouse asthma
model. Scand J Immunol. 75:599–605. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chen L and Guo D: The functions of tumor
suppressor PTEN in innate and adaptive immunity. Cell Mol Immunol.
14:581–589. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ghosh N and Katare R: Molecular mechanism
of diabetic cardiomyopathy and modulation of microRNA function by
synthetic oligonucleotides. Cardiovasc Diabetol. 17:432018.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Xue TM, Tao LD, Zhang M, Zhang J, Liu X,
Chen GF, Zhu YJ and Zhang PJ: Clinicopathological significance of
microRNA-20b expression in hepatocellular carcinoma and regulation
of HIF-1α and VEGF effect on cell biological behaviour. Dis
Markers. 2015:3251762015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Xue TM, Tao LD, Zhang M, Xu GC, Zhang J
and Zhang PJ: miR-20b overexpression is predictive of poor
prognosis in gastric cancer. Onco Targets Ther. 8:1871–1876. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ahmad A, Ginnebaugh KR, Sethi S, Chen W,
Ali R, Mittal S and Sarkar FH: miR-20b is up-regulated in brain
metastases from primary breast cancers. Oncotarget. 6:12188–12195.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Leidinger P, Brefort T, Backes C, Krapp M,
Galata V, Beier M, Kohlhaas J, Huwer H, Meese E and Keller A:
High-throughput qRT-PCR validation of blood microRNAs in non-small
cell lung cancer. Oncotarget. 7:4611–4623. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Borges NM, do Vale Elias M, Fook-Alves VL,
Andrade TA, de Conti ML, Macedo MP, Begnami MD, Campos AH, Etto LY,
Bortoluzzo AB, et al: Angiomirs expression profiling in diffuse
large B-Cell lymphoma. Oncotarget. 7:4806–4816. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Li J, Yen C, Liaw D, Podsypanina K, Bose
S, Wang SI, Puc J, Miliaresis C, Rodgers L, McCombie R, et al:
PTEN, a putative protein tyrosine phosphatase gene mutated in human
brain, breast, and prostate cancer. Science. 275:1943–1947. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Bertram J, Peacock JW, Tan C, Mui AL,
Chung SW, Gleave ME, Dedhar S, Cox ME and Ong CJ: Inhibition of the
phosphatidylinositol 3′-kinase pathway promotes autocrine
Fas-induced death of phosphatase and tensin homologue-deficient
prostate cancer cells. Cancer Res. 66:4781–4788. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Meng F, Henson R, Wehbe-Janek H, Ghoshal
K, Jacob ST and Patel T: MicroRNA-21 regulates expression of the
PTEN tumor suppressor gene in human hepatocellular cancer.
Gastroenterology. 133:647–658. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chung JH, Ginn-Pease ME and Eng C:
Phosphatase and tensin homologue deleted on chromosome 10 (PTEN)
has nuclear localization signal-like sequences for nuclear import
mediated by major vault protein. Cancer Res. 65:4108–4116. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Hjelmeland AB, Hjelmeland MD, Shi Q, Hart
JL, Bigner DD, Wang XF, Kontos CD and Rich JN: Loss of phosphatase
and tensin homologue increases transforming growth factor
beta-mediated invasion with enhanced SMAD3 transcriptional
activity. Cancer Res. 65:11276–11281. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Yang H, Kong W, He L, Zhao JJ, O'Donnell
JD, Wang J, Wenham RM, Coppola D, Kruk PA, Nicosia SV and Cheng JQ:
MicroRNA expression profiling in human ovarian cancer: miR-214
induces cell survival and cisplatin resistance by targeting PTEN.
Cancer Res. 68:425–433. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wu W, Yang J, Feng X, Wang H, Ye S, Yang
P, Tan W, Wei G and Zhou Y: MicroRNA-32 (miR-32) regulates
phosphatase and tensin homologue (PTEN) expression and promotes
growth, migration, and invasion in colorectal carcinoma cells. Mol
Cancer. 12:302013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Huse JT, Brennan C, Hambardzumyan D, Wee
B, Pena J, Rouhanifard SH, Sohn-Lee C, le Sage C, Agami R, Tuschl T
and Holland EC: The PTEN-regulating microRNA miR-26a is amplified
in high-grade glioma and facilitates gliomagenesis in vivo. Genes
Dev. 23:1327–1337. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Dai X, Fang M, Li S, Yan Y, Zhong Y and Du
B: miR-21 is involved in transforming growth factor β1-induced
chemoresistance and invasion by targeting PTEN in breast cancer.
Oncol Lett. 14:6929–6936. 2017.PubMed/NCBI
|
|
32
|
Chu P, Liang A, Jiang A and Zong L:
miR-205 regulates the proliferation and invasion of ovarian cancer
cells via suppressing PTEN/SMAD4 expression. Oncol Lett.
15:7571–7578. 2018.PubMed/NCBI
|
|
33
|
Luo B, Kang N, Chen Y, Liu L and Zhang Y:
Oncogene miR-106a promotes proliferation and metastasis of prostate
cancer cells by directly targeting PTEN in vivo and in vitro.
Minerva Med. 109:24–30. 2018.PubMed/NCBI
|
|
34
|
Chai Y1, Xiaoyu L and Haiyan W:
Correlation between expression levels of PTEN and p53 genes and the
clinical features of HBsAg-positive liver cancer. J BUON.
22:942–946. 2017.PubMed/NCBI
|