Introduction
Breast cancer (BC) is the most common malignant
tumor and the primary cause of cancer-associated mortality in women
worldwide (1). Although numerous
treatments, including chemotherapy, radiotherapy, endocrine therapy
and targeted therapy, are currently available for BC, the response
of patients greatly varies partly due to their own antitumor
immunity (2). Accumulating evidence
suggests that adaptive immunity mediated by T and B lymphocytes
provides the critical foundation for effective and sustained
antitumor responses. Tumor-infiltrating lymphocytes (TILs) are
likely to be the most relevant indicator of tumor immunity in solid
tumors, with prognostic value (3).
In BC, extensive tumor infiltration by cytotoxic CD8+ T
cells is markedly associated with patient survival (4,5) and
response to therapy (6).
Furthermore, the baseline expression of TILs can predict the
pathological complete response (pCR) result following neoadjuvant
chemotherapy in patients with BC (7), which is an important prognostic
indicator.
Previous studies revealed that the composition of
the gastrointestinal microbiome is a major environmental factor
that varies among individuals, which may affect systemic immunity
(8,9). The gastrointestinal microbiome has been
demonstrated to initiate the differentiation of T cells, the
expansion of specific molecular subsets (10,11) and
the activation state of innate antigen-presenting cells (APCs),
which may eventually affect priming of the systemic immune response
(12,13). In addition, the gastrointestinal
microbiome may improve the outcomes of cancer treatment by
impairing inflammatory activation in response to different
therapeutic protocols (14).
Numerous studies have confirmed the association between the
gastrointestinal microbiome and tumors, particularly in colon
carcinoma (15–17). However, for extraintestinal tumors,
to the best of our knowledge, such an association has not been
established. The present study aimed to assess whether the
diversity of the gastrointestinal microbiome was associated with
different expression patterns of TILs in patients with BC.
Materials and methods
Patients
Between March 2017 and October 2017, a total of 90
biopsy-confirmed female patients with BC were enrolled in the
present study at the Breast Center of The Fourth Hospital of Hebei
Medical University (Shijiazhuang, China). All patients were first
treated by chemotherapy, followed by surgical treatment, as
appropriate. Available clinicopathological data included age,
staging, menstrual state, estrogen receptor (ER) and/or
progesterone receptor (PgR) status, human epidermal growth factor
receptor 2 (HER2) status, TIL classification, and pCR cases. ER and
PgR were assessed as positive if ≥1% of tumor cells exhibited
nuclear staining (18).
HER2-positive status was defined as a score of 3+ based on
immunohistochemistry assay or HER2 gene amplification using
fluorescent in situ hybridization, as described previously
(19). The Miller-Payne grading
system was used to evaluate the pathological response in surgical
specimens (20), and pCR was defined
as the absence of residual invasive tumor cells in the breast and
axillary lymph nodes (ypT0/is + ypN0) in surgical specimens. All
procedures were supervised and approved by the Human Tissue
Research Committee of The Fourth Hospital of Hebei Medical
University, and informed consent was provided by all
participants.
Assessment of TILs using a three-grade
scale
Core needle biopsy was performed in the examination
room. Briefly, between three and five lump tissues were obtained
from different directions to obtain a suitable number of tissue
samples. Subsequently, the tissue strip was placed in 4% neutral
formalin solution and sent to the Pathology Department. Evaluation
of TILs on the core needle biopsy specimens was performed by two
experienced pathologists who were familiar with the evaluation
criteria recommended by the International TILs Working Group in
2014 (21). The whole slide was
screened using a low-power field, while an area with many
lymphocytes was identified as a ‘hotspot’. TILs were then evaluated
by light microscopy in a medium-power field (magnification, ×100).
The region of interest was restricted within the tumor borders as
described by Salgado et al (21).
TIL score was defined as the proportion of the area infiltrated by
lymphocytes within the tumor itself plus the adjacent stroma, and
the scores were classified as low (<10), intermediate (10–50)
and high (>50%), accordingly (22).
16S ribosomal DNA (rDNA)
amplification
Fresh fecal samples were collected from the 90
patients with BC and stored at −80°C. DNA was extracted from all
samples using a ProbeGene® Soil genomic DNA extraction
kit (ProbeGene, Jiangsu, China) according to the manufacturer's
protocol, and purified DNA was stored at −80°C prior to further
analysis.
The 16S rDNA V3-V4 region of the ribosomal RNA gene
was amplified by polymerase chain reaction using primers 341F
(5′-CCTACGGGNGGCWGCAG-3′) and 806R (5′-GGACTACHVGGGTATCTAAT-3′),
where the barcode was an eight-base sequence unique to each sample.
Polymerase chain reaction was performed in a 50-µl reaction system
consisting of 5 µl 10X KOD buffer, 5 µl 2.5 mM deoxyribonucleotide
triphosphates, 1.5 µl of each primer (5 µM), 1 µl KOD polymerase
(Toyobo (Shanghai) Co., Ltd., Shanghai, China) and 100 ng template
DNA. Briefly, following a denaturation step at 95°C for 2 min, the
amplifications were carried out with 27 cycles at a melting
temperature of 98°C for 10 sec, an annealing temperature of 62°C
for 30 sec, and an extension temperature of 68°C for 30 sec,
followed by a final extension step at 68°C for 10 min. Each
experiment was conducted in triplicate. Amplicons were subjected to
electrophoresis on 2% agarose gels, purified using the AxyPrep DNA
Gel Extraction kit (Axygen; Corning Inc., Corning, NY, US),
according to the manufacturer's protocol, and semi-quantified using
the QuantiFluor dsDNA system (Promega Corporation, Madison, WI,
US). Purified amplicons were pooled in equimolar concentrations and
underwent paired-end sequencing (2×250) on the Illumina HiSeq2500
platform (Illumina, Inc., San Diego, CA, USA) according to the
manufacturer's protocols.
Statistical analysis
Statistical analysis was performed using SPSS 23.0
software (SPSS, Inc., Chicago, IL, USA). Clinical characteristics,
including age, menopausal state, staging, level of HER2 and ER/PgR
expression, were analyzed using a χ2 test. P<0.05 was
considered to indicate a statistically significant difference.
Raw data from Illumina sequencing contain adapters
or low-quality reads that may affect subsequent data assembly and
analysis. Therefore, to obtain high-quality clean reads, raw reads
were filtered according to the following criteria: i) Reads
containing >10% unknown nucleotides were removed; and ii) reads
containing <80% high-quality bases (Q-value, >20) were
excluded.
Paired-end clean reads were merged as raw tags using
FLASH v1.2.11 (23) with a minimum
overlap of 10 bp and a mismatch error rate of 2%. Noisy sequences
of raw tags were filtered by QIIME v1.9.1 (24) pipeline under specific filtering
conditions (25). Clean tags were
searched against the reference database (http://drive5.com/uchime/uchime_download.html) to
perform reference-based chimera detection using the UCHIME
algorithm (http://www.drive5.com/usearch/manual/uchime_algo.html).
All chimeric tags were removed, and effective tags were finally
obtained for further analysis.
The effective tags were clustered into operational
taxonomic units (OTUs) of ≥97% similarity using the UPARSE
(26) pipeline. The tag sequence
with highest abundance was selected as a representative sequence
within each cluster. Venn analysis was performed among groups to
identify unique and common OTUs. The representative sequences were
classified into organisms by a naive Bayesian model using RDP
classifier v2.2 (27) based on the
Greengenes (28) database
(https://www.arb-silva.de/). Weighted and
unweighted UniFrac distance matrices were generated using QIIME for
β-diversity analysis. Between-group comparison of β-diversity was
performed using Welch's t-test and Wilcoxon rank test in R.
β-diversity comparison among groups was computed using Tukey's HSD
test and Kruskal-Wallis H test in R. Analysis of similarity
(ANOSIM) was used to test whether the differences among groups were
significantly greater than those within groups. Biomarker features
in each group were screened by Metastats software (v.20090414)
(29).
Results
Clinical characteristics
A total of 80 patients were included in the present
study (10 cases were unavailable since mass was removed in another
hospital or the biopsy section could not be found) and divided into
three groups as follows: High expression of TILs (TIL-H; n=21),
medium expression of TILs (TIL-M; n=34) and low expression of TILs
(TIL-L; n=25). Associations between TIL distribution and clinical
characteristics, including age, menstrual status, staging, HER2
expression and ER/PgR expression, were assessed (Table I). Only the expression status of HER2
was positively associated with TIL distribution (P=0.037). A total
of 58 patients who underwent surgery following chemotherapy (20
from TIL-H group, 22 from TIL-M group and 16 from TIL-L group) were
evaluated for chemotherapy efficiency (Table II).
 | Table I.Clinical characteristics associated
with TILs. |
Table I.
Clinical characteristics associated
with TILs.
|
| No. of cases |
|
|
|---|
|
|
|
|
|
|---|
|
Characteristics | TIL-low | TIL-medium | TIL-high | χ2 | P-value |
|---|
| Age (years) |
|
|
| 0.541 | 0.736 |
|
<45 | 7 | 14 | 7 |
|
|
|
45–59 | 6 | 12 | 12 |
|
|
|
≥60 | 7 | 8 | 6 |
|
|
| Staging |
|
|
| 2.701 | 0.259 |
| I | 0 | 0 | 0 |
|
|
| II | 7 | 5 | 7 |
|
|
|
III | 11 | 20 | 12 |
|
|
| IV | 3 | 9 | 6 |
|
|
| Menopausal
state |
|
|
| 0.269 | 0.874 |
|
Yes | 11 | 19 | 15 |
|
|
| No | 10 | 15 | 10 |
|
|
| HER2 |
|
|
| 6.597 | 0.037 |
|
Positive | 6 | 7 | 13 |
|
|
|
Negative | 15 | 26 | 12 |
|
|
| ER/PgR |
|
|
| 3.251 | 0.197 |
|
Positive | 18 | 25 | 15 |
|
|
|
Negative | 3 | 8 | 10 |
|
|
 | Table II.Comparison of pCR of patients in the
TIL-H group and patients in the other two groups. |
Table II.
Comparison of pCR of patients in the
TIL-H group and patients in the other two groups.
| Patients | TIL-low and
TIL-medium | TIL-H | χ2 | P-value |
|---|
| pCR |
2 |
4 | 3.015 | 0.082 |
| Non-pCR | 36 | 16 |
|
|
Analysis of species differences
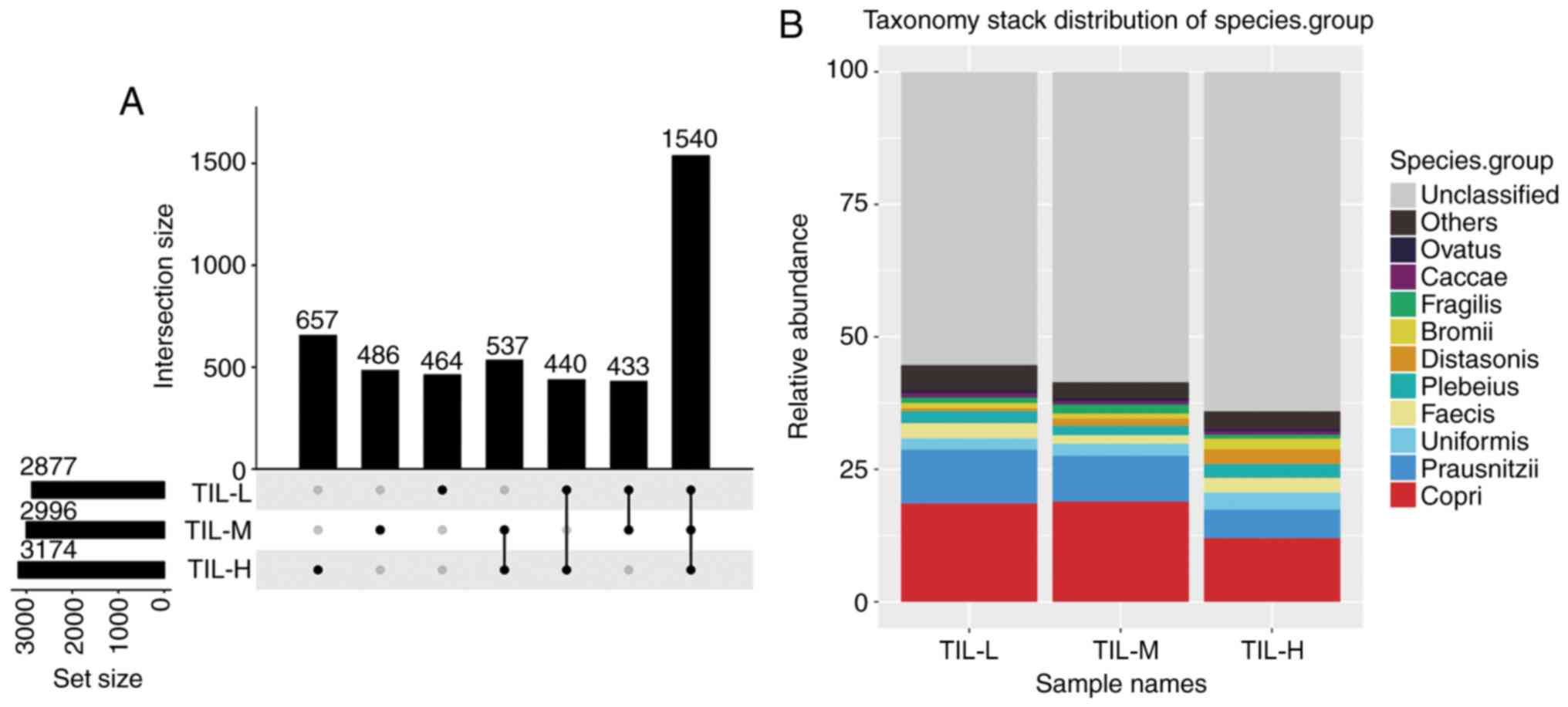
There were 3,174, 2,996 and 2,877 different OTUs in
the TIL-H, TIL-M and TIL-L groups, respectively. The number of
common and unique OTUs is shown in Fig.
1A, and Fig. 1B shows the top 10
species and their abundances. Tables
III and IV illustrated that the
gastrointestinal microbiome, when compared among the three groups
(TIL-L vs. TIL-M vs. TIL-H) or compared between the TIL-L and TIL-H
groups, exhibited significantly different β-diversities in weighted
and unweighted UniFrac analyses, which suggested that low
expression of TILs was associated with lower β-diversity
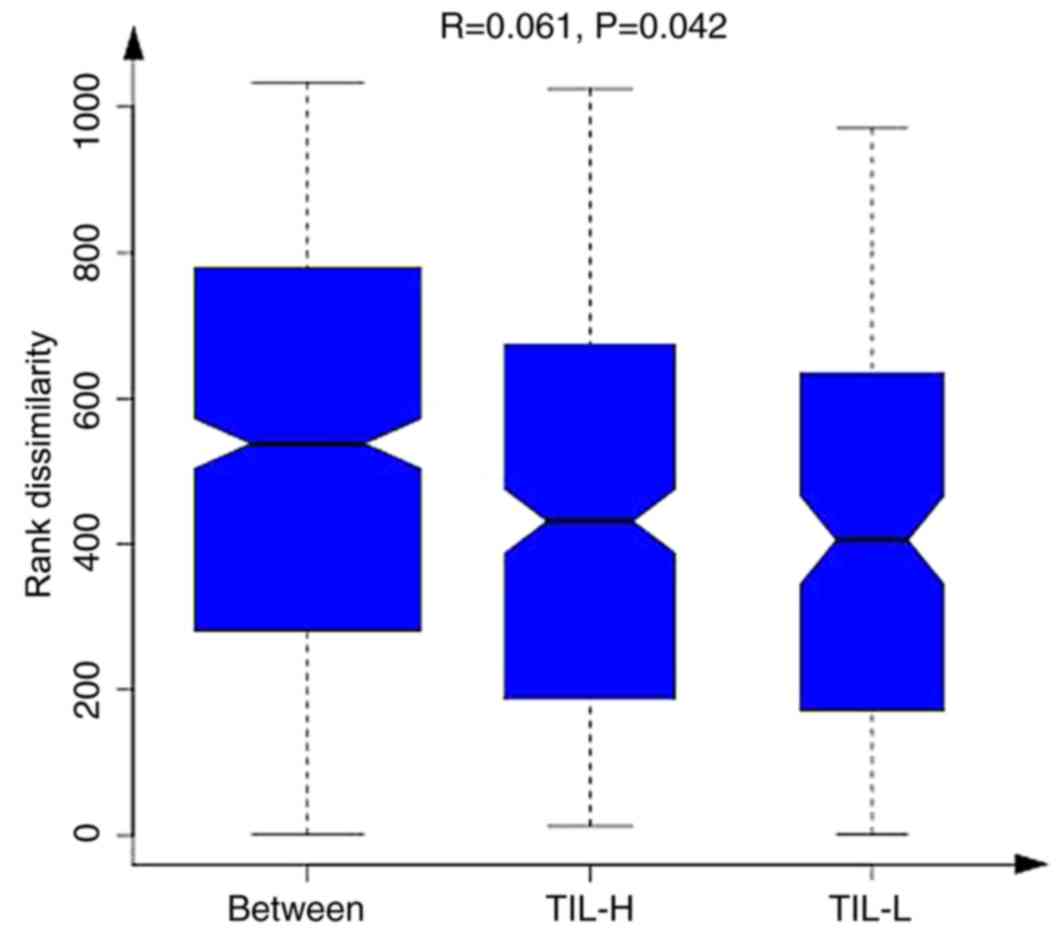
(P<0.01). Furthermore, ANOSIM revealed a greater intergroup
difference between the TIL-L and TIL-H groups compared with the
intragroup difference, which indicated that the grouping was
correct (P=0.042; Fig. 2).
 | Table III.Weighted UniFrac distance difference
analysis. |
Table III.
Weighted UniFrac distance difference
analysis.
| Groups | Test method | P-value |
|---|
| TIL-L vs.
TIL-M | t-test | 0.0004a |
| TIL-L vs.
TIL-M | Wilcoxon | 0.0001a |
| TIL-L vs.
TIL-H | t-test |
1.0147×10−6a |
| TIL-L vs.
TIL-H | Wilcoxon |
1.8884×10−7a |
| TIL-M vs.
TIL-H | t-test | 0.1227 |
| TIL-M vs.
TIL-H | Wilcoxon | 0.1245 |
| TIL-L vs. TIL-M vs.
TIL-H | Kruskal-Wallis |
7.2565×10−7a |
| TIL-L vs. TIL-M vs.
TIL-H | Tukey honest
significant difference |
7.0541×10−7a |
 | Table IV.Unweighted UniFrac distance
difference analysis. |
Table IV.
Unweighted UniFrac distance
difference analysis.
| Groups | Test method | P-value |
|---|
| TIL-L vs.
TIL-M | t-test | 0.4007 |
| TIL-L vs.
TIL-M | Wilcoxon | 0.1997 |
| TIL-L vs.
TIL-H | t-test |
1.2397×10−12a |
| TIL-L vs.
TIL-H | Wilcoxon |
7.5180×10−12a |
| TIL-M vs.
TIL-H | t-test |
4.9195×10−12a |
| TIL-M vs.
TIL-H | Wilcoxon |
2.9383×10−12a |
| TIL-L vs. TIL-M vs.
TIL-H | Kruskal-Wallis |
7.0570×10−14a |
| TIL-L vs. TIL-M vs.
TIL-H | Tukey honest
significant difference |
1.9653×10−15a |
Relative abundance of microbiota in
TIL-H and TIL-L groups
The different distributions of microbiota in TIL-H
and TIL-L groups were assessed using Metastats software. Table V demonstrated that At the genus
level, patients in the TIL-L group had higher abundances of
Mycobacterium, Rhodococcus, Catenibacterium, Bulleidia,
Anaerofilum, Sneathia, Devosia and TG5, but lower abundances of
Methanosphaera and Anaerobiospirillum compared with the TIL-H group
(P<0.05). At the species level, the abundances of stercoris,
barnesiae, coprophilus, flavefaciens and C21_c20 were greater in
the TIL-L group, while the abundances of producta and komagatae
were greater in the TIL-H group (P<0.05).
 | Table V.Bacterial species exhibiting
significant alterations in relative abundance between TIL-H and
TIL-L groups. |
Table V.
Bacterial species exhibiting
significant alterations in relative abundance between TIL-H and
TIL-L groups.
| Phylum | Class | Order | Family | Genus | Species | Mean, TIL-L
(%) | Mean, TIL-H
(%) | P-value | FDR |
|---|
| Actinobacteria | Actinobacteria |
Actinomycetales |
Mycobacteriaceae |
Mycobacterium |
|
1.35×10−5 | 0 |
7.97×10−6 | 0.001752485 |
| Actinobacteria | Actinobacteria |
Actinomycetales | Nocardiaceae |
Rhodococcus |
|
7.45×10−6 | 0 | 0.000872455 | 0.01998002 |
| Actinobacteria | Coriobacteriia |
Coriobacteriales |
Coriobacteriaceae |
Collinsella |
Stercoris |
7.11×10−5 | 0 | 0.000999001 | 0.01998002 |
| Bacteroidetes | Bacteroidia | Bacteroidales | Bacteroidaceae |
Bacteroides |
Barnesiae |
9.69×10−5 | 0 | 0.000999001 | 0.01998002 |
| Bacteroidetes | Bacteroidia | Bacteroidales | Bacteroidaceae |
Bacteroides |
Coprophilus | 0.012465402 |
5.90×10−5 | 0.002997003 | 0.038784745 |
| Euryarchaeota |
Methanobacteria |
Methanobacteriales |
Methanobacteriaceae |
Methanosphaera |
| 0 |
5.40×10−6 | 0.001388066 | 0.025447882 |
| Firmicutes |
Erysipelotrichi |
Erysipelotrichales |
Erysipelotrichaceae |
Catenibacterium |
|
7.09×10−5 | 0 | 0.000999001 | 0.01998002 |
| Firmicutes | Clostridia | Clostridiales |
Lachnospiraceae | Blautia |
Producta |
2.71×10−5 | 0.000354621 | 0.000999001 | 0.01998002 |
| Firmicutes |
Erysipelotrichi |
Erysipelotrichales |
Erysipelotrichaceae |
Bulleidia |
|
6.88×10−6 | 0 | 0.001908364 | 0.032295398 |
| Firmicutes | Clostridia | Clostridiales |
Ruminococcaceae |
Anaerofilum |
|
1.43×10−5 |
2.46×10−6 | 0.002518182 | 0.037610265 |
| Firmicutes | Clostridia | Clostridiales |
Ruminococcaceae |
Ruminococcus |
Flavefaciens |
1.04×10−5 |
9.19×10−7 | 0.002735292 | 0.037610265 |
| Fusobacteria | Fusobacteriia |
Fusobacteriales |
Leptotrichiaceae |
Sneathia |
|
2.31×10−5 | 0 | 0.000999001 | 0.01998002 |
| Proteobacteria |
Alphaproteobacteria | Rhizobiales |
Hyphomicrobiaceae | Devosia |
|
1.05×10−5 | 0 |
3.81×10−5 | 0.004192394 |
| Proteobacteria |
Deltaproteobacteria |
Desulfovibrionales |
Desulfovibrionaceae |
Desulfovibrio | C21_c20 |
9.42×10−6 | 0 | 0.00018235 | 0.013372361 |
| Proteobacteria |
Gammaproteobacteria | Aeromonadales |
Succinivibrionaceae |
Anaerobiospirillum |
| 0 |
1.25×10−5 | 0.000999001 | 0.01998002 |
| Proteobacteria |
Alphaproteobacteria | Rhizobiales |
Methylobacteriaceae |
Methylobacterium |
Komagatae | 0 |
4.99×10−6 | 0.00262006 | 0.037610265 |
| Synergistetes | Synergistia | Synergistales |
Dethiosulfovibrionaceae | TG5 |
|
7.16×10−6 |
8.63×10−7 | 0.000499914 | 0.01998002 |
Discussion
Recently, increasing attention has been paid to the
effects of the gastrointestinal microbiome on human diseases.
Diversity of the gastrointestinal microbiome is closely associated
with human health, including immunity, digestion, obesity (30), diabetes (31,32),
heart disease (33,34), acquired immune deficiency syndrome
(35) and cancer (36,37).
The present study demonstrated that the
gastrointestinal microbiome was distinctly diverse and
compositionally different among different TIL expression groups of
patients with BC. Higher TIL expression was associated with a
greater diversity of the gastrointestinal microbiome in the present
study. A previous study only suggested that patients with BC
possess statistically different microbiota composition compared
with controls (38), and the
gastrointestinal microbiome is associated with the clinical or
biological characteristics of patients with BC (38,39).
However, the present study revealed that microbiome diversity was
associated with TIL distribution. Additionally, differentially
expressed microbiota species among different TIL groups were
identified. Among the gastrointestinal microbiome, barnesiae
and coprophilus belong to the genus Bacteroides that
can modulate estrogen metabolism and function as a risk factor for
BC (40–43); in this study, higher abundance of
barnesiae was associated with the low expression of TILs.,
indicating barnesiae could be a risk factor for BC. The
mechanism underlying barnesiae-modulated estrogen metabolism
in response to immunity modification in BC remains unclear. The
state of TIL expression in situ exhibits a strong
association with the outcomes and treatment efficiency of patients
with BC, and the gastrointestinal microbiome can regulate immune
activation, following treatment with chemotherapeutic agents
(14,44–46). The
results of the present study revealed that greater quantity and
abundance of the gastrointestinal microbiome were positively
associated with the expression of TILs, demonstrating an internal
link between the microbiome and immunity in the pathogenesis of BC.
Therefore, the treatment efficiency should be assessed in a cohort
consisting of large-scale samples.
Patients with triple-negative BC and HER2-positive
BC may benefit from neoadjuvant chemotherapy (47), and the data of the present study
revealed that HER2 expression was positively associated with high
TIL expression. Furthermore, patients with high TIL expression
exhibited good outcomes following chemotherapy. All these findings
implied an inherent link among microbiome, immunity and treatment
efficiency in patients with BC.
The small sample size is the main limitation of the
present study. Additionally, further studies regarding the
mechanism need to be performed in the future. In these, a BC mouse
model will be established to verify the conclusions of the present
study by altering the gastrointestinal microbiome.
In conclusion, expression levels of TILs were
associated with the diversity of the gastrointestinal microbiome in
patients with BC. The results of the present study suggested that
the gastrointestinal microbiome may affect the prognosis of
patients with BC by interacting with TIL expression.
Acknowledgements
The authors would like to thank Professor Zhanjun
Guo (Rheumatic Immunology Department), Dr Xi Zhang and Dr Meng
Cheng (Breast Oncology Department) from The Fourth Hospital of
Hebei Medical University (Shijiazhuang, China) for their help in
reviewing this manuscript.
Funding
The present study was supported by grants from Hebei
Science and Technology Planning (grant no. 16967788D).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
CG designed the experiment, provided financial
support, revised the manuscript and gave final approval of the
version to be published. MS was responsible for the interpretation
of data. JS and WG performed the experiments, acquired the data and
wrote the paper. SL, SY and ZL made substantive contributions to
the work, including data collecting and manuscript revising.
Ethics approval and consent to
participate
Ethical approval for this project was obtained from
the Ethics Committee of The Fourth Hospital of Hebei Medical
University. Informed consent was provided by all participants. All
procedures involving human participants were performed in
accordance with the ethical standards of the institutional and/or
national research committee and with the 1964 Helsinki Declaration
and its later amendments or comparable ethical standards.
Patient consent for publication
The patients' data were anonymized, and hospital
numbers and associated data may be provided only for scientific
purposes. All patients provided written informed consent for their
data to be published.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Akram M, Iqbal M, Daniyal M and Khan AU:
Awareness and current knowledge of breast cancer. Biol Res.
50:332017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
de la Cruz-Merino L, Chiesa M, Caballero
R, Rojo F, Palazón N, Carrasco FH and Sánchez-Margalet V: Breast
cancer immunology and immunotherapy: Current status and future
perspectives. Int Rev Cell Mol Biol. 331:12017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Gooden MJ, de Bock GH, Leffers N, Daemen T
and Nijman HW: The prognostic influence of tumour-infiltrating
lymphocytes in cancer: A systematic review with meta-analysis. Br J
Cancer. 105:93–103. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Mahmoud SM, Paish EC, Powe DG, Macmillan
RD, Grainge MJ, Lee AH, Ellis IO and Green AR: Tumor-infiltrating
CD8+ lymphocytes predict clinical outcome in breast cancer. J Clin
Oncol. 29:1949–1955. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Liu S, Foulkes WD, Leung S, Gao D, Lau S,
Kos Z and Nielsen TO: Prognostic significance of FOXP3+ tumor
infiltrating lymphocytes in breast cancer depends on estrogen
receptor and human epidermal growth factor receptor-2 expression
status and concurrent cytotoxic T-cell infiltration. Breast Cancer
Res. 16:4322014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Seo AN, Lee HJ, Kim EJ, Kim HJ, Jang MH,
Lee HE, Kim YJ, Kim JH and Park SY: Tumour-infiltrating CD8+
lymphocytes as an independent predictive factor for pathological
complete response to primary systemic therapy in breast cancer. Br
J Cancer. 109:2705–2713. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Denkert C, Loibl S, Noske A, Roller M,
Muller BM, Komor M, Budczies J, Darb-Esfahani S, Kronenwett R,
Hanusch C, et al: Tumor-associated lymphocytes as an independent
predictor of response to neoadjuvant chemotherapy in breast cancer.
J Clin Oncol. 28:105–113. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Spranger S, Sivan A, Corrales L and
Gajewski TF: Tumor and host factors controlling antitumor immunity
and efficacy of cancer immunotherapy. Adv Immunol. 130:75–93. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Qin Q, Miao J, Wang S, Yu Q, Li M, He F
and Wang G: Association between intestinal flora and immunity in
middle-aged and aged people by PCR-DGGE. Wei Sheng Yan Jiu.
46:40–45. 2017.(In Chinese). PubMed/NCBI
|
|
10
|
Hooper LV, Littman DR and Macpherson AJ:
Interactions between the microbiota and the immune system. Science.
336:1268–1273. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ivanov II and Honda K: Intestinal
commensal microbes as immune modulators. Cell Host Microbe.
12:496–508. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Abt MC, Osborne LC, Monticelli LA, Doering
TA, Alenghat T, Sonnenberg GF, Paley MA, Antenus M, Williams KL,
Erikson J, et al: Commensal bacteria calibrate the activation
threshold of innate antiviral immunity. Immunity. 37:158–170. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ganal SC, Sanos SL, Kallfass C, Oberle K,
Johner C, Kirschning C, Lienenklaus S, Weiss S, Staeheli P, Aichele
P and Diefenbach A: Priming of natural killer cells by nonmucosal
mononuclear phagocytes requires instructive signals from commensal
microbiota. Immunity. 37:171–186. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Iida N, Dzutsev A, Stewart CA, Smith L,
Bouladoux N, Weingarten RA, Molina DA, Salcedo R, Back T, Cramer S,
et al: Commensal bacteria control cancer response to therapy by
modulating the tumor microenvironment. Science. 342:967–970. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Dingemanse C, Belzer C, van Hijum SA,
Günthel M, Salvatori D, den Dunnen JT, Kuijper EJ, Devilee P, de
Vos WM, van Ommen GB and Robanus-Maandag EC: Akkermansia
muciniphila and Helicobacter typhlonius modulate intestinal tumor
development in mice. Carcinogenesis. 36:1388–1396. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Omar Al-Hassi H, Ng O and Brookes M:
Tumour-associated and non-tumour-associated microbiota in
colorectal cancer. Gut. 67:3952018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Peled JU, Devlin SM, Staffas A, Lumish M,
Khanin R, Littmann ER, Ling L, Kosuri S, Maloy M, Slingerland JB,
et al: Intestinal microbiota and relapse after hematopoietic-cell
transplantation. J Clin Oncol. 35:1650–1659. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hammond ME, Hayes DF, Dowsett M, Allred
DC, Hagerty KL, Badve S, Fitzgibbons PL, Francis G, Goldstein NS,
Hayes M, et al: American society of clinical oncology/college of
American pathologists guideline recommendations for
immunohistochemical testing of estrogen and progesterone receptors
in breast cancer. J Clin Oncol. 28:2784–2795. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wolff AC, Hammond MEH, Allison KH, Harvey
BE, Mangu PB, Bartlett JMS, Bilous M, Ellis IO, Fitzgibbons P,
Hanna W, et al: Human epidermal growth factor receptor 2 testing in
breast cancer: American society of clinical oncology/college of
American pathologists clinical practice guideline focused update. J
Clin Oncol. 36:2105–2122. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ogston KN, Miller ID, Payne S, Hutcheon
AW, Sarkar TK, Smith I, Schofield A and Heys SD: A new histological
grading system to assess response of breast cancers to primary
chemotherapy: prognostic significance and survival. Breast.
12:320–327. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Salgado R, Denkert C, Demaria S, Sirtaine
N, Klauschen F, Pruneri G, Wienert S, Van den Eynden G, Baehner FL,
Penault-Llorca F, et al: The evaluation of tumor-infiltrating
lymphocytes (TILs) in breast cancer: recommendations by an
International TILs Working Group 2014. Ann Oncol. 26:259–271. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Hida AI and Ohi Y: Evaluation of
tumor-infiltrating lymphocytes in breast cancer; proposal of a
simpler method. Ann Oncol. 26:23512015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Magoč T and Salzberg SL: FLASH: Fast
length adjustment of short reads to improve genome assemblies.
Bioinformatics. 27:2957–2963. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Caporaso JG, Kuczynski J, Stombaugh J,
Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich
JK, Gordon JI, et al: QIIME allows analysis of high-throughput
community sequencing data. Nat Methods. 7:335–336. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Bokulich NA, Subramanian S, Faith JJ,
Gevers D, Gordon JI, Knight R, Mills DA and Caporaso JG:
Quality-filtering vastly improves diversity estimates from Illumina
amplicon sequencing. Nat Methods. 10:57–59. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Edgar and Robert C: UPARSE: highly
accurate OTU sequences from microbial amplicon reads. Nat Methods.
10:996–998. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wang Q, Garrity GM, Tiedje JM and Cole JR:
Naive Bayesian classifier for rapid assignment of rRNA sequences
into the new bacterial taxonomy. Appl Environ Microbiol.
73:5261–5267. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
DeSantis TZ, Hugenholtz P, Larsen N, Rojas
M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P and Andersen GL:
Greengenes, a chimera-checked 16S rRNA gene database and workbench
compatible with ARB. Appl Environ Microbiol. 72:5069–5072. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
White JR, Nagarajan N and Pop M:
Statistical methods for detecting differentially abundant features
in clinical metagenomic samples. PLoS Comput Biol. 5:e10003522009.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Cotillard A, Kennedy SP, Kong LC, Prifti
E, Pons N, Le Chatelier E, Almeida M, Quinquis B, Levenez F,
Galleron N, et al: Dietary intervention impact on gut microbial
gene richness. Nature. 500:585–588. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Larsen N, Vogensen FK, van den Berg FW,
Nielsen DS, Andreasen AS, Pedersen BK, Al-Soud WA, Sørensen SJ,
Hansen LH and Jakobsen M: Gut microbiota in human adults with type
2 differs from non-diabetic adults. PLoS One. 5:e908552000.
|
|
32
|
Markle JG, Frank DN, Mortin-Toth S,
Robertson CE, Feazel LM, Rolle-Kampczyk U, von Bergen M, McCoy KD,
Macpherson AJ and Danska JS: Sex differences in the gut microbiome
drive hormone-dependent regulation of autoim munity. Science.
339:1084–1088. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Koeth RA, Wang Z, Levison BS, Buffa JA,
Org E, Sheehy BT, Britt EB, Fu X, Wu Y, Li L, et al: Intestinal
microbiota metabolism of L carnitine, a nutrient in red meat
promotes atherosclerosis. Nat Med. 19:576–585. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wang T, Cai G, Qiu Y, Fei N, Zhang M, Pang
X, Jia W, Cai S and Zhao L: Structural segregation of gut
microbiota between colorectal cancer patients and healthy
volunteers. ISME J. 6:320–329. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Vujkovic-Cvijin I, Dunham RM, Iwai S,
Maher MC, Albright RG, Broadhurst MJ, Hernandez RD, Lederman MM,
Huang Y, Somsouk M, et al: Dysbiosis of the gut microbiota is
associated with HIV disease progression and tryptophan catabolism.
Sci Transl Med. 5:193ra912013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Fox JG, Feng Y, Theve EJ, Raczynski AR,
Fiala JLA, Doernte AL, Williams M, McFaline JL, Essigmann JM,
Schauer DB, et al: Gut microbes define liver cancer risk in mice
exposed to chemical and viral trans genic hepatocarcinogens. Gut.
59:88–97. 2009. View Article : Google Scholar
|
|
37
|
Wang Z, Klipfell E, Bennett BJ, Koeth R,
Levison BS, Dugar B, Feldstein AE, Britt EB, Fu X, Chung YM, et al:
Gut flora metabolism of phosphatidylcholine promotes cardiovascular
disease. Nature. 472:57–63. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Goedert JJ, Jones G, Hua X, Xu X, Yu G,
Flores R, Falk RT, Gail MH, Shi J, Ravel J and Feigelson HS:
Investigation of the association between the fecal microbiota and
breast cancer in postmenopausal women: a population-based
case-control pilot study. J Natl Cancer Inst. 107:djv1472015.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Luu TH, Michel C, Bard JM, Dravet F, Nazih
H and Bobin-Dubigeon C: Intestinal proportion of Blautia sp. is
associated with clinical stage and histoprognostic grade in
patients with early-stage breast cancer. Nutr Cancer. 69:267–275.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Fuhrman BJ, Feigelson HS, Flores R, Gail
MH, Xu X, Ravel J and Goedert JJ: Associations of the fecal
microbiome with urinary estrogens and estrogen metabolites in
postmenopausal women. J Clin Endocrinol Metab. 99:4632–4640. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Flores R, Shi J, Fuhrman B, Xu X, Veenstra
TD, Gail MH, Gajer P, Ravel J and Goedert JJ: Fecal microbial
determinants of fecal and systemic estrogens and estrogen
metabolites: A cross-sectional study. J Transl Med. 10:2532012.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Estrogen excretion patterns and plasma
levels in vegetarian and omnivorous women. Nutr Rev. 41:180–183.
1983.PubMed/NCBI
|
|
43
|
Goldin BR, Adlercreutz H, Gorbach SL,
Warram JH, Dwyer JT, Swenson L and Woods MN: Estrogen excretion
patterns and plasma levels in vegetarian and omnivorous women. N
Engl J Med. 307:1542–1547. 1982. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Savas P, Salgado R, Denkert C, Sotiriou C,
Darcy PK, Smyth MJ and Loi S: Clinical relevance of host immunity
in breast cancer: from TILs to the clinic. Nat Rev Clin Oncol.
13:228–241. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Viaud S, Saccheri F, Mignot G, Yamazaki T,
Daillère R, Hannani D, Enot DP, Pfirschke C, Engblom C, Pittet MJ,
et al: The intestinal microbiota modulates the anticancer immune
effects of cyclophosphamide. Science. 342:971–976. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Galon J, Costes A, Sanchez-Cabo F,
Kirilovsky A, Mlecnik B, Lagorce-Pagès C, Tosolini M, Camus M,
Berger A, Wind P, et al: Type, density, and location of immune
cells within human colorectal tumors predict clinical outcome.
Science. 313:1960–1964. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Rastogi P, Anderson SJ, Bear HD, Geyer CE,
Kahlenberg MS, Robidoux A, Margolese RG, Hoehn JL, Vogel VG, Dakhil
SR, et al: Preoperative chemotherapy: Updates of national surgical
adjuvant breast and bowel project protocols B-18 and B-27. J Clin
Oncol. 26:778–785. 2008. View Article : Google Scholar : PubMed/NCBI
|