Introduction
Programmed death ligand 1 (PD-L1) is a protein
expressed by activated antigen-presenting cells, the majority of
normal body cells and certain types of cancer cells. PD-L1 is an
immune checkpoint protein that protects against autoimmunity
(1). Certain types of cancer,
including non-small cell lung cancer (NSCLC), are able to express
this molecule on the cell surface. Interactions between tumor PD-L1
and programmed cell death protein 1 (PD-1) localized to the T-cell
surface, cause lymphocyte anergy and the resistance of cancer cells
to immune surveillance (1,2). Inhibition of this signaling pathway
using anti-PD-1 or anti-PD-L1 monoclonal antibodies results in
mobilization of the immune system, and the subsequent recognition
and destruction of cancer cells (3–7).
The effectiveness of anti-PD-1 (nivolumab and
pembrolizumab) and anti-PD-L1 (atezolizumab and durmvalumab)
antibody therapy in patients with NSCLC depends on the expression
of PD-L1 on tumor and immune cells, which is detected using
immunohistochemistry (IHC) (8–12).
Immunotherapy, based on the inhibition of immune checkpoints
improved overall survival in patients expressing PD-L1, but certain
NSCLC patients without expression may also benefit from
immunotherapy (13,14). There are 2 platforms to assess PD-L1
expression in tumor cells and/or in tumor-infiltrating immune cells
(9). During clinical trials with
pembrolizumab and nivolumab (which bind the extracellular domain of
PD-L1) the antibodies 22C3 and 28-8 were used with the Autostainer
Link platform to determine the PD-L1 status of participants.
Inclusion criteria for pembrolizumab therapy were >50% of tumor
cells with PD-L1 expression for first line treatment, and any
expression of PD-L1 (PD-L1-positive tumor cells ≥1%) for second
line treatment (11). However,
nivolumab may be used for second line therapy in patients without
PD-L1-positive tumor cells, and testing of PD-L1 expression with
the 28-8 antibody is not required to qualify patients for nivolumab
therapy (9). The SP142 antibody,
used with the Benchmark platform, was employed in clinical trials
for atezolizumab. This clone can bind to the intracellular domain
of PD-L1 (15). The scoring of PD-L1
status is complicated, due to the requirement of assessing
expression on stromal immune cells that have infiltrated the tumor.
However, atezolizumab may also be used as a second line of
treatment in patients without PD-L1-expressing tumor or immune
cells. The antibody clone SP263, used with the Benchmark platform,
has the ability to bind the intracellular domain of PD-L1; this was
examined in clinical trials with durmvalumab (13).
Different kinds of antibodies used in the assessment
of PD-L1 give incompatible scores. This may be due to the
properties of particular clones and their affinity for different
PD-L1 domains. There is a requirement to supplement the diagnosis
of PD-L1 with additional factors, facilitating the qualification of
patients for immunotherapy. Genetic factors that influence the
effectiveness of this method may be identified. One such factor may
be the tumor mutation burden (TMB). There are no studies using
epigenetic factors as predictors for immunotherapy. However,
microRNAs (miRs) present as potential candidates. miRs are small
molecules (~20 nt in length) that regulate gene expression on a
post-transcriptional level. These molecules act by either promoting
mRNA degradation or inhibiting translation (15,16).
There are few reports concerning the regulation of PD-L1 expression
by miRs in lung cancer (17–19). Therefore, the purpose of the present
study was to assess the expression of miRs with the ability to
regulate PD-L1 expression in patients with NSCLC, and to determine
the association between these factors and the expression of PD-L1
mRNA and protein in tumor cells.
Materials and methods
Patients
A total of 43 formalin-fixed paraffin-embedded
(FFPE; 91.49%) tissues from patients with NSCLC, and 4 cell blocks
(8.51%) were assessed for the expression of PD-L1 protein, mRNA and
miR. Adenocarcinoma was diagnosed in 31 (65.9%) cases and squamous
cell carcinoma was detected in 13 (27.7%). In 3 (6.4%) cases, NSCLC
not otherwise specified (NOS) was diagnosed. There were 36 (76.60%)
specimens from primary tumors, 5 (10.64%) from distant metastases
and 6 (12.76%) from metastases to the lymph node. Samples were
collected between September 2017 and April 2018 in the Departments
of Pneumonology, Oncology and Allergology, Medical University of
Lublin (Lublin, Poland). The study group included 33 (70.21%)
patients at stage I–IIIA, 8 (17.02%) at stage IIIB, and 6 (12.77%)
at stage IV. The median age was 65 years (±9.03 years), and the
group consisted of 20 male (42.55%) and 27 female patients
(57.45%), of which 10 were non-smokers (21.28%). Due to the lack of
reimbursement for anti-PD-1 or anti-PD-L1 antibodies in Poland at
the time, information regarding the efficacy of immunotherapy was
not available. Therefore patients were not treated with anti-PD-1
or anti-PD-L1 monoclonal antibodies. Furthermore, surgically
resected patients were enrolled, and immunotherapy in such patients
was not available at the time (with the exception of those in
clinical trials).
IHC analysis
During IHC analysis, the monoclonal antibody clones
SP142 (Ventana Medical Systems, Inc., Tucson, AZ, USA) and 22C3
(Dako; Agilent Technologies, Inc., Santa Clara, CA, USA) were
employed. Briefly, 3-µm paraffinized tissue sections were mounted
using SuperFrost™ Plus slides (Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) on a hot-plate in 60°C for at least one hour.
Deparaffinization and antigen retrieval were performed prior to
staining, using the Dako PT Link device (Dako; Agilent
Technologies, Inc.). Slides were washed and dehydrated twice in 96%
ethanol and xylene, and mounted on coverslips. The slides were
pre-heated to 59°C for at least 3 h, and IHC with the SP142
antibody was conducted using the Ventana Benchmark GX platform
(Roche Diagnostics GmbH, Mannheim, Germany), with the Ventana test
certified for in vitro diagnostics (CE-IVD). The OptiView
Amplification kit was incorporated for signal amplification, and
the OptiView DAB IHC Detection kit was used (Roche Diagnostics). As
the negative control, a rabbit monoclonal antibody (standard
negative control by Ventana Medical Systems, Inc., Tucson, AZ, USA;
cat. no. 790-4795) was applied. IHC conducted with the 22C3
antibody was performed using the Dako Autostainer Link 48
instrument (Dako; Agilent Technologies, Inc.) incorporating the
CE-IVD IHC 22C3 PharmDx PD-L1 Kit and the EnVision FLEX
visualization system (Agilent Technologies, Inc.). The
aforementioned staining procedures were conducted according to the
manufacturer's protocols. Counterstaining with hematoxylin was also
conducted according to the manufacturer's protocol. The slides were
later observed by the pathologist using an Olympus BX41
microscope.
RNA isolation
Total RNA isolation was conducted using 5-µm
sections of FFPE tissues or cell blocks using an miRNeasy FFPE Kit
(Qiagen GmbH, Hilden, Germany), according to the manufacturers'
instructions. RNA was stored at −80°C until the synthesis of
cDNA.
Quantification of PD-L1 mRNA
expression level
The relative level of PD-L1 mRNA expression was
determined using reverse transcription-quantitative polymerase
chain reaction (RT-qPCR) in reference to the internal control,
GAPDH. Reverse transcription was conducted using a High-Capacity
RNA-to-cDNA™ kit (Thermo Fisher Scientific, Inc.) according to the
manufacturer's instruction. qPCR was performed using TaqMan Fast
Advanced Master Mix (Thermo Fisher Scientific, Inc.) and the
Illumina Eco Real-Time PCR System (Illumina Inc, San Diego, CA,
USA). The 20 µl PCR mixture contained the following: 10 µl TaqMan
Fast Advanced Master Mix, 1 µl TaqMan Gene Expression Assay mix
(assay ID Hs00204257 for PD-L1 and Hs02786624 for GAPDH; Thermo
Fisher Scientific, Inc.), 5 µl RNase free water and 4 µl cDNA. The
thermocycling conditions were as follows: 95°C for 20 sec, 40
cycles at 95°C for 3 sec, and 60°C for 30 sec. Analysis was
performed using the 2−ΔΔCq method (20).
Quantification of miR expression
Expression of 8 miRs, complementary to the 3′
untranslated region (UTR) of PD-L1 mRNA was assessed: miR-141-3p
(478501_mir,), miR-200a-3p (478490_mir), miR-200b-3p (477963_mir),
miR-200c-3p (478351_mir), miR-429 (477849_mir), miR-508-3p
(478961_mir), miR-1184 (478629_mir) and miR-1255a (478661_mir) were
all acquired from Thermo Fisher Scientific, Inc. The targets for
miRs in 3′UTR PD-L1 mRNA were predicted using the TargetScan
(version 7.1; www.targetscan.org) and miRBase (release no. 22;
www.mirbase.org) systems. miR-191-5p (477952_mir)
was used as an internal control. Reverse transcription was
conducted using the TaqMan Advanced miRNA cDNA Synthesis kit
(Thermo Fisher Scientific, Inc.) according to the manufacturer's
instructions. qPCR was performed using the Illumina Eco Real-Time
PCR System (Illumina, Inc.). The 20-µl PCR mixture contained the
following: 10 µl TaqMan Fast Advanced Master Mix, 1 µl TaqMan Fast
Advanced miRNA Assay mix, 4 µl RNase free water and 5 µl cDNA. The
reactions were conducted as follows: 95°C for 20 sec, 40 cycles at
95°C for 5 sec and 60°C for 30 sec. Analysis was performed using
the 2−ΔΔCq method (20).
Statistical analysis
Statistical analysis was performed using Statistica
software (version 13.1; TIBCO® Software Inc., Palo Alto,
CA, USA). The Spearman's rank test was used to examine the
correlation between the expression of miRs and PD-L1 mRNA and
protein expression. The Mann-Whitney U test was used to compare
PD-L1 and miR expression in different patient groups (stratified by
age, material for analysis, histopathological diagnosis, and PD-L1
expression on tumor and immune cells). Data are presented as the
median ± standard deviation. P<0.05 was considered to indicate a
statistically significant difference.
Results
The median percentage of PD-L1-postive tumor cells
assessed using the 22C3 antibody was greater than that gained using
the SP142 clone (35.00±39.95 vs. 2.5±37.96%). The median percentage
of PD-L1-positive areas of the tumor infiltrated with immune cells,
as tested with SP142, was 5±22.27%. Due to the degradation of mRNA,
only 33 cases were assessed for PD-L1 mRNA expression. A positive
correlation was observed between the expression of PD-L1 mRNA and
the percentage of tumor cells with PD-L1 expression (R=0.35;
P=0.046 tested using 22C3, and R=0.419; P=0.015 using SP142), and
the percentage of the tumor area infiltrated by immune cells with
PD-L1 expression (R=0.44; P=0.013).
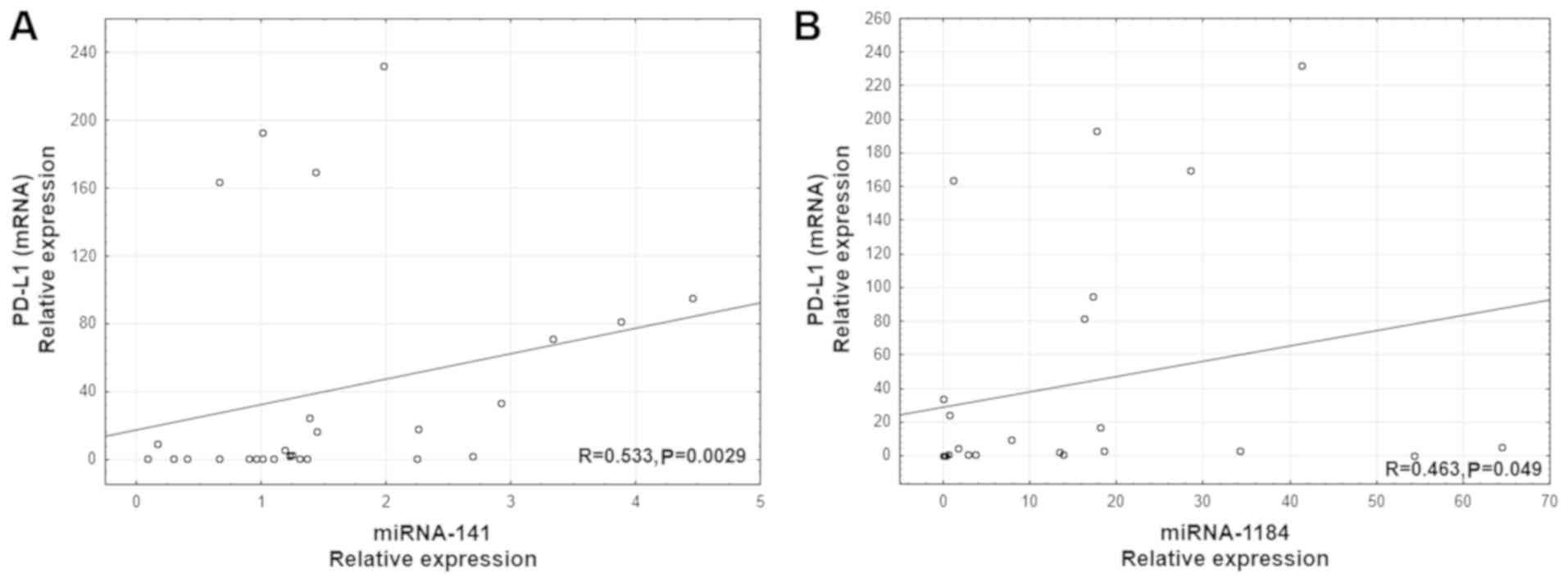
A positive correlation was revealed between PD-L1
mRNA level and the expression of two miRs: miR-141 (R=0.533;
P=0.0029; Fig. 1A) and miR-1184
(R=0.463, P=0.049, Fig. 1B).
Additionally, a positive correlation was observed between the
percentage of PD-L1-positive tumor cells (IHC using SP142) and the
expression of three miRs: miR-141 (R=0.441; P=0.0024; Fig. 2A), miR-200b (R=0.372; P=0.011;
Fig. 2B), and miR-429 (R=0.430;
P=0.0028; Fig. 2C). Furthermore,
there was a positive correlation between the percentage of the
tumor area with immune cell infiltration and the expression of two
miRs: miR-141 (R= 0.333; P=0.03; Fig.
2D) and miR-200b (R=0.312; P=0.046; Fig. 2E). The percentage of PD-L1-positive
tumor cells assessed using the 22C3 antibody positively correlated
with miR-141 expression (R=0.407; P=0,0055; Fig. 2F).
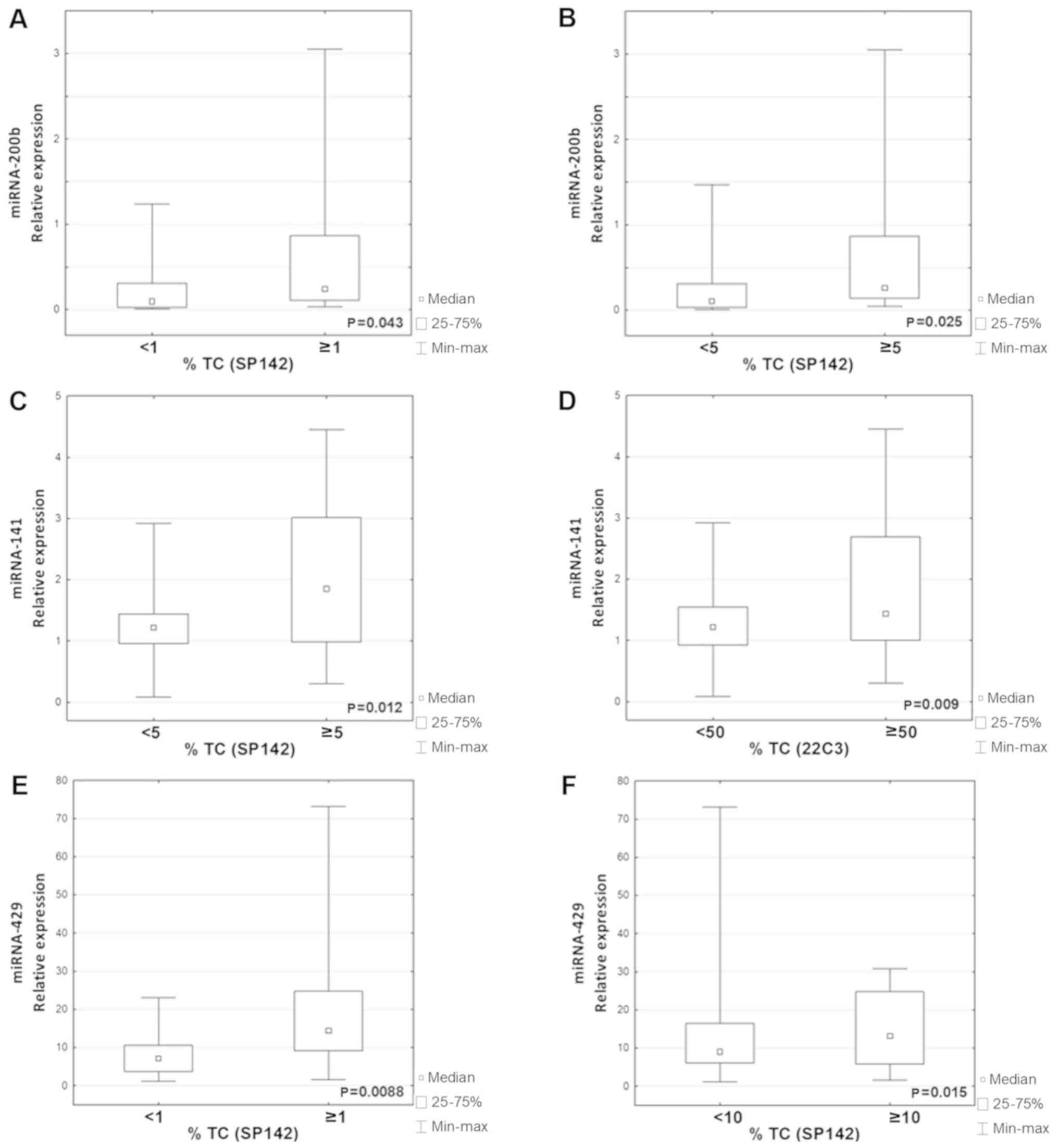
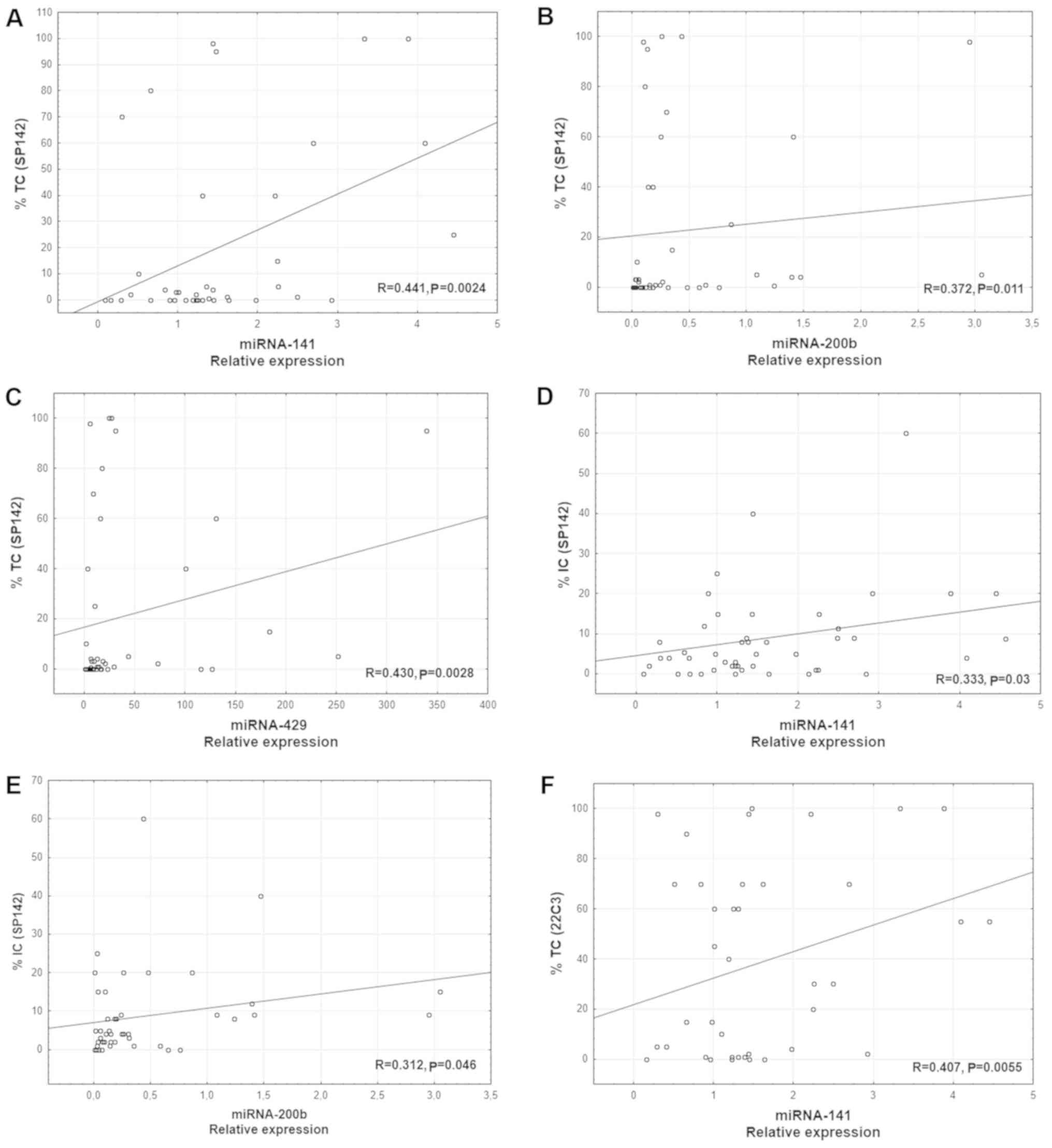 | Figure 2.Correlation between the percentage of
PD-L1-positive tumor cells in IHC using the SP142 antibody, and
miR-141, miR-200b and miR-429 expression. (A) miR-141, n=43; (B)
miR-200b, n=42; (C) miR-429, n=41. Correlation between the
percentages of the tumor area infiltrated with PD-L1-positive
immune cells in IHC using the SP142 antibody, and miR-141 and
miR-200b expression. (D) miR-141, n=40; (E) miR-200b, n=40. (F)
Correlation between the percentage of PD-L1-positive tumor cells in
IHC using the 22C3 antibody, and miR-141 expression; n=39. PD-L1,
programmed death ligand 1; miR/miRNA, microRNA; TC, tumor cells;
IC, immune cells; IHC, immunohistochemistry. |
Patients were stratified according to PD-L1
expression. Tumor cell (TC)1/2/3 or immune cell (IC)1/2/3 was
defined as PD-L1 expression on ≥1% of tumor cells, or 1% of the
area of tumor infiltrated by immune cells; TC2/3 or IC2/3 were
defined as PD-L1 expression on 5% of the tumor cells or the tumor
area; TC3 or IC3 were defined as PD-L1 expression on 10% of these
cells or the tumor area; and TC4 was defined as PD-L1 expression on
≥50% of all tumor cells. TC0 was used to define PD-L1-negative
samples.
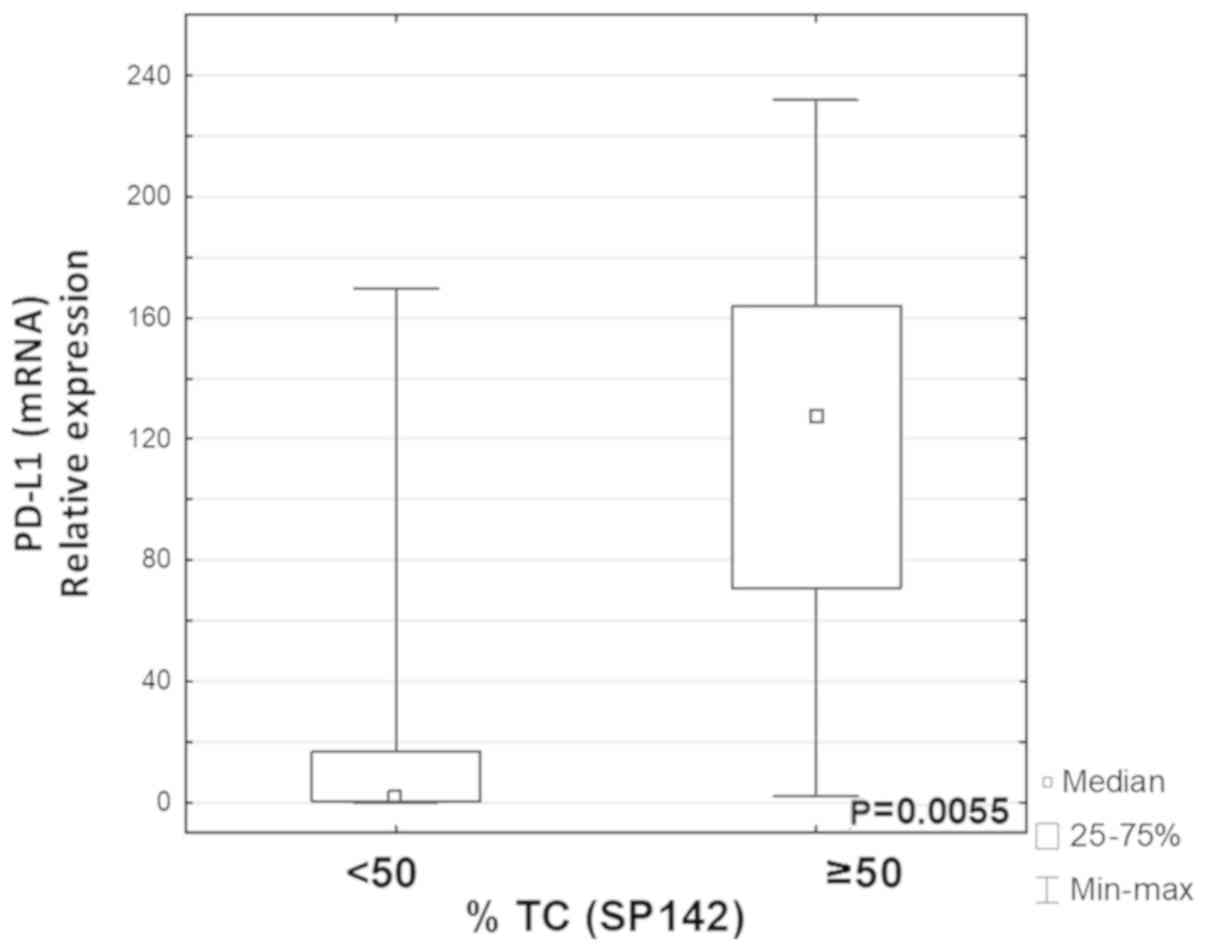
Groups of patients that differed by their expression
levels of PD-L1 (determined by IHC using SP142) were analyzed. The
expression level of PD-L1 mRNA was significantly higher in the TC4
group compared with the TC0/1/2/3 group (P=0.0055; Fig. 3). It was also observed that the
expression levels of miR-200b and miR-429 were significantly higher
in the TC1/2/3 group with any PD-L1 protein, compared with the
PD-L1-negative group, TC0 (P=0.043; P=0.0088 respectively; Fig. 4A and E). The expression levels of
miR-200b and miR-141 were significantly higher in the TC2/3 group
compared with the TC0/1 group (P=0.025; P=0.012 respectively;
Fig. 4B and C). Furthermore, the
expression level of miR-429 was also significantly higher in the
TC3 group compared with the TC0/1/2 group (P=0.015; Fig. 4F). There were no differences in the
expression of miRs in the groups that differed in the expression of
PD-L1 on immune cells.
Only one association was revealed between the
expression of miR and PD-L1 on tumor cells analyzed using the 22C3
antibody; miR-141 expression was significantly higher in the TC4
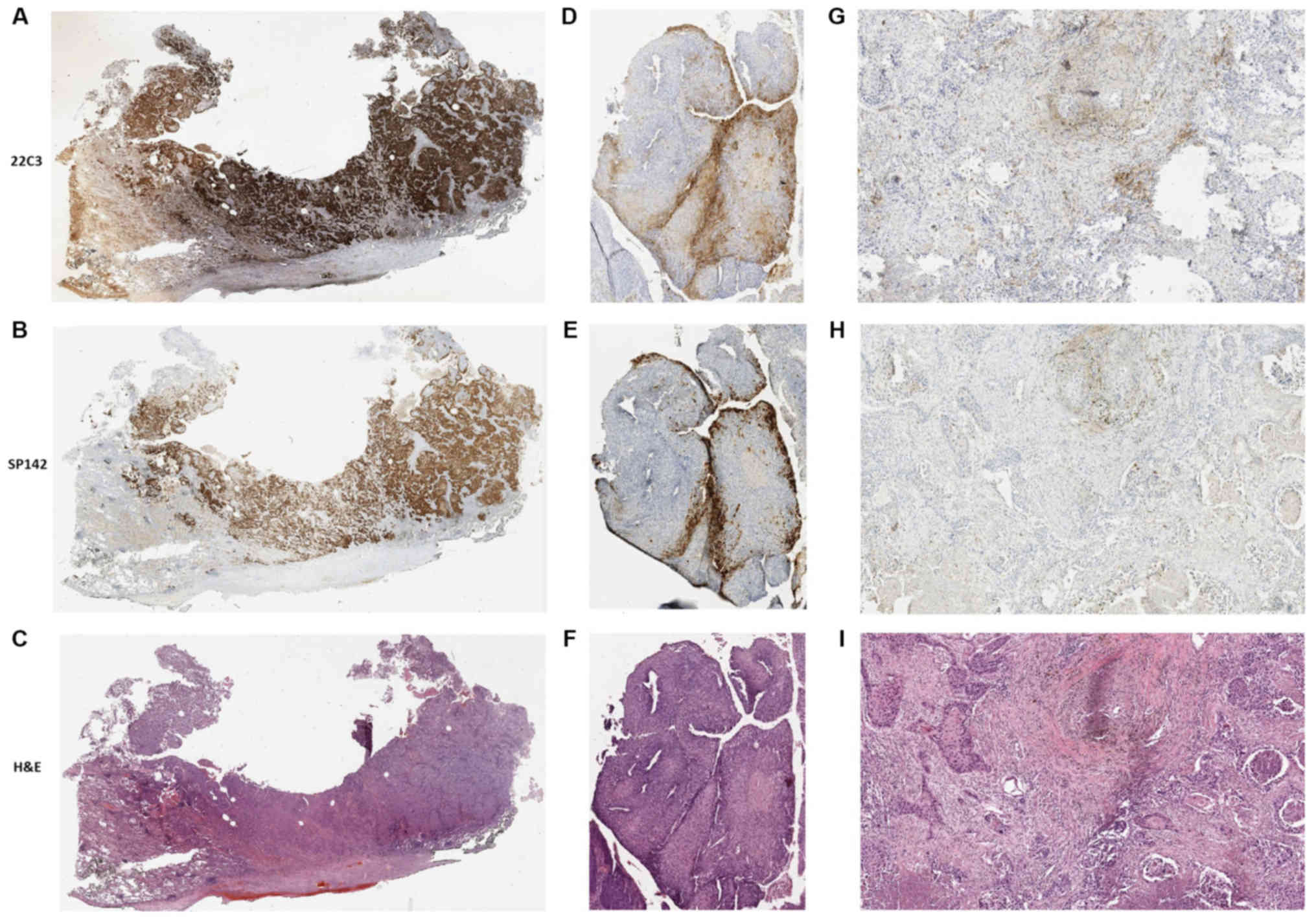
group compared with in the TC0/1/2/3 group (P=0.009; Fig. 4D). Examples of stained NSCLC tissues
visualized using the SP142 or 22C3 antibodies are displayed in
Fig. 5. The expression of miRs was
similar in groups of patients stratified by age, sex,
pathomorphological diagnosis, status of metastasis or disease stage
(P<0.05).
Discussion
Immune checkpoint inhibitors are able to prolong
patient survival, and their use has been associated with fewer side
effects comparison to the chemotherapy (21). These side effects are also
differently classified (Immune-Related Adverse Events) (22). The greatest benefit of PD-1/PD-L1
inhibitors was observed among patients whose tumor cells expressed
PD-L1 (9–11). Pembrolizumab and chemotherapy as
first line treatments for patients with advanced NSCLC with PD-L1
expression on >50% of tumor cells (scored using the 22C3
antibody) were compared. This revealed a significantly higher
response rate and longer progression-free and overall survival
rates in patients treated with pembrolizumab, compared with those
who received platinum-based chemotherapy (6). However, the association between the
benefits of second line immunotherapy and the expression of PD-L1
on tumor cells is not so obvious. Brahmer et al (10) demonstrated that overall survival,
response rate and progression-free survival were significantly
greater when using nivolumab, compared with docetaxel, in patients
with squamous-cell cancer, regardless of tumor cell PD-L1
expression. Similarly, Rittmeyer et al (23) revealed that atezolizumab treatment
resulted in a clinically relevant improvement in overall survival,
compared with docetaxel in previously treated patients with NSCLC,
regardless of PD-L1 expression. Patients without PD-L1 expression
on tumor cells or immune cells infiltrating the tumor achieved a
median overall survival of 12.6 months, following treatment with
atezolizumab, and only 8.9 months following docetaxel treatment.
The authors indicated that the lack of an association between PD-L1
expression and immunotherapy efficacy was probably due to the
complex interactions between tumors and the immune system. There
are a number of studies that suggest genetic factors may also be
associated with the response to immunotherapeutic treatment, in
spite of PD-L1-negative tumor cell status (10,24,25).
Shukuya et al (26) indicated that miR may influence the
efficiency of immunotherapy. Next-generation sequencing was used to
determine the expression of specific miRs in the plasma, and the
miR content of extracellular vesicles (EVs). A total of 26
circulating miRs and four EV-associated miRs exhibited significant
differences in concentration between responders and non-responders
to immunotherapy with anti-PD-1 or anti-PD-L1 antibodies.
Furthermore, these miRs were potential predictive biomarkers for
the response to treatment.
In the present study, 8 miRs with the ability to
regulate the expression of PD-L1 transcripts in tumor cells were
analyzed. A positive correlation between PD-L1 mRNA and protein
expression levels and miR expression (particularly miR-141 and
miR-1184) was identified. This indicates that these miRs may
regulate the expression of PD-L1 at a post-transcriptional level,
and thus participate in tumor-cell escape from immune surveillance.
However, these miRs are not involved in mRNA degradation, which can
be stored in the cell and activated if necessary. Positive
correlations may indicate that these two miRs (which exhibit
increased expression levels) may be able to silence PD-L1
expression in cancerous cells, but that this is restricted by the
carcinogenetic characteristics of these cells, allowing them to
bypass epigenetic regulation. This hypothesis evidently requires
experimental confirmation, potentially with a larger patient cohort
and in vitro experimentation using cell lines. Furthermore,
no relevant publications on the subject were identified, thus
extended studies including biochemical and molecular biological
experiments will increase knowledge of the regulatory functions of
miR-141 and miR-1184 in association with PD-L1. Due to the small
size of the patient group, the present study may be considered as a
pilot study to indicate subsequent genetic and epigenetic
experimentation surrounding PD-L1 immunotherapy.
miRs are able to act as oncogenes or tumor
suppressors, depending on the cancer type in question (27). There is limited evidence of miR-1184
expression in different types of cancer. Knyazev et al
(28) demonstrated the potential of
miR-1184 to differentiate between prostate cancer and benign
prostatic hyperplasia, indicating higher levels of miR-1184
expression in the peripheral blood of patients with prostate
cancer. Feng et al (29)
revealed that miR-1184 was able to regulate the expression of
certain oncogenes and transcription factors, including SMAD family
member 5, signal transducer and activator of transcription 3 and
glioma-associated oncogene family zinc finger 3. Farina et
al (30) identified lower
expression levels of miR-1184 in patients with breast cancer,
compared with healthy controls.
Mei et al (31) demonstrated that miR-141 was
upregulated in NSCLC, which was associated with aggressive disease
course and accelerated tumor growth. It was indicated to regulate
PH domain and leucine-rich repeat protein phosphatase 1 (PHLPP1)
and PHLPP2, which are involved in the AKT serine/threonine kinase 1
pathway and oncogenesis. In turn, Zuo et al (32) identified miR-141 as a tumor
suppressor in gastric cancer; it was concluded that decreased
expression of this miR was correlated with a more aggressive
phenotype, compared with the healthy controls. Further analysis
demonstrated that miR-141 targeted transcriptional co-activator
with PDZ-binding motif, a transcriptional coactivator that promotes
cell proliferation and epithelial-mesenchymal transition.
Furthermore, ectopic overexpression of miR-141 suppressed tumor
growth and pulmonary metastasis in nude mice.
There is no clear evidence of the regulation of the
immune response and PD-L1 expression by miRs. However, Huang et
al (33) indicated that miR-141
targets C-X-C motif chemokine ligand 12, and thus participates in
the control of colonic leukocyte trafficking during Crohn's
disease-associated intestinal inflammation.
Another molecule identified as a potential regulator
of PD-L1 is miR-200b. A correlation between the expression levels
of miR-200b and PD-L1 on tumor and immune cells was indicated.
miR-200b belongs to the family which also includes miR-141,
miR-200a, miR-429 and miR-200c, though its role in NSCLC
development is not well understood. Xiao et al (34) postulated that the overexpression of
this miR may inhibit NSCLC cell migration and invasion. It was
indicated that miR-200b regulates fascin actin bundling protein 1
and therefore, is involved in the regulation of cell migration,
motility, adhesion and other cellular interactions. Gibbons et
al (35) stated that
epithelial-mesenchymal transition (ETM) is entirely dependent on
the expression of the miR-200 family, which decreased during EMT. A
high miR-200 expression level inhibits tumor cell migration and
metastasis.
In the present study, expression of another member
of the miR-200 family, miR-429, was correlated with PD-L1 protein
expression. Xiao et al (36)
indicated that overexpression of miR-429 resulted in increased
NSCLC cell proliferation, while knockdown of miR-429 attenuated the
proliferation of H1229 cells. This was partially due to the
regulation of tumor suppressor deleted in liver cancer 1 (DLC-1)
expression by miR-429, suggesting that miR-429 may have an
oncogenic role in the regulation of cell proliferation via the
direct inhibition of DLC-1 protein expression in tumor cells.
In the present study, a significant association
between PD-L1 mRNA level and PD-L1 protein expression on tumor
cells was also demonstrated. Erber et al scored PD-L1 mRNA
and PD-L1 protein expression on tumor cells, where a notable
association between the expression level of PD-L1 mRNA and protein
was identified in NSCLC (37). This
suggested that in patients with PD-L1 protein expression on >50%
of tumor cells, PD-L1 mRNA may be reliably detected by RT-qPCR, and
that quantitative PD-L1 mRNA scoring may be considered as an
alternative to IHC (37). On the
other hand, Sepesi et al (38) suggested that the value of PD-L1 mRNA
expression analysis in the diagnosis and prognosis of lung cancer
is limited. It was demonstrated that PD-L1 mRNA expression in lung
cancer was higher compared with in normal tissues and other tumor
types. Furthermore, mRNA expression level was significantly higher
in lung squamous cell carcinoma compared with adenocarcinoma. Other
than PD-L1 mRNA, the authors suggested that further studies were
required to identify novel prognostic biomarkers that are
associated with improved patient survival and may be useful in
identifying patients suitable for immunotherapy (38).
There is a requirement for further studies to assess
the association between the expression of miR and PD-L1, as such
interactions may partially explain the varied responses to
immunotherapy between patients. In the present study, the positive
correlation between the expression level of PD-L1 mRNA and the
miR-200 family indicated no transcript degradation. The mechanism
for regulation of PD-L1 expression on tumor cells remains unclear.
However, miRs may serve an important role in this process, and the
study of their expression may result in the discovery of novel
predictors for immunotherapy.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
AG, MN, MSz and PK contributed to conception and
design of the study. JP, MSa, JS, PK and JM acquired the data, and
MN, TK and BJ conducted the laboratory experiments. MN, AG, BJ, JP
and PB analyzed and interpreted the data, and AG, PK and TK were
involved in drafting the manuscript or revising it critically for
important intellectual content. AG, MN, MSz, PK, BJ, JP, MSa JS, PB
and JM gave final approval of the published manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of the Medical University of Lublin (Lublin, Poland) (no.
KE-0254/169/2014). All participants provided written informed
consent to participate.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Muenst S, Soysal SD, Tzankov A and Hoeller
S: The PD-1/PD-L1 pathway: biological background and clinical
relevance of an emerging treatment target in immunotherapy. Expert
Opin Ther Targets. 19:201–211. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Sharma P and Allison JP: The future of
immune checkpoint therapy. Science. 348:56–61. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zou W, Wolchok JD and Chen L: PD-L1
(B7-H1) and PD-1 pathway blockade for cancer therapy: mechanisms,
response biomarkers, and combinations. Sci Transl Med.
8:328rv42016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Weber JS, D'Angelo SP, Minor D, Hodi FS,
Gutzmer R, Neyns B, Hoeller C, Khushalani NI, Miller WH Jr, Lao CD,
et al: Nivolumab versus chemotherapy in patients with advanced
melanoma who progressed after anti-CTLA-4 treatment (CheckMate
037): a randomised, controlled, open-label, phase 3 trial. Lancet
Oncol. 16:375–384. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Horn L, Spigel DR, Vokes EE, Holgado E,
Ready N, Steins M, Poddubskaya E, Borghaei H, Felip E, Paz-Ares L,
et al: Nivolumab versus docetaxel in previously treated patients
with advanced non-small-cell lung cancer: two-year outcomes from
two randomized, open-Label, phase III trials (CheckMate 017 and
CheckMate 057). J Clin Oncol. 35:3924–3933. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Reck M, Rodríguez-Abreu D, Robinson AG,
Hui R, Csőszi T, Fülöp A, Gottfried M, Peled N, Tafreshi A, Cuffe
S, et al KEYNOTE-024 Investigators, : pembrolizumab versus
chemotherapy for PD-L1-positive non-small-cell lung cancer. N Engl
J Med. 375:1823–1833. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Gandhi L, Rodríguez-Abreu D, Gadgeel S,
Esteban E, Felip E, De Angelis F, Domine M, Clingan P, Hochmair MJ,
Powell SF, et al KEYNOTE-189 Investigators, : pembrolizumab plus
chemotherapy in metastatic non-small-cell lung cancer. N Engl J
Med. 378:2078–2092. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Sharma P and Allison JP: Immune checkpoint
targeting in cancer therapy: toward combination strategies with
curative potential. Cell. 161:205–214. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Borghaei H, Paz-Ares L, Horn L, Spigel DR,
Steins M, Ready NE, Chow LQ, Vokes EE, Felip E, Holgado E, et al:
Nivolumab versus docetaxel in advanced nonsquamous non-small-cell
lung cancer. N Engl J Med. 373:1627–1639. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Brahmer J, Reckamp KL, Baas P, Crinò L,
Eberhardt WE, Poddubskaya E, Antonia S, Pluzanski A, Vokes EE,
Holgado E, et al: Nivolumab versus docetaxel in advanced
squamous-cell non-small-cell lung cancer. N Engl J Med.
373:123–135. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Garon EB, Rizvi NA, Hui R, Leighl N,
Balmanoukian AS, Eder JP, Patnaik A, Aggarwal C, Gubens M, Horn L,
et al KEYNOTE-001 Investigators, : pembrolizumab for the treatment
of non-small-cell lung cancer. N Engl J Med. 372:2018–2028. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Gadgeel S, Kowanetz M, Zou W, Hirsch FR,
Kerr KM, Gandara DR, Barlesi F, Park K, McCleland M, Koeppen H, et
al: Clinical efficacy of atezolizumab (Atezo) in PD-L1 subgroups
defined by SP142 and 22C3 IHC assays in 2L+NSCLC: results from the
randomized OAK study. Ann Oncol. 28 (Suppl 5):v460–v496. 2017.
View Article : Google Scholar
|
|
13
|
Mino-Kenudson M: Immunohistochemistry for
predictive biomarkers in non-small cell lung cancer. Transl Lung
Cancer Res. 6:570–587. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Brogden KA, Parashar D, Hallier AR, Braun
T, Qian F, Rizvi NA, Bossler AD, Milhem MM, Chan TA, Abbasi T and
Vali S: Genomics of NSCLC patients both affirm PD-L1 expression and
predict their clinical responses to anti-PD-1 immunotherapy. BMC
Cancer. 18:2252018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Pillai RS: MicroRNA function: Multiple
mechanisms for a tiny RNA? RNA. 11:1753–1761. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Parker R and Sheth U: P bodies and the
control of mRNA translation and degradation. Mol Cell. 25:635–646.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang Q, Lin W, Tang X, Li S, Guo L, Lin Y
and Kwok HF: The roles of microRNAs in regulating the expression of
PD-1/PD-L1 immune checkpoint. Int J Mol Sci. 18:E25402017.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Xie WB, Liang LH, Wu KG, Wang LX, He X,
Song C, Wang YQ and Li YH: MiR-140 expression regulates cell
proliferation and targets PD-L1 in NSCLC. Cell Physiol Biochem.
46:654–663. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Gibbons DL, Chen L, Goswami S, Cortez MA,
Ahn YH, Byers LA, Lin W, Diao L, Wang J, Roybal J, et al:
Regulation of tumor cell PD-L1 expression by microRNA-200 and
control of lung cancer metastasis. J Clin Oncol. 32 (Suppl
15):8063. 2015. View Article : Google Scholar
|
|
20
|
Schmittgen TD and Livak KJ: Analyzing
real-time PCR data by the comparative C(T) method. Nat Protoc.
3:1101–1108. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Rudzki JD: Management of adverse events
related to checkpoint inhibition therapy. Memo. 11:132–137. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Postow MA, Sidlow R and Hellmann MD:
Immune-related adverse events associated with immune checkpoint
blockade. N Engl J Med. 378:158–168. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Rittmeyer A, Barlesi F, Waterkamp D, Park
K, Ciardiello F, von Pawel J, Gadgeel SM, Hida T, Kowalski DM, Dols
MC, et al OAK Study Group, : Atezolizumab versus docetaxel in
patients with previously treated non-small-cell lung cancer (OAK):
A phase 3, open-label, multicentre randomised controlled trial.
Lancet. 389:255–265. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Herbst RS, Soria JC, Kowanetz M, Fine GD,
Hamid O, Gordon MS, Sosman JA, McDermott DF, Powderly JD, Gettinger
SN, et al: Predictive correlates of response to the anti-PD-L1
antibody MPDL3280A in cancer patients. Nature. 515:563–567. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Rizvi NA, Hellmann MD, Snyder A, Kvistborg
P, Makarov V, Havel JJ, Lee W, Yuan J, Wong P, Ho TS, et al: Cancer
immunology. Mutational landscape determines sensitivity to PD-1
blockade in non-small cell lung cancer. Science. 348:124–128. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Shukuya T, Amann J, Ghai V, Wang K and
Carbone DP: Circulating miRNA and extracellular vesicle containing
miRNA as response biomarkers of anti PD-1/PD-L1 therapy in
non-small-cell lung cancer. J Clin Oncol 36 (Suppl 15):.
3058:2018.
|
|
27
|
Bracken CP, Gregory PA, Kolesnikoff N,
Bert AG, Wang J, Shannon MF and Goodall GJ: A double-negative
feedback loop between ZEB1-SIP1 and the microRNA-200 family
regulates epithelial-mesenchymal transition. Cancer Res.
68:7846–7854. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Knyazev EN, Fomicheva KA, Mikhailenko DS,
Nyushko KM, Samatov TR, Alekseev BY and Shkurnikov MY: Plasma
levels of hsa-miR-619-5p and hsa-miR-1184 differ in prostatic
benign hyperplasia and cancer. Bull Exp Biol Med. 161:108–111.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Feng F, Wu J, Gao Z, Yu S and Cui Y:
Screening the key microRNAs and transcription factors in prostate
cancer based on microRNA functional synergistic relationships.
Medicine (Baltimore). 96:e56792017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Farina NH, Ramsey JE, Cuke ME, Ahern TP,
Shirley DJ, Stein JL, Stein GS, Lian JB and Wood ME: Development of
a predictive miRNA signature for breast cancer risk among high-risk
women. Oncotarget. 8:112170–112183. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Mei Z, He Y, Feng J, Shi J, Du Y, Qian L,
Huang Q and Jie Z: MicroRNA-141 promotes the proliferation of
non-small cell lung cancer cells by regulating expression of PHLPP1
and PHLPP2. FEBS Lett. 588:3055–3061. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zuo QF, Zhang R, Li BS, Zhao YL, Zhuang Y,
Yu T, Gong L, Li S, Xiao B and Zou QM: MicroRNA-141 inhibits tumor
growth and metastasis in gastric cancer by directly targeting
transcriptional co-activator with PDZ-binding motif, TAZ. Cell
Death Dis. 6:e16232015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Huang Z, Shi T, Zhou Q, Shi S, Zhao R, Shi
H, Dong L, Zhang C, Zeng K, Chen J, et al: miR-141 Regulates
colonic leukocytic trafficking by targeting CXCL12β during murine
colitis and human Crohn's disease. Gut. 63:1247–1257. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Xiao P, Liu W and Zhou H: miR-200b
inhibits migration and invasion in non-small cell lung cancer cells
via targeting FSCN1. Mol Med Rep. 14:1835–1840. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Gibbons DL, Lin W, Creighton CJ, Rizvi ZH,
Gregory PA, Goodall GJ, Thilaganathan N, Du L, Zhang Y,
Pertsemlidis A, et al: Contextual extracellular cues promote tumor
cell EMT and metastasis by regulating miR-200 family expression.
Genes Dev. 23:2140–2151. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Xiao P, Liu W and Zhou H: miR-429 promotes
the proliferation of non-small cell lung cancer cells via targeting
DLC-1. Oncol Lett. 12:2163–2168. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Erber R, Stöhr R, Herlein S, Giedl C,
Rieker RJ, Fuchs F, Ficker JH, Hartmann A, Veltrup E, Wirtz RM, et
al: Comparison of PD-L1 mRNA expression measured with the
CheckPoint Typer® Assay with PD-L1 protein expression
assessed with immunohistochemistry in non-small cell lung cancer.
Anticancer Res. 37:6771–6778. 2017.PubMed/NCBI
|
|
38
|
Sepesi B, Nelson DB, Mitchell KG, Gibbons
DL, Heymach JV, Vaporciyan AA, Swisher SG and Roszik J: Prognostic
value of PD-L1 mRNA sequencing expression profile in non-small cell
lung cancer. Ann Thorac Surg. 105:1621–1626. 2018. View Article : Google Scholar : PubMed/NCBI
|