Introduction
As a type of malignancy that develops in the hard
palate, lips, the anterior two-thirds of the tongue, the upper and
lower alveolar ridges, buccal mucosa, retromolar trigone,
sublingual region and floor of the mouth, oral cancer affected
>0.0001% of people between 2000 and 2010 in the USA (1,2). The
incidence rate of oral cancer has exhibited an increasing trend in
recent years (3). The occurrence of
oral cancer has been demonstrated to be significantly associated
with smoking, alcohol drinking, poor diet, poor oral hygiene and
human papilloma virus infection (4).
However, to the best of our knowledge, the molecular mechanism of
the pathogenesis of oral cancer remains unknown (5), which hinders the development of
effective treatment strategies.
Tumor cells are characterized by abnormally
accelerated energy metabolism (6).
Therefore, inhibition of energy metabolism is considered a
promising treatment target for cancer therapy (7). Glucose transporter 1 (GLUT1), also
termed facilitated glucose transporter member 1, is a uniporter
protein that facilitates the transport of glucose to mammalian
cells (8). A number of studies have
reported that GLUT1 is abnormally upregulated in human cancer and
promotes cancer development and progression by regulating cancer
cell glucose metabolism (9,10). MicroRNAs (miRNAs) are critical
factors in cancer biology (11).
MicroRNAs in certain cases participate in cancer biology by
affecting energy metabolism (12),
particularly by regulating the expression of GLUT1 (13). miRNA-10a has been characterized as an
oncogenic miRNA in lung cancer (14). In lung cancer, miRNA-10a is
upregulated and the overexpression of miRNA-10a promotes cancer
development and progression via interactions with phosphatase and
tensin homolog signaling (14).
However, to the best of our knowledge, no study has investigated
the involvement of miRNA-10a in energy metabolism or the regulation
of GLUT1 expression. The present study revealed that miRNA-10a may
promote cancer cell proliferation in oral squamous cell carcinoma
(OSCC), a major type of oral cancer, by serving as an upstream
activator of GLUT1 and promoting glucose metabolism.
Materials and methods
Human materials and cell lines
Tumor tissue and adjacent healthy tissue samples
were obtained from 52 patients with OSCC who were treated at
Nanjing Stomatological Hospital (Nanjing, China) between July 2014
and July 2018. The inclusion criteria were as follows: i) Patients
were diagnosed with OSCC by pathological biopsies; ii) patients
with complete medical records; iii) patients with no history of
another type of malignancy; and iv) patients provided written
informed consent. The exclusion criteria were as follows: i)
Patients who had been diagnosed with multiple diseases; and ii)
patients who had received treatment prior to admission at Nanjing
Stomatological Hospital. In total, the present study included 29
males and 23 females, with an age range of 33–65 years and a mean
age of 45.3±4.4 years. The current study was approved by the Ethics
Committee of Nanjing Stomatological Hospital (Nanjing, China).
The OSCC cell lines SCC090 and SCC25 were purchased
from American Type Culture Collection (ATCC; Manassas, VA, USA).
Cells were cultured in Eagle's Minimum Essential Medium (ATCC)
containing 2 mM L-glutamine (Sangon Biotech Co., Ltd., Shanghai,
China) and 10% fetal bovine serum (FBS, Sangon Biotech Co., Ltd.)
at 37°C with 5% CO2.
RNA extraction and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
GenElute™ Total RNA Purification kit (Sigma-Aldrich;
Merck KGaA, Darmstadt, Germany) was used to extract total RNA from
tumor tissues, adjacent healthy tissues and in vitro
cultured cells. For miRNA extraction, TaqMan miRNA Isolation kit
(Applied Biosystems; Thermo Fisher Scientific, Inc., Waltham, MA,
USA) was used. RevertAid RT Reverse Transcription kit for total RNA
reverse transcription (Thermo Fisher Scientific, Inc.) and TaqMan
microRNA Reverse Transcription kit (Applied Biosystems; Thermo
Fisher Scientific, Inc.) was used for miRNA reverse transcription.
qPCR was performed using a Luna® Universal One-Step
RT-qPCR kit (catalog no. E3005; New England BioLabs, Inc., Ipswich,
MA, USA) or miScript SYBR Green PCR kit (Qiagen GmbH, Hilden,
Germany), according to the manufacturers' protocols. Primers for
miRNA-10a, GLUT1 and the endogenous control U6 were designed and
synthesized by Sangon Biotech Co., Ltd. The following primer
sequences were used: miRNA-10a forward,
5′-GGAGGGGTACCAGAATCCCATTTTGGCCA-3′ and reverse,
5′-GGAGGAAGCTTGCGGAGTGTTTATGTCAACT-3′; GLUT1 forward,
5′-CATCCTTATTGCCCAGGTGTTT-3′ and reverse,
5′-GAAGACGACACTGAGCAGCAGA-3′; and U6 forward,
5′-GCTTCGGCAGCACATATACTAAAAT-3′ and reverse,
5′-CGCTTCACGAATTTGCGTGTCAT-3′. All PCR reactions were performed on
a StepOnePlus real-time PCR system (Applied Biosystems; Thermo
Fisher Scientific, Inc.) with the following conditions: 95°C for 55
sec, 40 cycles at 95°C for 18 sec and 60.5°C for 35 sec. Ct values
were normalized using 2−∆∆Cq method (15).
Cell transfection
MISSION® microRNA Mimic hsa-miR-10a
(3′-CAAAUUCGUAUCUAGGGGAAUA-5′) and scrambled negative control miRNA
(cat. no., SIC001-1NMOL) were purchased from Sigma-Aldrich; Merck
KGaA. Vectors expressing GLUT1, GLUT1 small interferring RNA
(siRNA) (5′-CCUCUUUGUUAAUCGCUUU-3′) and scrambled control-sense
(5′-UUCUCCGAACGUGUCACGU-3′) were designed and synthesized by
Shanghai GenePharma Co., Ltd. (Shanghai, China).
Lipofectamine® 2000 reagent (catalog no. 11668-019;
Invitrogen; Thermo Fisher Scientific, Inc.) was used to perform
cell transfection with siRNA or miRNA at a dose of 50 nM and
vectors at a dose of 15 nM. Cells transfected with scrambled
negative control miRNA, scrambled control-sense or empty vector
were used as negative control cells. Cells treated with
Lipofectamine 2000 only were used as control cells. Overexpression
rates of miRNA-10a and GLUT1 >200% and a GLUT1-knockdown rate
<50% were reached 24 h after transfection, as revealed by
RT-qPCR.
Cell proliferation assay
Cell proliferation was evaluated using Cell Counting
Kit-8 (CCK-8; Beyotime Institute of Biotechnology, Haimen, China)
24 h after transfection. Briefly, cell suspensions with a cell
density of 4×104 cells/ml were prepared in Eagle's
Minimum Essential Medium containing 2 mM L-glutamine and 10% FBS,
and cells were transferred to a 96-well plate with 0.1 ml cell
suspension in each well. Cells were cultured at 37°C in a 5%
CO2 incubator, followed by the addition of 10 µl CCK-8
solution at 24, 48, 72 and 96 h. Subsequently, cells were cultured
for a further 6 h and optical density values at 450 nm were
measured using a Fisherbrand™ accuSkan™ GO UV/Vis Microplate
Spectrophotometer (Thermo Fisher Scientific, Inc.).
Glucose uptake assay
Glucose uptake abilities were measured by a glucose
uptake assay 24 h after transfection. A total of 6×105
cells were harvested and washed twice with Krebs-Ringer-HEPES (KRH)
buffer (25 mM Hepes, pH 7.4, 120 mM NaCl, 1.2 mM MgSO4,
5 mM KCl 1.3 mM CaCl2 and 1.3 mM
KH2PO4) supplemented with 1 µCi
[3H]-2-deoxyglucose (PerkinElmer, Inc., Waltham, MA, USA). Glucose
uptake was initiated by incubating cells at 37°C for 25 min.
Subsequently, cells were washed twice with ice-cold KRH buffer to
stop glucose uptake. A liquid scintillation spectrometer was used
to measure radioactivity and the [3H]-2-deoxyglucose content in
cells was indicated by disintegrations per minute.
Total protein extraction and western
blot analysis
The effect of miRNA-10a on GLUT1 expression was
detected by western blot analysis. A Total Protein Extraction kit
(catalog no. NBP2-37853; Novus Biologicals, Ltd., Cambridge, UK)
was used to extract total protein from cells, according to the
manufacturer's protocol. Electrophoresis was performed to separate
denatured proteins using 10% SDS-PAGE gel with 20 µg protein per
lane. Following transfer to PVDF membranes, the membranes were
blocked with 5% non-fat milk for 2 h at room temperature. Western
blot analysis was performed by incubation with rabbit anti-human
GLUT1 (1:1,500; catalog no. ab15309; Abcam, Cambridge, UK) and
rabbit anti-human GAPDH (1:1,300; catalog no. ab8245; Abcam) at 4°C
overnight. The membranes were then incubated with goat anti-rabbit
IgG-horseradish peroxidase secondary antibody (1:1,000; catalog no.
MBS435036; MyBioSource, San Diego, CA, USA) at room temperature for
2 h. An ECL™ Western Blotting Analysis system (Sigma-Aldrich; Merck
KGaA) was used to develop signals. Signals were normalized using
Image J v1.46 software (National Institutes of Health, Bethesda,
MD, USA).
Statistical analysis
All experiments were performed three times. Data are
presented as the mean ± standard deviation and were processed using
GraphPad Prism 6 software (GraphPad Software, Inc., La Jolla, CA,
USA). Correlation analysis between the expression levels of
miRNA-10a and GLUT1 was performed using Pearson's correlation
coefficient. Comparisons of the expression levels of miRNA-10a and
GLUT1 between tumor tissues and adjacent healthy tissues were
performed using a paired Student's t-test. Comparisons among
multiple groups were performed by one-way analysis of variance
followed by Tukey's test. P<0.05 was considered to indicate a
statistically significant difference.
Results
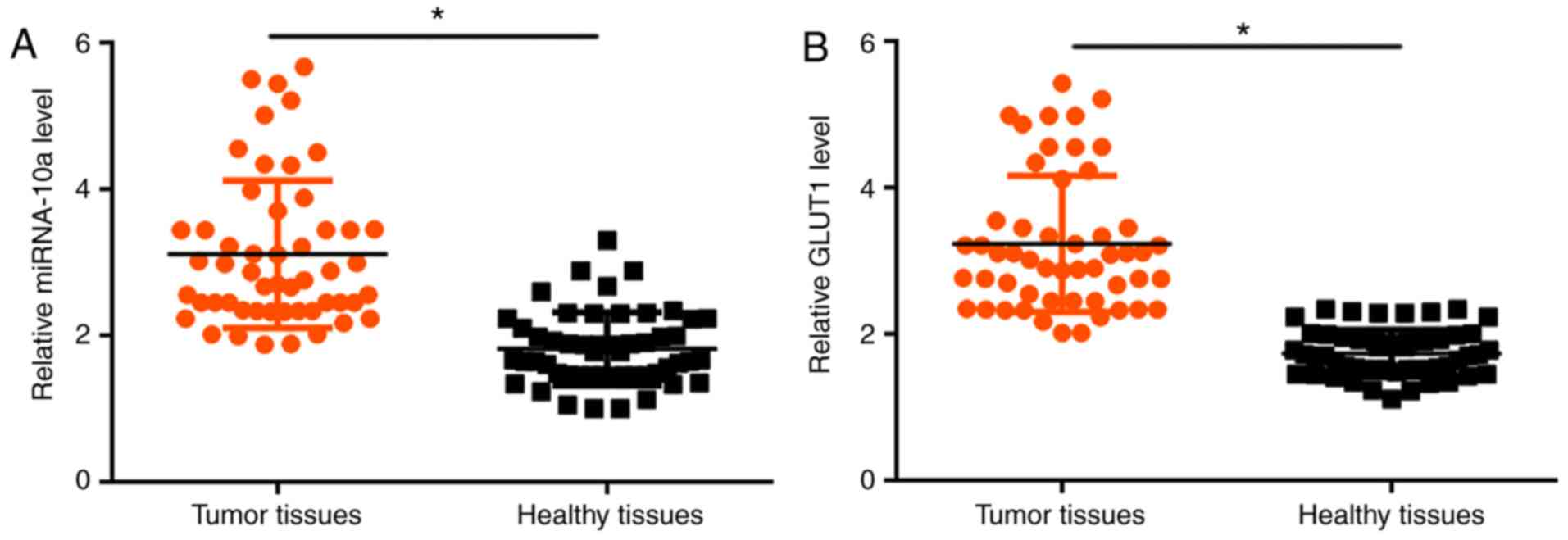
Expression levels of miRNA-10a and
GLUT1 are upregulated in tumor tissues compared with adjacent
healthy tissues
The expression levels of miRNA-10a and GLUT1 in
tumor tissues and adjacent healthy tissues obtained from patients
with OSCC were detected by RT-qPCR. Compared with adjacent healthy
tissues, the expression level of miRNA-10a was significantly
increased in tumor tissues (P<0.05; Fig. 1A). In addition, the expression of
GLUT1 was significantly upregulated in tumor tissues compared with
adjacent healthy tissues (P<0.05; Fig. 1B).
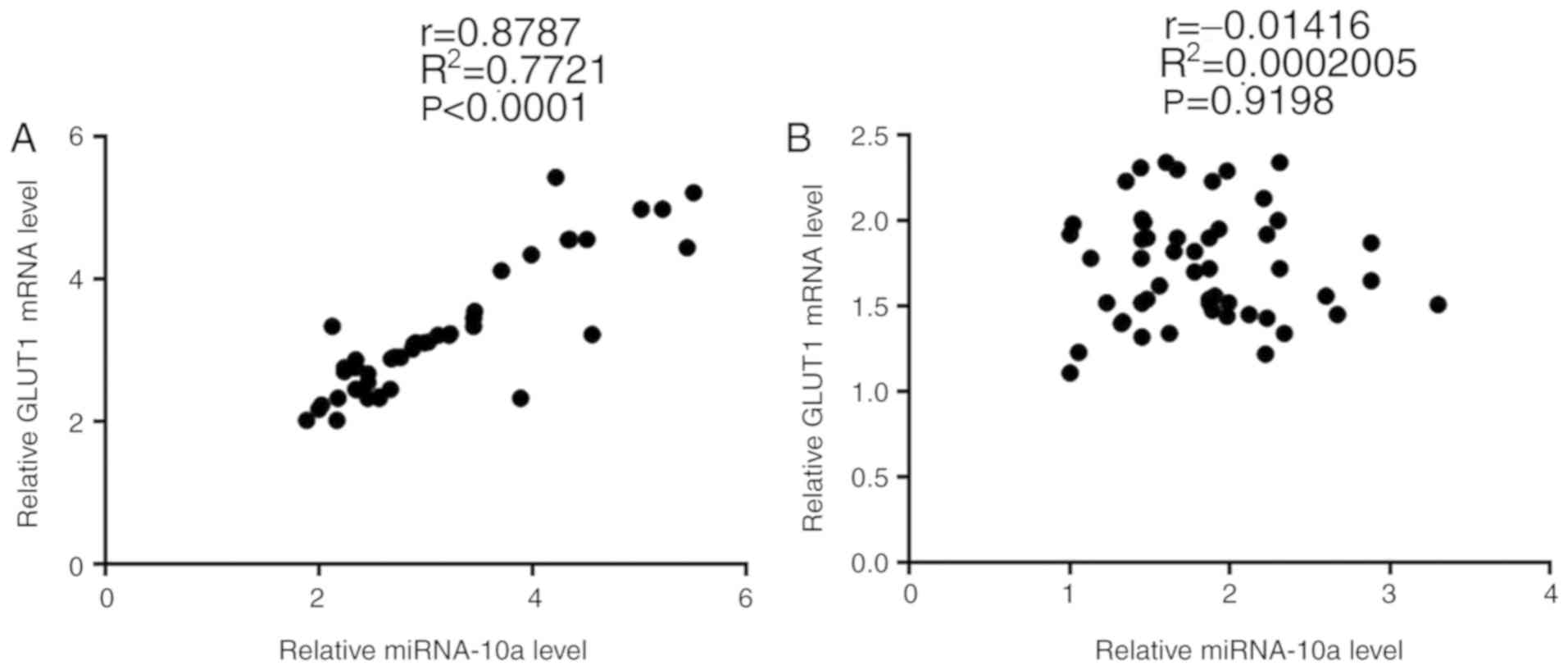
Expression levels of miRNA-10a and
GLUT1 are positively correlated in tumor tissues but not in
adjacent healthy tissues
Correlation analyses between the expression levels
of miRNA-10a and GLUT1 were performed using Pearson's correlation
coefficient. A significant positive correlation was revealed
between the expression levels of miRNA-10a and GLUT1 in tumor
tissues (P<0.001; Fig. 2A). By
contrast, a significant correlation was not identified between the
expression levels of miRNA-10a and GLUT1 in adjacent healthy
tissues (P=0.9198; Fig. 2B).
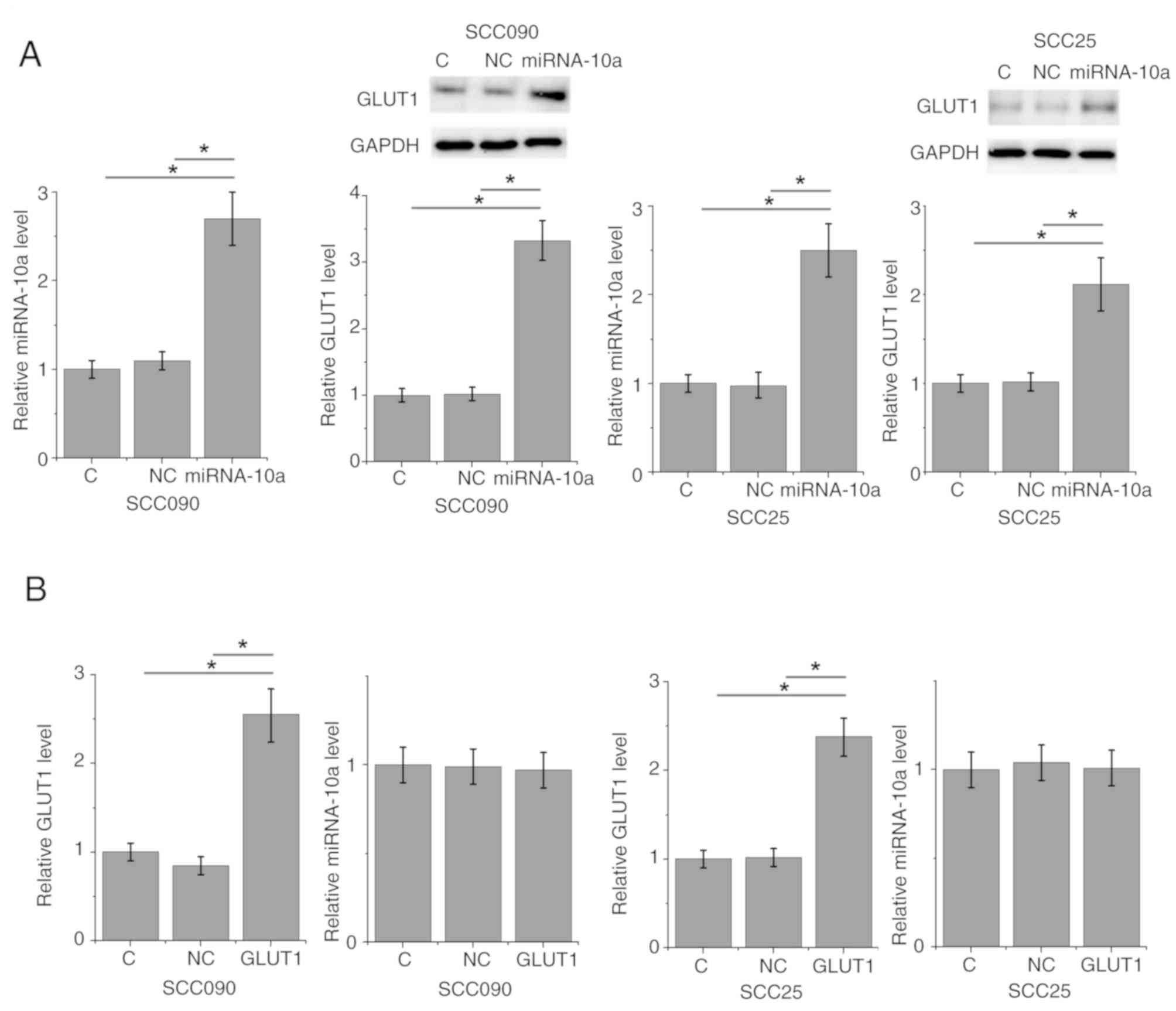
miRNA-10a overexpression upregulates
GLUT1 in the OSCC SCC090 and SCC25 cell lines
To further investigate the association between
miRNA-10a and GLUT1, miRNA-10a mimic and a GLUT1 expressing vector
were transfected into SCC090 and SCC25 OSCC cell lines, followed by
the detection of GLUT1 protein and miRNA-10a expression levels by
RT-qPCR and western blot, respectively. Compared with the control
and negative control cells, miRNA-10a and GLUT1 were significantly
increased at 24 h after transfections (Fig. 3), indicating the transfections were
successful. miRNA-10a overexpression significantly upregulated the
expression of GLUT1 in SCC090 and SCC25 cells (P<0.05; Fig. 3A). By contrast, GLUT1 overexpression
did not significantly affect the expression of miRNA-10a in SCC090
and SCC25 cells (Fig. 3B).
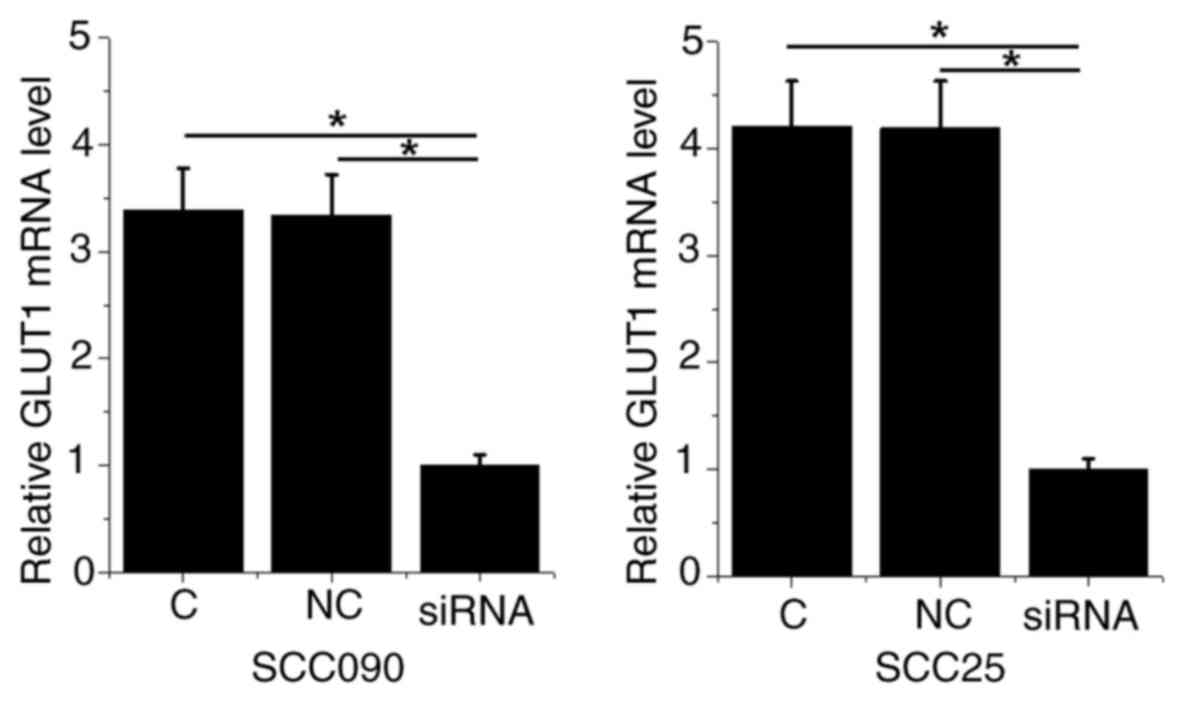
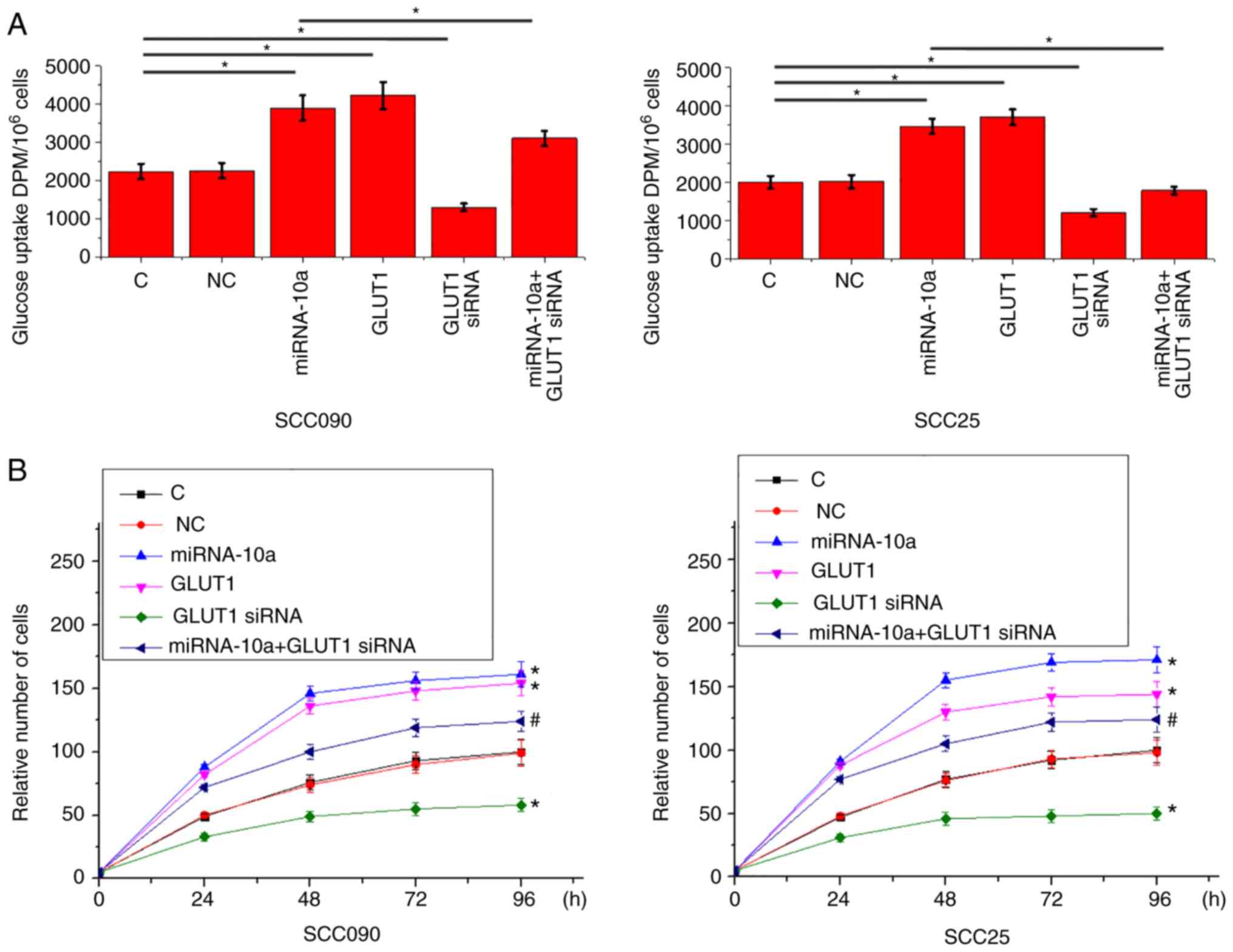
miRNA-10a overexpression promotes
glucose uptake and cell proliferation via GLUT1
The expression levels of GLUT1 in SCC090 and SCC25
cells transfected with GLUT1 siRNA were significantly decreased
compared with the control and negative control cells (Fig. 4). Compared with the control and
negative control cells, overexpression of miRNA-10a and GLUT1
significantly promoted glucose uptake, while GLUT1-knockdown
significantly inhibited glucose uptake in SCC090 and SCC25 cells
(P<0.05l Fig. 5A). In addition,
GLUT1-knockdown significantly attenuated the enhancing effects of
miRNA-10a overexpression on glucose uptake (P<0.05).
Furthermore, GLUT1 overexpression significantly promoted the
proliferation of SCC090 and SCC25 cells, while GLUT1-knockdown
significantly inhibited the proliferation of SCC090 and SCC25 cells
(P<0.05; Fig. 5B). Additionally,
GLUT1-knockdown significantly attenuated the enhancing effects of
miRNA-10a overexpression on cell proliferation (P<0.05).
Discussion
A previous study characterized miRNA-10a as an
oncogenic miRNA in lung cancer (14); however, to the best of our knowledge,
the role of miRNA-10a in other human diseases remains unknown. The
present study demonstrated that miRNA-10a is also likely an
oncogenic miRNA in OSCC, which is a major type of oral cancer that
accounted for <95% of oral cancer cases worldwide in recent
years (4,5). The role of miRNA-10a in the
proliferation of OSCC cells is likely achieved via an upregulation
of GLUT1 and accelerated glucose uptake.
An upregulation of GLUT1 is frequently observed
during the development of different types of human cancer,
including OSCC (16). Overexpression
of GLUT1 promotes glucose metabolism, which provides energy for
cancer cell division and proliferation (17). Therefore, inhibition of GLUT1 is
considered as a potential therapeutic target for the treatment of
different types of cancer (18).
Upregulation of GLUT1 also promotes the growth of oral tumors
(19). Consistent with previous
studies, the present study demonstrated that the expression level
of GLUT1 was significantly higher in tumor tissues compared with
adjacent healthy tissues of patients with OSCC. In vitro
cell experiments also revealed that overexpression of GLUT1
promoted cancer cell proliferation and glucose uptake, while
silencing of GLUT1 with siRNA inhibited cancer cell proliferation
and glucose uptake. These data further suggested an oncogenic role
of GLUT1 in oral cancer.
miRNAs serve key roles in the regulation of glucose
metabolism in cancer cells (20,21). As
an oncogenic miRNA, miRNA-10a is upregulated in lung cancer
(14); however, to the best of our
knowledge, its expression pattern in OSCC remains unknown. The
present study demonstrated that miRNA-10a was upregulated in tumor
tissues compared with adjacent healthy tissues of patients with
OSCC. In addition, in vitro cell experiments revealed that
miRNA-10a promoted cancer cell proliferation and glucose uptake.
Therefore, miRNA-10a may serve an oncogenic role in OSCC by
upregulating glucose uptake and accelerating cell
proliferation.
The expression of GLUT1 is regulated by miRNAs
during cancer progression (13). The
present study identified a significant positive correlation between
the expression levels of miRNA-10a and GLUT1 in tumor tissues. The
in vitro experiments revealed that overexpression of
miRNA-10a could significantly mediate the upregulation of GLUT1,
while overexpression of GLUT1 did not significantly alter the
expression of miRNA-10a. Furthermore, silencing of GLUT1 with siRNA
significantly attenuated the enhancing effects of miRNA-10a
overexpression on cancer cell proliferation and glucose uptake.
Therefore, miRNA-10a may promote cancer cell proliferation and
glucose uptake in OSCC by serving as an upstream activator of
GLUT1. Previous studies have also reported that miRNAs participate
in the growth, development and progression of cancer by regulating
the expression of GLUT1 (13,22). In
summary, the current study identified a novel miRNA regulator of
GLUT1 in cancer biology.
The present study is limited by the small sample
size. Therefore, studies with a larger sample size and further
correlation analyses are required. Notably, there is no promising
binding site of miRNA-10a in GLUT1 based on a local blast analysis.
In addition, the correlation between miRNA-10a and GLUT1 expression
levels was not significant in adjacent normal tissues. Therefore,
miRNA-10a may be not able to directly regulate GLUT1 and the
interaction between miRNA-10a and GLUT1 may be mediated by certain
pathological factors, such as tumor suppressive or oncogenic
pathways. The present study failed to perform miR-10a silencing
experiments due to a low knockdown efficiency; therefore, this
should be improved in future studies.
In conclusion, miRNA-10a was identified to be
upregulated in OSCC. miRNA-10a may promote cancer cell
proliferation and glucose uptake in OSCC by acting as an upstream
activator of GLUT1.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The analyzed datasets generated during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
YC and WC designed experiments. YC and YS performed
experiments. YY and XT analyzed data. WC drafted the manuscript and
all authors approved this manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of Nanjing Stomatological Hospital (Nanjing, China). All
patients provided written informed consent prior to their inclusion
in the study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Manikandan M, Deva Magendhra Rao AK,
Arunkumar G, Manickavasagam M, Rajkumar KS, Rajaraman R and
Munirajan AK: Oral squamous cell carcinoma: microRNA expression
profiling and integrative analyses for elucidation of
tumourigenesis mechanism. Mol Cancer. 15:282016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Weatherspoon DJ, Chattopadhyay A,
Boroumand S and Garcia I: Oral cavity and oropharyngeal cancer
incidence trends and disparities in the United States: 2000–2010.
Cancer Epidemiol. 39:497–504. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Tota JE, Anderson WF, Coffey C, Califano
J, Cozen W, Ferris RL, St John M, Cohen EE and Chaturvedi AK:
Rising incidence of oral tongue cancer among white men and women in
the United States, 1973–2012. Oral Oncol. 67:146–152. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Alnuaimi AD, Wiesenfeld D, O'Brien-Simpson
NM, Reynolds EC and McCullough MJ: Oral Candida colonization in
oral cancer patients and its relationship with traditional risk
factors of oral cancer: A matched case-control study. Oral Oncol.
51:139–145. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Williams HK: Molecular pathogenesis of
oral squamous carcinoma. Mol Pathol. 53:165–172. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zheng J: Energy metabolism of cancer:
Glycolysis versus oxidative phosphorylation. Oncol Lett.
4:1151–1157. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Rodríguez-Enríquez S, Marín-Hernández A,
Gallardo-Pérez JC, Carreño-Fuentes L and Moreno-Sánchez R:
Targeting of cancer energy metabolism. Mol Nutr Food Res. 53:29–48.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Olson AL and Pessin JE: Structure,
function, and regulation of the mammalian facilitative glucose
transporter gene family. Annu Rev Nutr; 16. pp. 235–256. 1996,
PubMed/NCBI
|
|
9
|
Krzeslak A, Wojcik-Krowiranda K, Forma E,
Jozwiak P, Romanowicz H, Bienkiewicz A and Brys M: Expression of
GLUT1 and GLUT3 glucose transporters in endometrial and breast
cancers. Pathol Oncol Res. 18:721–728. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Carvalho KC, Cunha IW, Rocha RM, Ayala FR,
Cajaíba MM, Begnami MD, Vilela RS, Paiva GR, Andrade RG and Soares
FA: GLUT1 expression in malignant tumors and its use as an
immunodiagnostic marker. Clinics. 66:965–972. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hayes J, Peruzzi PP and Lawler S:
MicroRNAs in cancer: Biomarkers, functions and therapy. Trends Mol
Med. 20:460–469. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chen B, Li H, Zeng X, Yang P, Liu X, Zhao
X and Liang S: Roles of microRNA on cancer cell metabolism. J
Transl Med. 10:2282012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Liu M, Gao J, Huang Q, Jin Y and Wei Z:
Downregulating microRNA-144 mediates a metabolic shift in lung
cancer cells by regulating GLUT1 expression. Oncol Lett.
11:3772–3776. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yu T, Liu L, Li J, Yan M, Lin H, Liu Y,
Chu D, Tu H, Gu A and Yao M: MiRNA-10a is upregulated in NSCLC and
may promote cancer by targeting PTEN. Oncotarget. 6:30239–30250.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2-ΔΔCT method. Methods. 25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wang J, Ye C, Chen C, Xiong H, Xie B, Zhou
J, Chen Y, Zheng S and Wang L: Glucose transporter GLUT1 expression
and clinical outcome in solid tumors: A systematic review and
meta-analysis. Oncotarget. 8:16875–16886. 2017.PubMed/NCBI
|
|
17
|
Oh S, Kim H, Nam KS and Shin I: Glut1
promotes cell proliferation, migration and invasion by regulating
epidermal growth factor receptor and integrin signaling in
triple-negative breast cancer cells. BMB Rep. 50:132–137. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Shibuya K, Okada M, Suzuki S, Seino M,
Seino S, Takeda H and Kitanaka C: Targeting the facilitative
glucose transporter GLUT1 inhibits the self-renewal and
tumor-initiating capacity of cancer stem cells. Oncotarget.
6:651–661. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kraus D, Reckenbeil J, Wenghoefer M, Stark
H, Frentzen M, Allam JP, Novak N, Frede S, Götz W, Probstmeier R,
et al: Ghrelin promotes oral tumor cell proliferation by modifying
GLUT1 expression. Cell Mol Life Sci. 73:1287–1299. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chen B, Liu Y, Jin X, Lu W, Liu J, Xia Z,
Yuan Q, Zhao X, Xu N and Liang S: MicroRNA-26a regulates glucose
metabolism by direct targeting PDHX in colorectal cancer cells. BMC
Cancer. 14:4432014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Lv X, Yao L, Zhang J, Han P and Li C:
Inhibition of microRNA-155 sensitizes lung cancer cells to
irradiation via suppression of HK2-modulated glucose metabolism.
Mol Med Rep. 14:1332–1338. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Li P, Yang X, Cheng Y, Zhang X, Yang C,
Deng X, Li P, Tao J, Yang H, Wei J, et al: MicroRNA-218 increases
the sensitivity of bladder cancer to cisplatin by targeting Glut.
Cell Physiol Biochem. 41:921–932. 2017. View Article : Google Scholar : PubMed/NCBI
|