Introduction
The treatment outcomes of non-metastatic tumors are
frequently satisfactory (1);
however, if the tumor cells become metastatic, the survival rate
for patients with cancer is significantly decreased (2). Therefore, preventing and inhibiting the
metastasis of cancer is a principal challenge in clinical
practices. Liver cancer is the second most frequently diagnosed
malignancy. Hepatocellular carcinoma (HCC) is the most commonly
diagnosed liver cancer, which primarily affects middle-aged and
older adults resulting in >700,000 mortalities every year
(3). Chronic viral hepatitis
infection is the principal cause of HCC worldwide (4). With an increase in the rate of viral
hepatitis infections, the incidence of HCC is predicted to increase
in certain regions, including China (5). Therefore, effective treatment
strategies are required to improve the survival of patients with
HCC.
Transforming growth factor-β1 (TGF-β1) signaling is
a central pathway involved in the metastasis of different tumors
(6). Activation of TGF-β1 signaling
mediates epithelial-mesenchymal transition (EMT), which facilitates
metastasis (7). Inhibition of the
signaling pathways initiated by TGF-β1 has been considered as a
potential approach for cancer treatment (8). Components of the TGF-β1 signaling
pathways interact with various signaling molecules, including
different long non-coding RNAs (lncRNAs) (9,10), which
are a group of non-coding RNAs that are involved in physiological
and pathological processes (11).
Accumulating evidence has demonstrated the importance of lncRNAs in
understanding cancer biology (12).
lncRNA small NF90-associated RNA (snaR) is a previously identified
lncRNA, which exhibited tumor suppression activity in human colon
cancer (13), whereas, its
involvement in other diseases is unknown. In the present study,
snaR was upregulated in HCC and may be involved in the regulation
of HCC metastasis through interactions with TGF-β1 signaling.
Patients and methods
Participants
A total of 233 patients with HCC were treated at The
Second Affiliated Hospital of Kunming Medical University (Kunming,
China) between January 2015 and January 2018. From the 233
patients, 56 patients were enrolled based on strict inclusion and
exclusion criteria. The inclusion criteria were: i) Patients who
were diagnosed and treated for the first time at The Second
Affiliated Hospital of Kunming Medical University; ii) patients who
fully understood the experimental procedure; iii) patients younger
than 70 years old; and iv) patients diagnosed by liver biopsy. The
exclusion criteria were: i) Patients with other malignancies; ii)
patients with other liver diseases and chronic diseases; and iii)
patients who were treated prior to their admission at The Second
Affiliated Hospital of Kunming Medical University. The final cohort
of patients included 32 males and 24 females, age range between 30
and 68 years with an average age of 49.2±6.4 years. Liver biopsies
and plasma of those patients were collected. During the same
period, a total of 102 individuals with suspected liver lesions
were subjected to liver biopsy and liver lesions were not present
in 46 of these cases. Amongst those 46 cases, 30 were included in
the control group, of which 17 were male and 13 female. The average
age was 50.4±6.9 years (range, 32–69 years). These control patients
were selected to match the age and sex distribution of the
patients. Cases with a previous history of malignancies were
excluded. Liver biopsies and plasma were additionally collected
from the control group. Approval was obtained from the Ethics
Committee of The Second Affiliated Hospital of Kunming Medical
University and all patients signed informed consent.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was extracted from biopsies, plasma and
cells using TRIzol® reagent (Invitrogen; Thermo Fisher
Scientific, Inc., Waltham, MA, USA). RT was performed using
SuperScript III Reverse Transcriptase kit (Thermo Fisher
Scientific, Inc.) to synthesize cDNA at following temperature
conditions: 25°C for 5 min, 55°C for 15 min and 80°C for 10 min.
SYBR-Green Real-Time PCR Master Mixes (Thermo Fisher Scientific,
Inc.) was used to prepare all PCR reactions. The PCR reaction
conditions were 95°C for 1 min, followed by 40 cycles of 95°C for
10 sec and 60°C for 40 sec. Primers used in the PCR reactions were:
snaR forward, 5′-TGGAGCCATTGTGGCTCCGGCC-3′ and reverse,
5′-CCCATGTGGACCAGGTTGGCCT-3′; and GAPDH forward,
5′-CAGGAGGCATTGCTGATGAT-3′ and reverse, 5′-GAAGGCTGGGGCTCATTT-3.′
The data were analyzed using the 2−∆∆Cq method (14).
Cell lines and cell transfection
HEP G2 and C33A were provided by American Type
Culture Collection (ATCC; Manassas, VA, USA). Cells were cultured
with RPMI-1640 medium (ATCC) containing 10% fetal bovine serum
(FBS; ATCC) at 37°C in an incubator with atmosphere of 5%
CO2. Full-length snaR was reverse transcribed into cDNA
using the aforementioned method. snaR was amplified and inserted
into a pcDNA3.1 vector (Sangon Biotech Co., Ltd., Shanghai, China)
to make an snaR expression vector. The vectors were transfected
into cells at a dose of 50 nM using Lipofectamine 2000 (Invitrogen;
Thermo Fisher Scientific, Inc.). Expression of snaR was detected by
RT-qPCR. Subsequent experiments were performed 24 h after
transfection only in cases where snaR overexpression was increased
>200% compared with the control untransfected cells and
mock-transfected negative control cells (cells transfected with
empty vector). For treatment with exogenous TGF-β1 (Sigma-Aldrich;
Merck KGaA, Darmstadt, Germany) and TGF-β inhibitor SD 208 (R&D
Systems China Co., Ltd., Shanghai, China), cells (105
cells/ml) were treated for 24 h at 37°C prior to use. TGF-β1 was
used at concentrations of 10 and 30 ng/ml and TGF-β inhibitor was
used at a dose of 10 ng/ml, based on the manufacturers'
protocols.
ELISA
Plasma TGF-β1 was detected using a human TGF-β1
Quantikine ELISA kit (cat. no. DB100B; R&D Systems China Co.,
Ltd.) according to the manufacturer's protocol.
Transwell migration and Matrigel
invasion assays
Cells were collected and cell suspensions with a
cell density of 4×104 cells/ml were prepared using
RPMI-1640 medium containing 1% FBS. From this cell suspension, 0.1
ml was added into the upper chamber, and the lower chamber was
filled with RPMI-1640 medium containing 20% FBS. Cells were
cultured for 12 h and the membranes were collected, cleaned using a
cotton swab and stained with 0.5% crystal violet (Sigma-Aldrich;
Merck KGaA) for 15 min at room temperature. Invasion assays were
performed according to the same method with the exception that the
upper chamber was coated with Matrigel (EMD Millipore, Billerica,
MA, USA) prior to the addition of the cells. A light microscope was
used to count stained cells.
Western blotting
Cell lysis buffer (Beyotime Institute of
Biotechnology, Haimen, China) was used to extract the total protein
from the cells. A bicinchoninic acid assay was used to determine
the protein concentration. Following denaturing at 95°C for 12 min,
protein samples were loaded (30 µg per well) on a 10% SDS-PAGE gel
for separation and transferred to a polyvinylidene fluoride
membrane. Membranes were blocked in 5% skimmed milk for 1 h at room
temperature. The primary antibodies used were rabbit anti-human
primary antibodies against TGF-β1 (1:1,500; cat. no. ab92486) and
GAPDH (1:1,500; cat. no. ab37168; both Abcam, Cambridge, MA, USA)
overnight at 4°C. The secondary antibody used was goat anti-rabbit
immunoglobulin-G conjugated with horseradish peroxidase (1:1,000;
cat. no. MBS435036; MyBioSource, Inc., San Diego, CA, USA) at room
temperature for 1 h. Signal development was performed using
enhanced chemiluminescence reagent (Sigma-Aldrich; Merck KGaA).
ImageJ v1.6 (National Institutes of Health, Bethesda, MD, USA) was
used for densitometric analysis.
Statistical analysis
GraphPad Prism 6 (GraphPad Software, Inc., La Jolla,
CA, USA) was used for all the data analysis. Data from 3 biological
replicates are presented as the mean ± standard deviation. A
Student's t-test was used to compare between two groups or a
one-way analysis of variance followed by a post hoc Fisher's least
significant difference test for comparisons between multiple
groups. Correlations between plasma levels of snaR and TGF-β1 were
analyzed using Pearson's correlation coefficient. Associations
between the clinicopathological data of patients and expression
levels of snaR were analyzed using a χ2-test. P<0.05
was considered to indicate a statistically significant
difference.
Results
Expression levels of snaR and TGF-β1
are significantly increased in patients with HCC compared with
healthy controls
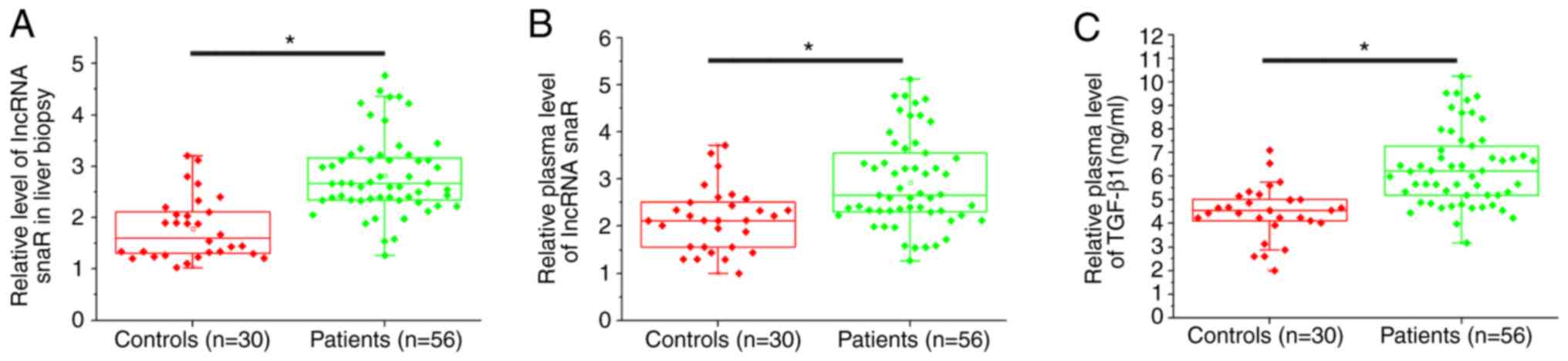
The expression level of snaR in liver biopsies and
plasma was measured by RT-qPCR. ELISA was used to detect the plasma
expression levels of TGF-β1. The expression levels of snaR in liver
biopsies (Fig. 1A) and plasma
(Fig. 1B) were significantly
increased in patients with HCC compared with the healthy controls
(P<0.05). Additionally, the serum expression levels of TGF-β1
were significantly increased in patients with HCC compared with the
healthy controls (P<0.05).
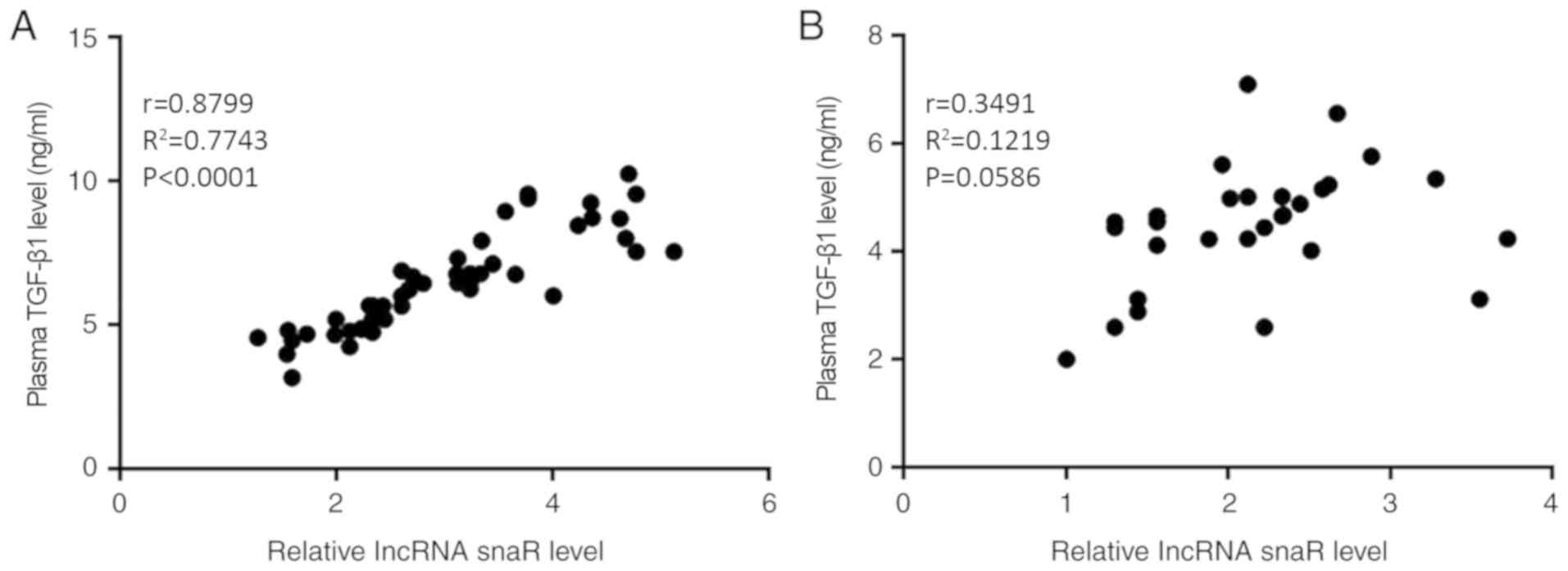
Plasma expression levels of snaR and
TGF-β1 are positively correlated in patients with HCC
Correlations between the plasma expression levels of
snaR and TGF-β1 were analyzed using Pearson's correlation
coefficient. A significantly positive correlation was identified
between the plasma expression levels of snaR and TGF-β1 in the
patients with HCC (Fig. 2A;
P<0.05). In contrast, there was no correlation between the
plasma expression levels of snaR and TGF-β1 in the healthy controls
(Fig. 2B; P>0.05).
snaR expression is significantly
associated with tumor metastasis; however, not with primary tumor
diameter
Patients were divided into high (n=28) and low
(n=28) snaR expression groups, according to the median expression
levels of snaR in the liver biopsies (median, 2.87) and plasma
(median, 2.72). Associations between the clinicopathological data
of patients and expression levels of snaR were analyzed using a
χ2-test. The results demonstrated that the expression
levels of snaR in liver biopsies (Table
I) and plasma (Table II) were
significantly associated with the existence of tumor metastasis
(P<0.05); however, were not associated with the primary tumor
diameter, age, sex or smoking and drinking habits.
 | Table I.Association between
clinicopathological feature of patients and expression levels of
small NF90-associated RNA in cancer tissue. |
Table I.
Association between
clinicopathological feature of patients and expression levels of
small NF90-associated RNA in cancer tissue.
| Clinicopathological
features | Cases | High-expression | Low-expression |
χ2-value | P-value |
|---|
| Age |
|
|
| 0.64 | 0.42 |
| >50
years | 29 | 13 | 16 |
|
|
| <50
years | 27 | 15 | 12 |
|
|
| Sex |
|
|
| 0.29 | 0.59 |
| Male | 32 | 15 | 17 |
|
|
|
Female | 24 | 13 | 11 |
|
|
| Smoking |
|
|
| 0.29 | 0.59 |
| Yes | 26 | 14 | 12 |
|
|
| No | 30 | 14 | 16 |
|
|
| Drinking |
|
|
| 0.07 | 0.79 |
| Yes | 33 | 17 | 16 |
|
|
| No | 23 | 11 | 12 |
|
|
| Primary tumor
diameter |
|
|
| 1.20 | 0.27 |
| >5
cm | 34 | 19 | 15 |
|
|
| <5
cm | 22 | 9 | 13 |
|
|
| Tumor distant
metastasis |
|
|
| 5.79 | 0.02 |
| Yes | 29 | 19 | 10 |
|
|
| No | 27 | 9 | 18 |
|
|
 | Table II.Association between
clinicopathological data of patients and expression levels of small
NF90-associated RNA in plasma. |
Table II.
Association between
clinicopathological data of patients and expression levels of small
NF90-associated RNA in plasma.
| Clinicopathological
features | Cases | High-expression | Low-expression |
χ2-value | P-value |
|---|
| Age |
|
|
| 0.07 | 0.59 |
| >50
years | 29 | 14 | 15 |
|
|
| <50
years | 27 | 14 | 13 |
|
|
| Sex |
|
|
| 1.17 | 0.28 |
|
Male | 32 | 14 | 18 |
|
|
|
Female | 24 | 14 | 10 |
|
|
| Smoking |
|
|
| 0.29 | 0.59 |
|
Yes | 26 | 12 | 14 |
|
|
| No | 30 | 16 | 14 |
|
|
| Drinking |
|
|
| 0.07 | 0.59 |
|
Yes | 33 | 17 | 16 |
|
|
| No | 23 | 11 | 12 |
|
|
| Primary tumor
diameter |
|
|
| 0.30 | 0.58 |
| >5
cm | 34 | 18 | 16 |
|
|
| <5
cm | 22 | 10 | 12 |
|
|
| Tumor distant
metastasis |
|
|
| 5.79 | 0.02 |
|
Yes | 29 | 19 | 10 |
|
|
| No | 27 | 9 | 18 |
|
|
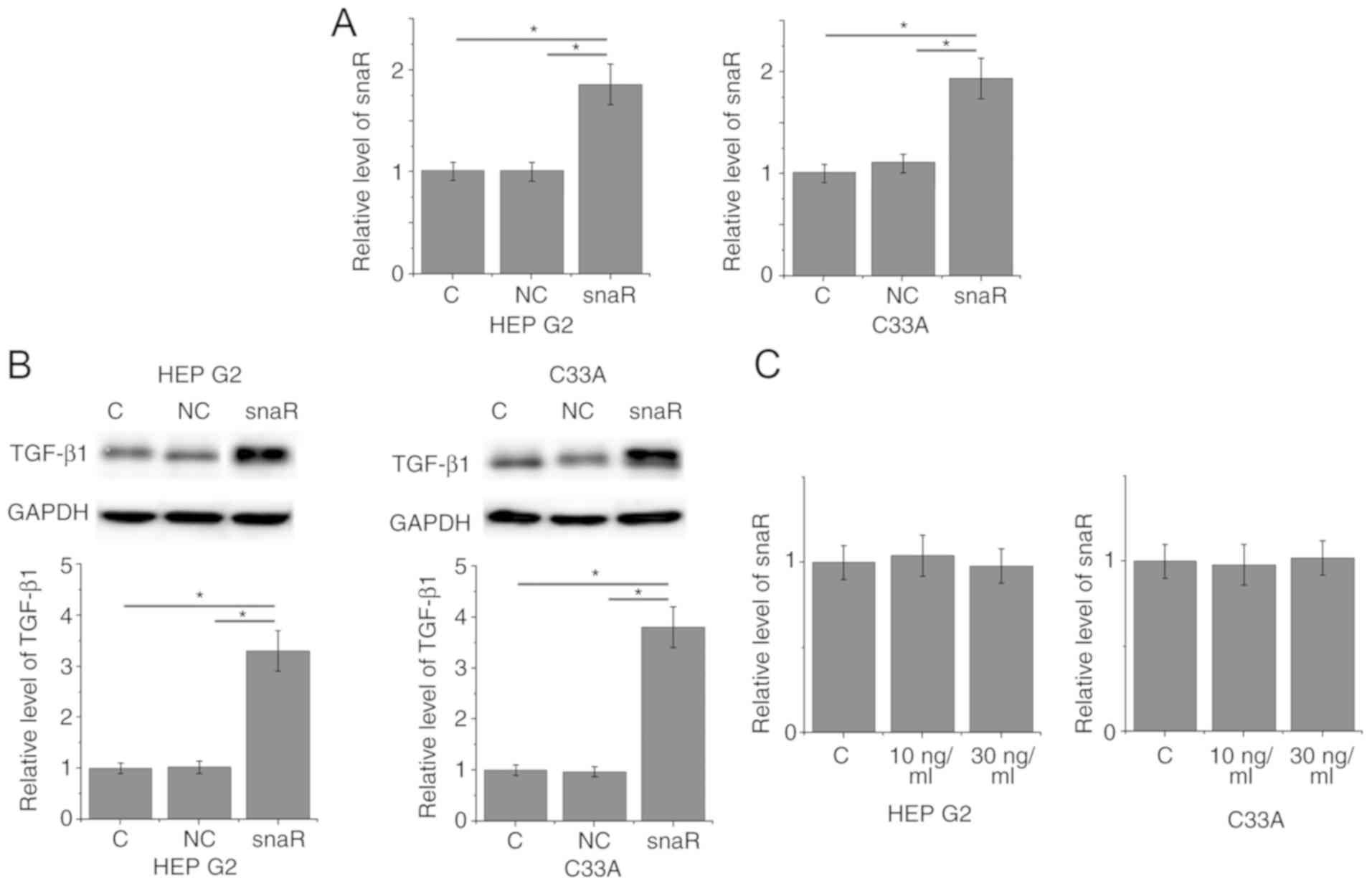
snaR is an upstream activator of
TGF-β1 in patients with HCC
To further investigate the correlation between snaR
and TGF-β1, snaR was overexpressed in cancer cells and the effects
on TGF-β1 expression were examined by western blotting.
Overexpression of snaR following transfection was demonstrated in
two cell lines, HEP G2 and C33A (Fig.
3A; P<0.05). Compared with the control cells and negative
control cells, cells with snaR overexpression demonstrated
significantly upregulated TGF-β1 expression (Fig. 3B; P<0.05). In contrast, treatment
with exogenous TGF-β1 at concentrations of 10 and 30 ng/ml
demonstrated no significant effects on snaR expression (Fig. 3C; P>0.05).
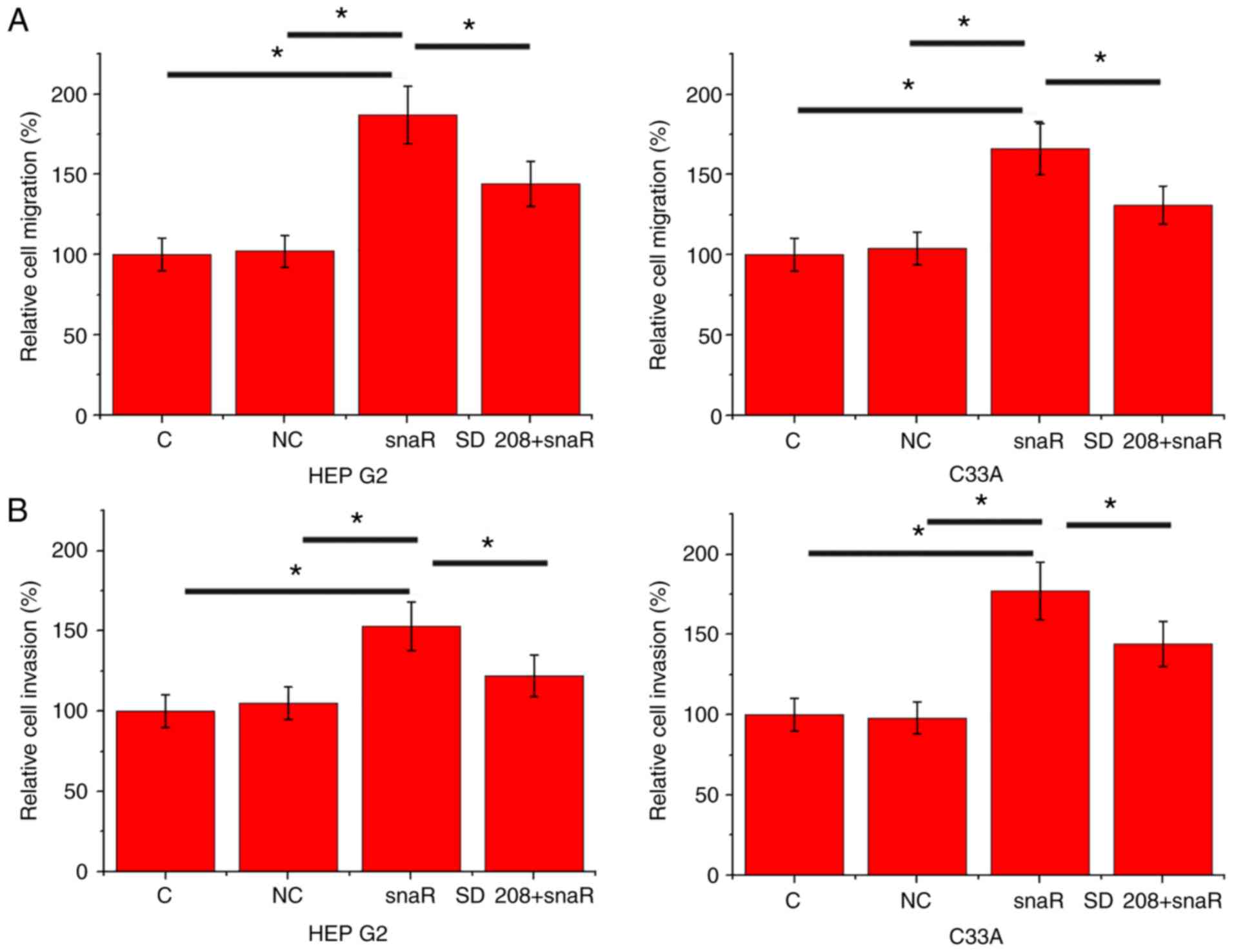
snaR overexpression promotes cell
migration and invasion
The data in Tables I
and II suggested the potential
involvement of snaR in the regulation of HCC metastasis. To
investigate this hypothesis, cell migration and invasion were
detected by Transwell migration and Matrigel invasion assays,
respectively, following snaR overexpression. Compared with the
control cells and negative control cells, cells with snaR
overexpression demonstrated significantly increased migration
(Fig. 4A; P<0.05) and invasion
(Fig. 4B; P<0.05). However,
treatment with TGF-β inhibitor SD 208 (R&D Systems China Co.,
Ltd.) at a dose of 10 ng/ml significantly attenuated the effects of
lncRNA snaR overexpression on cell migration (Fig. 4A; P<0.05) and invasion (Fig. 4B; P<0.05).
Discussion
The present study is the first study investigating
the involvement of snaR in human HCC, to the best of the author's
knowledge. snaR may be involved in the regulation of tumor
metastasis and the snaR mediated increase in HCC metastasis may be
achieved through TGF-β signaling.
Human hepatitis B virus (HBV) and/or hepatitis C
virus (HCV) infection is the principal cause of HCC (15). The present study did not analyze the
association between lncRNA snaR expression and HBV/HCV infection as
53 of the 56 patients in the present study were infected by HBV/HCV
and HBV/HCV-negative patients are rare. However, altered expression
of snaR in the plasma of HBV/HCV-positive patients without HCC was
not observed (data not shown). Therefore, the upregulation of snaR
in patients with HCC is likely a by-product of HCC rather than a
by-product of HBV/HCV infection specifically.
TGF-β1 serves as a tumor suppressor or
pro-metastatic factor depending on the stage of cancer (16). The activation of TGF-β1 signaling
inhibits tumor cell growth at the very early stage of tumor
development (17). However, TGF-β1
signaling additionally promotes tumor cell metastasis during the
later stages (18). In the present
study, a significantly increased plasma expression level of TGF-β1
in patients with HCC, compared with healthy controls, was observed.
However, one limitation of the present study is that the majority
of the patients included in the present study were in relatively
advanced stages and thus, it remains unclear whether snaR
expression is associated with the progression of cancer.
Although the functionality of a considerable number
of lncRNAs has been characterized in HCC (19–21),
those lncRNAs are either induced by HBV/HCV infection or are
involved in the whole process of cancer development, including
tumor growth and metastasis (19–21).
lncRNAs that are specifically involved in the metastasis of HCC are
rare. In the present study, a significant association between
plasma expression levels of snaR and the existence of tumor
metastases in patients with HCC was observed. However, no
significant association between plasma expression levels of snaR
and tumor size was observed, suggesting a potential involvement of
snaR in tumor metastasis. The cell migration and invasion assays
demonstrated that upregulation of snaR promoted the migration and
invasion of cells.
TGF-β1 signaling interacts with different lncRNAs in
different pathological processes (9,10). In
the present study, a positive correlation between plasma TGF-β1 and
snaR expression levels in patients with HCC was observed.
Furthermore, snaR may be an upstream activator of TGF-β1 signaling.
This conclusion is based on the following observations: i) snaR
overexpression led to increased expression of TGF-β1 in cells; ii)
treatment with exogenous TGF-β1 did not alter snaR expression in
cells; and iii) treatment with a TGF-β1 inhibitor attenuated the
enhancing effect of snaR overexpression on cancer cell migration
and invasion. However, there may be disease-associated factors,
which affect TGF-β1 and snaR expression, as there was no
correlation between plasma snaR and TGF-β1 in the healthy controls.
Another possibility is that the low expression levels of plasma
snaR and TGF-β1 in the healthy controls may make a correlation
between these two factors more difficult to determine.
Due to limited resources, it was not possible to
detect the expression of TGF-β1 mRNA. Another limitation of the
present study is that the majority of the patients were in
relatively advanced stages and the number of patients at early
stages was small. Therefore, it remains to be determined if there
is an association between the disease stage and expression levels
of TGF-β1. In conclusion, lncRNA snaR was upregulated in patients
with HCC and it may promote the metastasis of HCC through the
upregulation of TGF-β1. Therefore, lncRNA snaR is a potential
therapeutic target for HCC.
Acknowledgements
Not applicable.
Funding
The present study was supported by The National
Natural Science Foundation of China (grant no. 81660399).
Availability of data and materials
The datasets used and analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
ZS and LW designed experiments. ZS, DW, HW, JG and
XL performed experiments. ZG, RZ, SX, TW, RM and RA analyzed data.
LW drafted the manuscript. All authors approved the manuscript.
Ethics approval and consent to
participate
Approval was obtained from the Ethics Committee of
The Second Affiliated Hospital of Kunming Medical University and
all patients signed informed consent.
Patient consent for publication
All patients provided consent for publication of the
present study.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Chang SS, Bochner BH, Chou R, Dreicer R,
Kamat AM, Lerner SP, Lotan Y, Meeks JJ, Michalski JM, Morgan TM, et
al: Treatment of non-metastatic muscle-invasive bladder cancer:
AUA/ASCO/ASTRO/SUO guideline. J Urol. 198:552–559. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Khan MS, Kirkwood AA, Tsigani T, Lowe H,
Goldstein R, Hartley JA, Caplin ME and Meyer T: Early changes in
circulating tumor cells are associated with response and survival
following treatment of metastatic neuroendocrine neoplasms. Clin
Cancer Res. 22:79–85. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
White DL, Kanwal F, Jiao L and El Serag
HB: Epidemiology of hepatocellular carcinoma. Hepatocellular
Carcinoma Diagnosis and Treatment. Carr BI: Springer; Cham,
Switzerland: pp. 3–24. 2016, View Article : Google Scholar
|
|
4
|
Kummar S and Shafi NQ: Metastatic
hepatocellular carcinoma. Clin Oncol (R Coll Radiol). 15:288–294.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhu RX, Seto WK, Lai CL and Yuen MF:
Epidemiology of hepatocellular carcinoma in the Asia-Pacific
region. Gut Liver. 10:332–339. 2016. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Derynck R, Akhurst RJ and Balmain A:
TGF-beta signaling in tumor suppression and cancer progression. Nat
Genet. 29:117–129. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Katsuno Y, Lamouille S and Derynck R:
TGF-β signaling and epithelial-mesenchymal transition in cancer
progression. Curr Opin Oncol. 25:76–84. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wei W and Birrer MJ: Abstract 5401: TGF-β
signaling inhibition as a potential approach to target suboptimally
debulked ovarian tumors. Cancer Res. 75:5401. 2015. View Article : Google Scholar
|
|
9
|
Yuan J, Yang F, Wang F, Ma JZ, Guo YJ, Tao
QF, Liu F, Pan W, Wang TT, Zhou CC, et al: A long noncoding RNA
activated by TGF-β promotes the invasion-metastasis cascade in
hepatocellular carcinoma. Cancer Cell. 25:666–681. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Li W and Kang Y: A new Lnc in metastasis:
Long noncoding RNA mediates the prometastatic functions of TGF-β.
Cancer Cell. 25:557–559. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Mercer TR, Dinger ME and Mattick JS: Long
non-coding RNAs: Insights into functions. Nat Rev Genet.
10:155–159. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
12
|
Gutschner T and Diederichs S: The
hallmarks of cancer: A long non-coding RNA point of view. RNA Biol.
9:703–719. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lee H, Kim C, Ku JL, Kim W, Yoon SK, Kuh
HJ, Lee JH, Nam SW and Lee EK: A long non-coding RNA snaR
contributes to 5-fluorouracil resistance in human colon cancer
cells. Mol Cells. 37:540–546. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
El-Serag HB: Epidemiology of viral
hepatitis and hepatocellular carcinoma. Gastroenterology.
142:1264–1273.e1. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Akhurst RJ and Derynck R: TGF-beta
signaling in cancer-a double-edged sword. Trends Cell Biol.
11:S44–S51. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Markowitz SD and Roberts AB: Tumor
suppressor activity of the TGF-beta pathway in human cancers.
Cytokine Growth Factor Rev. 7:93–102. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Pardali K and Moustakas A: Actions of
TGF-beta as tumor suppressor and pro-metastatic factor in human
cancer. Biochim Biophys Acta. 1775:21–62. 2007.PubMed/NCBI
|
|
19
|
Huang JF, Guo YJ, Zhao CX, Yuan SX, Wang
Y, Tang GN, Zhou WP and Sun SH: Hepatitis B virus X protein
(HBx)-related long noncoding RNA (lncRNA) down-regulated expression
by HBx (Dreh) inhibits hepatocellular carcinoma metastasis by
targeting the intermediate filament protein vimentin. Hepatology.
57:1882–1892. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yang X, Xie X, Xiao YF, Xie R, Hu CJ, Tang
B, Li BS and Yang SM: The emergence of long non-coding RNAs in the
tumorigenesis of hepatocellular carcinoma. Cancer Lett.
360:119–124. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chen Z, Yu C, Zhan L, Pan Y, Chen L and
Sun C: LncRNA CRNDE promotes hepatic carcinoma cell proliferation,
migration and invasion by suppressing miR-384. Am J Cancer Res.
6:2299–2309. 2016.PubMed/NCBI
|