Introduction
Endometrial cancer (EC) arises from the uterine
endometrium and accounts for the most common complex gynecological
malignant tumor in the female reproductive system worldwide
(1,2). Statistical data have shown that the
incidence of endometrial carcinoma dramatically increased in the
past decades, and the morbidity of EC ranks second among female
reproductive tract tumors (3–5). It is
well established that the incidence of EC is predicted
statistically to increase by 50–100% in 20 years according to
parallel growth (6,7). Several studies have shown dysregulated
expression and function of genes in endometrial carcinoma (8–10). For
example, LncRNA-FER1L4 notably decreases in EC and can suppress the
proliferation of EC cells by targeting PTEN directly (11). In clinic, early and accurate
prognosis is critical to the treatment and prognosis of patients
with EC (12). Therefore, research
discoveries of reliable and potential biomarkers and therapeutic
candidates for early prognosis and preoperative identification of
patients with endometrial carcinoma is urgent.
It is reported that incidence and development
process of endometrial carcinoma involves various types of genes
(13–16). Transformer 2 protein homolog beta
(TRA2B), which has also been commonly known as SFRS10, is one of
the SR protein family members (17).
It is reported that TRA2B containing 419bp UCR to identify RNA
motifs and has neighboring regions of serine residues as well as
arginine residues resembling well characterized trans protein
factors (18–20). In addition, TRA2B is highly and
specially conserved across species including human, mouse, and
others (21–23). Many studies have made clear that
TRA2B plays a momentous role in a number of human cancers,
including breast, ovarian, lung and cervical cancer (24–28).
TRA2B has been shown to be connected with the viability,
carcinogenesis and chemotherapeutic sensitivity of human cancer
cells (29). Nevertheless, there are
very few reports on the expression levels of TRA2B in the tissues
and serum samples of patients with endometrial carcinoma and the
roles of TRA2B in EC cells. Given the fact that TRA2B is implicated
in various cancers, it is plausible to hypothesize that TRA2B may
participate in the pathogenesis of endometrial carcinoma.
The biological and clinical relevance of TRA2B in EC
progression and tumor metastasis is largely unknown. Therefore, the
objective of this research was to characterize the expression
levels of TRA2B in the tissues and blood collected from the
patients with EC and the function feature of TRA2B in HEC-1B, an EC
cell line. Our initial investigation found that the expression of
TRA2B was dysregulated in EC patients. We hypothesized that the
abnormal levels of TRA2B may lead to the incidence and course of
EC. In order to further elucidate the function and effect of TRA2B
in EC cells, we transfected EC cell line HEC-1B with synthetic
TRA2B plasmid, siRNA-TRA2B#1, siRNA-TRA2B#2 and siRNA-TRA2B#3.
These results implied that the expression of TRA2B markedly
increases in the tissues and serum of EC and TRA2B exhibited
stimulative effects on the multiplication capacity and invasion
ability of EC cells. Our findings therefore provide the first
evidence that TRA2B is a critical tumor promoter in the development
or progression of EC and supply a reliable biomarker and
therapeutic targets for patients who suffer from EC. The potential
role of TRA2B as a prognostic target needs to be further detected
on a larger number of samples over longer time.
Patients and methods
Patients and tumor specimens
In total, 68 female patients admitted to the
Department of Gynecology of Tongji Hospital Affiliated to Tongji
University (Shanghai, China) from January, 2016 to December, 2017
with the pathological diagnosis of the EC tissues were selected as
subjects, and the adjacent tissue samples were regarded as
controls, which were acquired from the Department of Pathology of
Tongji Hospital Affiliated to Tongji University. The age of
patients ranged from 30 to 60 years (average age, 45.45±11.25
years). All the procedures carried out conformed to the criteria of
the International Federation of Gynecology and Obstetrics (FIGO
staging system for uterine cancer, 2009) (30). The subjects had undergone surgical
resections or biopsies at Tongji Hospital Affiliated to Tongji
University, and the diagnoses were decided and determined by two or
more gynaecologic pathologists. For patients with endometrial
carcinoma, the EC tissues and normal tissues adjacent to cancer
from the same patient were collected immediately in the operating
theater from the removed uterus. The resected tissue samples were
promptly treated with RNAlater (Vazyme) and frozen in liquid
nitrogen container and ultimately stored in −80°C freezer. All the
experiments were repeated three times by using the tissues from
donors.
In this study, patients who had corpus uteri or
cervical inflammation, long term use of hormone drugs, other
gynecological malignant tumors or suffered from preoperative
chemotherapy, radiotherapy or biological targeted therapy were
excluded from this investigation.
The present study was approved by the Ethics
Committee of Tongji Hospital Affiliated to Tongji University.
Patients who participated in this research had complete clinical
data. The signed informed consents were obtained from the patients
or the guardians.
Serum samples
The sera were collected from the 68 above-mentioned
patients who were diagnosed with EC. The serum was age matched with
healthy volunteers from Tongji Hospital Affiliated to Tongji
University and were regarded as controls for statistical purpose.
Healthy controls were women who were confirmed to have no history
of cancer. Both patients and healthy volunteers were born in China,
lived in the province of Heilongjiang, and aged 30–60 years.
Approximately 15 ml fresh blood from each subject was collected by
using vacuum blood collection tube, and the serum was isolated and
frozen immediately at −80°C. The serum of patients and control
subjects was collected in parallel when possible. Clinical
information was retrieved from medical records.
Cell culture
The human EC cell line HEC-1B (GDC129) was obtained
from the China Center for Type Culture Collection. Human
endometrial epithelial cell (HEEC) line (BSC-5110479756-01) was
purchased from BioMart. The HEC-1B cells were transported in a box
with dye ice and stored in a liquid nitrogen container or −80°C
freezer. When used HEC-1B was placed in a 37°C water bath (Prima)
until 90% of ice thawed. The cells were then resuspended in 10 ml
volume of warm Dulbecco's modified Eagle's medium (DMEM; HyClone)
in a 15 ml centrifuge (300 × g at 4°C for 6 min) (Thermo Fisher
Scientific, Inc.). Afterwards, the cell pellet was plated and was
allowed to attach in DMEM containing 10% fetal bovine serum
(Invitrogen; Thermo Fisher Scientific, Inc.), 100 U/ml penicillin,
100 µg/ml streptomycin and optimal calcium ion. The resuscitated
cells were grown in a 25 cm2 culture flask and placed at
37°C in a 5% CO2 and 95% air humidified incubator
(Thermo Fisher Scientific, Inc.). The culture solution was changed
every two or three days. When the confluence reached ~80-90%,
HEC-1B cells were detached with 0.25% trypsin based on the
recommended instructions. HEC1-B cells in good condition were
strictly plated in 25 cm2 cell culture flasks or 6-well
plates (Nest) at a density of 5.0×105 by using Countstar
software (Thermo Fisher Scientific, Inc.). When the HEC1-B cells
grew to 60–70% confluence, the cells were used for further
experiments. The ESC cells were cultured in the specific culture
medium (X-Y Technology) and maintained at 37°C in a 5%
CO2 and 95% air humidified incubator (Thermo Fisher
Scientific, Inc.).
Plasmid transfection
Full-length cDNA of human TRA2B was subcloned into
pHR′-puro vector, and the packaged TRA2B overexpressing plasmid and
negative control (NC) lentivirus vector were obtained from
GenePharma Co. To generate TRA2B overexpressing cells, BMSCs were
treated with TRA2B plasmid. After cell plating in 6-well plates,
and reaching 40–50% confluence transfection was performed. In most
cases, plasmid vectors were used at 4 µg in this study.
Transfection of plasmid was strictly performed according to the
protocol manual. Synthesized TRA2B plasmid, Lipofectamine 2000 and
appropriate volume of fresh Opti-MEM Reduced Serum Medium (both
from Thermo Fisher Scientific, Inc.) were used during transfection.
After 6 h transfection of plasmid, the solution was changed with 2
ml fresh cell medium with 10% FBS followed by further experiments.
The expression of TRA2B in the EC cells after transfection of TRA2B
plasmid for 24 was detected by RT-qPCR analysis.
Transfection of small interfering
RNA
Lipofectamine 2000 transfection reagent (Thermo
Fisher Scientific, Inc.) was used to transfect small interfering
RNA TRA2B (siRNA-TRA2B) and was used for knocking down TRA2B and
the NC into EC cells respectively. SiRNA-TRA2B and its NC was
conducted (obtained from GenePharma Co). The protocol was performed
in accordance with the manual instructions.
The HEC-1B were seeded into 6-well plates and
transfection was conducted when the cell density reached 50–60%.
The cells were treated with Opti-MEM Reduced Serum Medium (Thermo
Fisher Scientific, Inc.) containing siRNA-TRA2B at concentration of
50 nM and 10 µl volume of Lipofectamine 2000 for 6 h. The cell
supernatant was replaced with fresh medium and cells were cultured
for 48 h, then the transfected cells were harvested and used for
further experiments. The control group was treated with NC
following the same method. Three siRNAs, siRNA-TRA2B-1,
siRNA-TRA2B-2 and siRNA-TRA2B-3, were designed to ensure the
efficiency of the transfection. The following sequences of
siRNA-TRA2B were used in this study; For TRA2B-1 forward:
5′-CCAGGCGUUCCAGAUCAAATT-3′ and reverse:
5′-UUUGAUCUGGAACGCCUGGTT-3′; for TRA2B-2, forward:
5′-GCUAUGAUGAUCGGGACUATT-3′ and reverse:
5′-UAGUCCCGAUCAUCAUAGCTT-3′; TRA2B-3, forward:
5′-CCAUUGCCGAUGUGUCUAUTT-3′ and reverse:
5′-AUAGACACAUCGGCAAUGGTT-3′. The expression level of TRA2B in the
EC cells treated with siRNA-TRA2B was assessed by RT-qPCR
technology. We obtained three siRNAs that obviously knocked down
TRA2B in HEC-1B cells.
RNA extraction
We validated the expression of TRA2B gene between
the EC tissues and normal tissues from 68 participants with EC. The
tissues were treated with RNAlater (Vazyme) and stored at −80°C
after isolation from patients. The serum of patients with EC and
healthy volunteers were stored at −80°C. Total RNA was extracted
from the tissues and serum into 1.5 ml tubes containing 1 ml volume
of TRIzol reagent (Life Technology). The purity and the
concentrations of the extracted RNA was analyzed and calculated by
a NanoDrop 2000 machine (Thermo Fisher Scientific, Inc.), and only
highly adequate and pure RNA (>1.8 A260/A280 <2.0) and RNA of
good quality (>200 ng/ml) was used for additional analysis.
Reverse transcription
The reverse transcription procedure was carried out
by using the High Capacity cDNA Reverse Transcription Kit (ABI).
For reverse transcription, 0.5 µg of total RNA was first treated
with 1 µl 10×RT buffer, 1 µl random primer, 0.4 µl deoxynucleoside
Triphosphates (dNTPs) and 0.5 µl reverse transcriptase based on
published protocol. Autoclaved MilliQ water was added to 10 µl
volume. The cDNA was generated with the reaction condition of 95°C
for 10 min, 2 cycles at 37°C for 1 h, 85°C for 5 min, and cooling
at 4°C. The generated cDNA was placed at −20°C for further
experiments.
Quantitative polymerase chain reaction
(qPCR)
The resulting cDNA synthesis was then amplified by
RT-qPCR method by using SYBR-Green PCR Master Mix (Vazyme). RT-qPCR
analysis was set up by a Real-Time PCR Detection System (Roche
Systems) by using a 20 µl reaction composed of 1 µl reverse
transcription product, 10 µl SYBR, 2 µl Universal PCR Primer for
specific genes and 7 µl double distilled water. The conditions of
the RT-qPCR were as follows: Denaturation at 95°C for 10 sec, 40
cycles at 95°C for 10 sec and 55°C for 30 sec, finally cooling at
4°C. The mRNA expression level of 18S was used as internal controls
for quantification of mRNA level of TRA2B gene. β-actin was used as
reference, and the sequence of β-actin was: Forward,
5′-GGGAAATCGTGCGTGACATT-3′ and reverse, 5′-GGAACCGCTCATTGCCAAT-3′.
The primers were designed and obtained from GenePharma Co. The
following primers were used to amplify specific genes: For TRA2B,
forward: 5′-GCTCAGCCCAAATACTCCAAG-3′ and reverse:
5′-CATTCTCCCATGTCTACTCGC-3′. For 18S, forward:
5′-GACCAGAGCGAAAAGCAT-3′ and reverse: 5′-TCGGAACTACGACGGTATC-3′.
The data were analyzed using the 2−ΔΔCq (31) relative expression method to calculate
the level of relative TRA2B mRNA. The results were analyzed based
on the sample threshold cycle (Cq) values from the experimentations
which were repeated at least three times.
Western blot analysis
The western blot analysis was performed as
previously reported (32). In short,
the protein extracted from the EC tissues and adjacent normal
tissues were obtained and resuspended in precooled
radioimmunoprecipitation (RIPA) lysis buffer solution
(Sigma-Aldrich; Merck KGaA) containing 200 mM NaCl, 2.5 mM
MgCl2, 20 mM Tris, 60 U/ml Superase-In, 1 mM DTT and
protease inhibitors (Roche Systems). BCA protein assay kit
(Beyotime) was applied to determine the proteins. A total of 40 µg
of proteins were added per lane. After centrifugation at 2,000 × g,
at 4°C for ~30 min, the liquid supernatant was transferred to 1.5
ml sterilized tubes. Protein extracts (50 µg) were separated by 12%
sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis
(PAGE; Beyotime). Next, the proteins were slowly transferred to an
appropriate nitrocellulose membrane (EMD Millipore). The membrane
was blocked in 5% dry fat-free milk in tris-buffered saline (TBS)
at room temperature for ~2 h. After milk saturation, the
aforementioned membrane was incubated in diluted primary antibody
against TRA2B (cat. no. sc-166829; dil, 1:500; Santa Cruz
Biotechnology, Inc.) overnight with gentle shaking at 4°C. The
membrane was washed three times in TBS-Tween-20 buffer for 20 min
on a shaking table, and subsequently incubated with appropriate
secondary antibody goat anti-rabbit IgG (cat. no. A0286; dil,
1:1,000; Beyotime Institute of Biotechnology) at dil, 1:1,000 for 1
h at 4°C. The protein bands were quantified using enhanced
chemiluminescence western blot kit (ECL; Amersham Biosciences).
TRA2B levels were normalized to β-actin (cat. no. sc-47778; dil,
1:500; Santa Cruz Biotechnology, Inc.). The expression of protein
was visualized by ECL system (Cell Signaling Technology, Inc.). The
level of proteins was analyzed using ImageJ software 1.50 (National
Institutes of Health).
Immunohistochemistry
Immunohistochemistry was performed as described in a
previous study (33). The paraffin
embedded EC tissues and normal tissues from the same donors who met
the requirements were collected. First, each tissue was fixed in 4%
paraformaldehyde (PBS; Solarbio) for 48 h. Next, ~4 µm consecutive
paraffin sections were cut, heated, dewaxed and dehydrated with
immunohistochemical method in accordance with the protocol. EDTA
solution of 1 mM (pH 8.9–9.1) was used for antigen retrieval and
then endogenous peroxidase of tissues was removed. Then, sections
were incubated in primary antibody dilution (antibody: PBS 1:500)
against TRA2B (cat. no. sc-166829; dil, 1:500; Santa Cruz
Biotechnology, Inc.) overnight at 4°C and washed by sterilized PBS
at least 3 times. The tissues were then incubated in the presence
of peroxidase-conjugated goat anti-rabbit antibody IgG which served
as a secondary antibody (mouse, monoclonal, cat. no. A0286; dil,
1:1,000; Beyotime Institute of Biotechnology) at 37°C for 2 h.
Then, the sections were stained with diaminobenzidine (ZLI-9031)
and counterstained with hematoxylin solution (ZLI-9643) (both from
ZSGB-Bio). Ten different fields per group were randomly selected
and recorded under a light microscope (Olympus Corp.).
Cell viability
The influence of TRA2B on the viability of HEC-1B
cells was quantified by Cell Counting Kit-8 (CCK-8; Biotech) assay,
and the procedure was performed as described previously (34). Briefly, HEC1-B cells were grown in
96-well plates (Labserv) at a density of 2.0×103 and
maintained in DMEM medium. At 24 h after transfection, the cells in
96-well plates were incubated in 10 µl CCK-8 solution in 100 µl
DMEM medium without FBS at 37°C for 1, 2, 3 and 4 h. After gentle
shaking for 1 min, the absorbance values were detected and analyzed
by using a microplate reader (BioTek Instruments) at 405 nm
wavelength.
Cell proliferation
The proliferation of TRA2B was measured by
Tetrazolium (MTT) method. 3.0×103 HEC1-B cells per well
were plated in 96-well plates (Corning, Inc.) with 500 µl culture
medium composed of 10% FBS and incubated in a cell incubator. At 0,
12, 24 and 48 h after seeding, the cells were treated with 20 µl
MTT solution (Solarbio) at a concentration of 5 mg/ml for ~4 h.
After 4 h incubation, the solution was discharged, and precipitated
formazan was dissolved in 150 µl volume of dimethyl sulfoxide
(DMSO). After gentle shaking for 2 min, the OD values at 490 nm
were assessed by a microplate reader (BioTek Instruments). Blank
values obtained from wells containing only media were
subtracted.
TUNEL staining
The anti-apoptotic effects of TRA2B on the EC cell
were analyzed by TUNEL staining. In Situ cell death
detection kit (POD)/TUNEL was purchased from Roche Corporation. The
following steps were performed referring to the protocol. Briefly,
EC cells were rinsed with cool PBS three times and fixation was
performed in 4% PFA solution for 20–30 min. Then, the cells were
thoroughly permeabilized by 0.1% Triton X-100 (Biosharp). Cells
were then incubated in 500 µl TUNEL reaction mixture for 2 h at
37°C in the dark. Finally, the nucleus were stained by DAPI
solution (Solarbio) for 30 min in the dark. The number of apoptotic
cells was determined under an optical microscope (Olympus Corp.).
For each well, more than ten random areas of TUNEL-stained cells
were captured and counted. The percentage of apoptotic cells was
determined from TUNEL-positive cells relative to DAPI-stained
cells. The sections were fixed in 4% paraformaldehyde (PBS;
Solarbio) at room temperature for 48 h. The staining concentration
was 1:500, at room temperature.
Transwell cell invasion assay
The in vitro invasion ability of the EC cell
line was analyzed as previously described (35). In brief, we used 8-µm pore Transwell
cell culture chambers (Corning, Inc.) coated with 30 µl specific
Matrigel (BD Biosciences) which served as a reconstituted basement
membrane for the invasion assay. Cells (2×104) were
plated in upper chamber. The cells were then resuspended in 200 µl
DMEM medium without FBS and placed in the top compartment of the
Transwell chambers. Then, optimal volume of standard culture medium
(10% FBS), which was regarded as chemoattractant, was added into
the lower chamber and the staining was performed at room
temperature. After 24 h, EC cells on the upper surface were
carefully removed by a clean cotton swab. After removing the
non-invasive cells, the cells at the bottom of the lower chamber
were harvested and fixed in 4% PFA (PBS; Solarbio) for 30 min.
After PBS washing, the cells were stained with 0.1% crystal violet
solution (Biosharp) for ~30 min. After rinsed in PBS for 10 min,
the total number of invasive cells was accurately detected under an
Olympus fluorescence microscope (Olympus Corp.). The total number
was counted by using AIS software.
Statistical analysis
Data were analyzed using SPSS 21.0 software (IBM).
The data were representative of similar results which were repeated
at least three times. Results were expressed as mean ± SD. The
differences among multiple groups were analyzed by ANOVA followed
by the Fisher's test, and between two groups were compared by
Student's t-test. Paired t-test was used for the comparisons
between paired samples. One-way analysis of variance was used for
the comparisons between multiple samples. A value of P<0.05 was
considered as statistically significant.
Results
Identification of differentially
expressed genes
To discover a key gene which plays a critical role
in the progression of EC, we chose several cancer related genes
from previous studies (36–38), and identified the expression of these
genes in 68 pairs of EC tissues and paired normal tissues. The
genes were recently reported to be differentially expressed in
tumor tissues and associated to the progression and metastasis of
cancer. However, the relationship between these genes and EC is not
fully explored. In this study, RT-qPCR was performed to detect the
relative mRNA levels of these genes. The results showed that SNCA,
PLZF, AZGP1 and PHD2 were commonly downregulated and D-dimer,
PD-L1, TRA2B, HIF-1α and CYR61 increased in the tissue of patients
with ECs (Fig. 1A and B). However,
the expression of SR-BI, cytokeratin 14, CXCL7, MUC13, CD83 was not
associated with EC (Fig. 1C). Based
on these differentially expressed genes in EC, we finally selected
3 upregulated genes including TRA2B, CYR61 and HIF-1α for further
analysis.
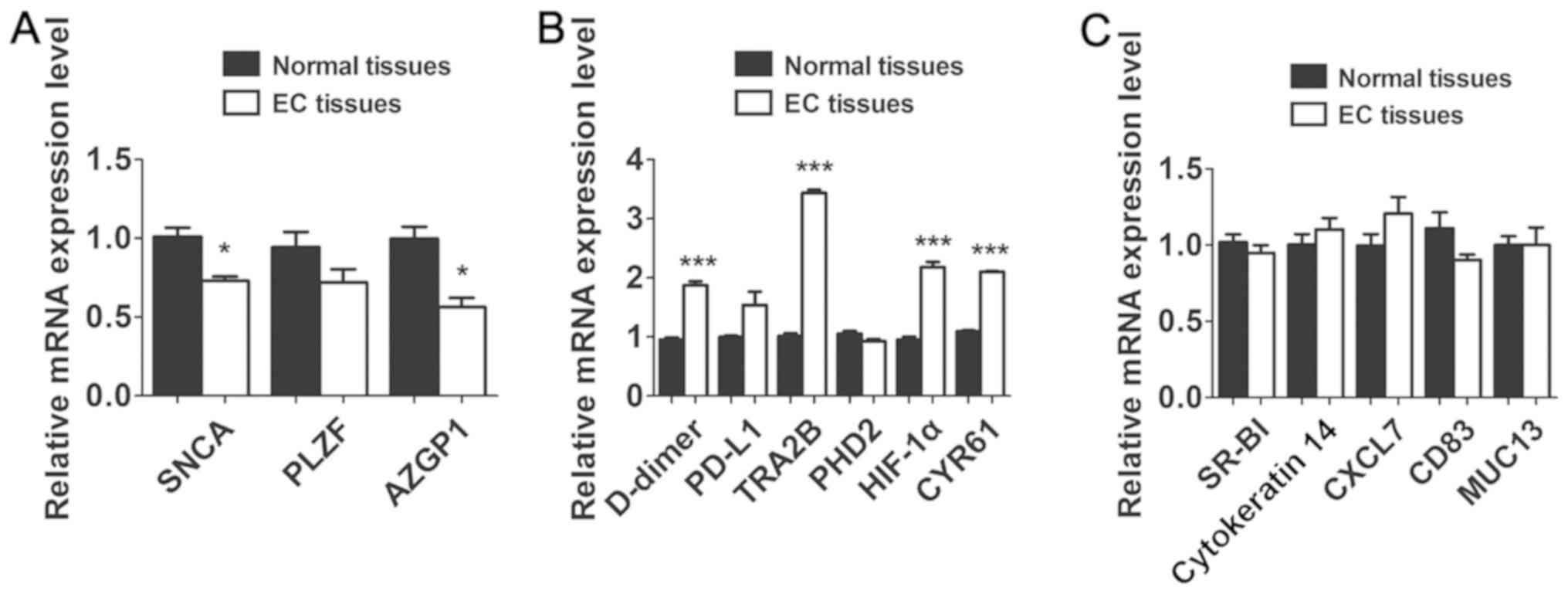 | Figure 1.The expression of cancer related
genes in the tissues of patients with EC. The relative expression
of cancer related genes in EC tissues compared with controls.
Results are presented as the mean ± standard deviation. *P<0.05,
***P<0.001 compared with the control group. (A) SNCA, PLZF,
AZGP1 and PHD2 were commonly downregulated in EC tissue compared
with control group. *P<0.05. (B) D-dimer, PD-L1, TRA2B, HIF-1α
and CYR61 increased in the tissue of patients with ECs compared
with control group. ***P<0.001. (C) The difference of expression
of SR-BI, cytokeratin 14, CXCL7, MUC13, CD83 between ECs and
control group were not significantly different (P>0.05). |
Expression of TRA2B in endometrial
carcinoma tissues
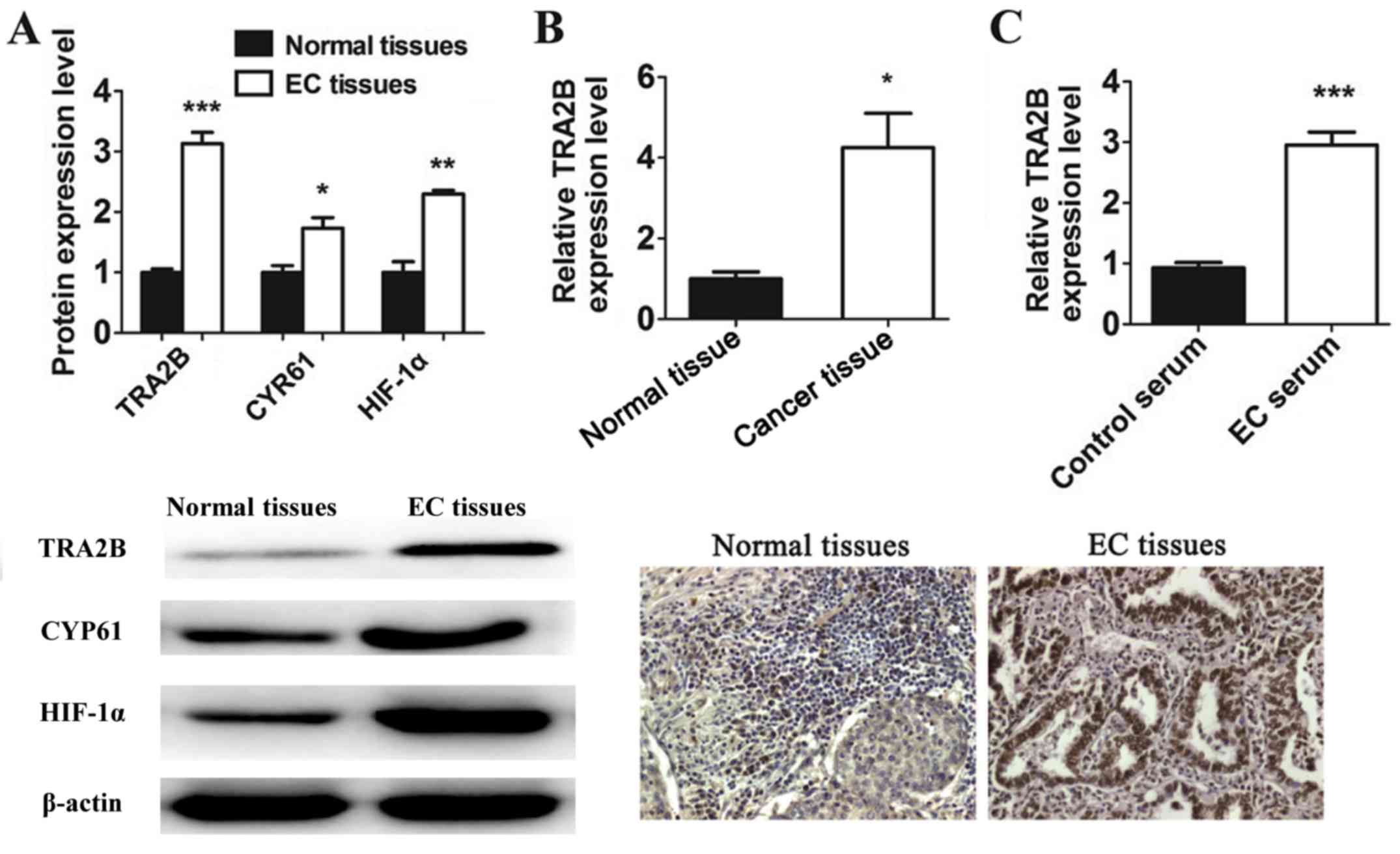
To investigate the protein expression of TRA2B,
CYR61 and HIF-1α, western blot (WB) assay was applied to determine
the protein expression of the three genes that exhibited
dysregulated expression between EC tissues and normal tissues. As
shown in the results, the expression of TRA2B, CYR61 and HIF-1α was
significantly increased in the EC tissues relative to normal
tissues (Fig. 2A). Specifically,
TRA2B showed at least a 3-fold change in expression between these
groups. Therefore, among them, TRA2B was the most significantly
upregulated gene in EC tissues.
Immunohistochemistry results indicated that 68 cases
of EC subjects showed positive expression of TRA2B. The expression
of TRA2B in EC tissue was greatly higher than that in normal tissue
(P<0.05) (Fig. 2B).
We also detected the expression of TRA2B in the
serum sample of patients with EC and healthy volunteers. The
increased levels of TRA2B in the patients with EC was measured by
RT-qPCR (Fig. 2C). Taken together,
our results therefore indicated that TRA2B was seen to be closely
related to EC and had the potential of oncogenicity.
Knockdown of TRA2B inhibits the
viability and proliferation of EC cells
The aforementioned data suggest that TRA2B
expression was positively associated with EC. Based on the
above-mentioned experimental results, we hypothesized that TRA2B
may affect the proliferation and invasion of EC cells. Thus, we
continue to detect the roles of TRA2B on the progression of EC
cells to test this hypothesis.
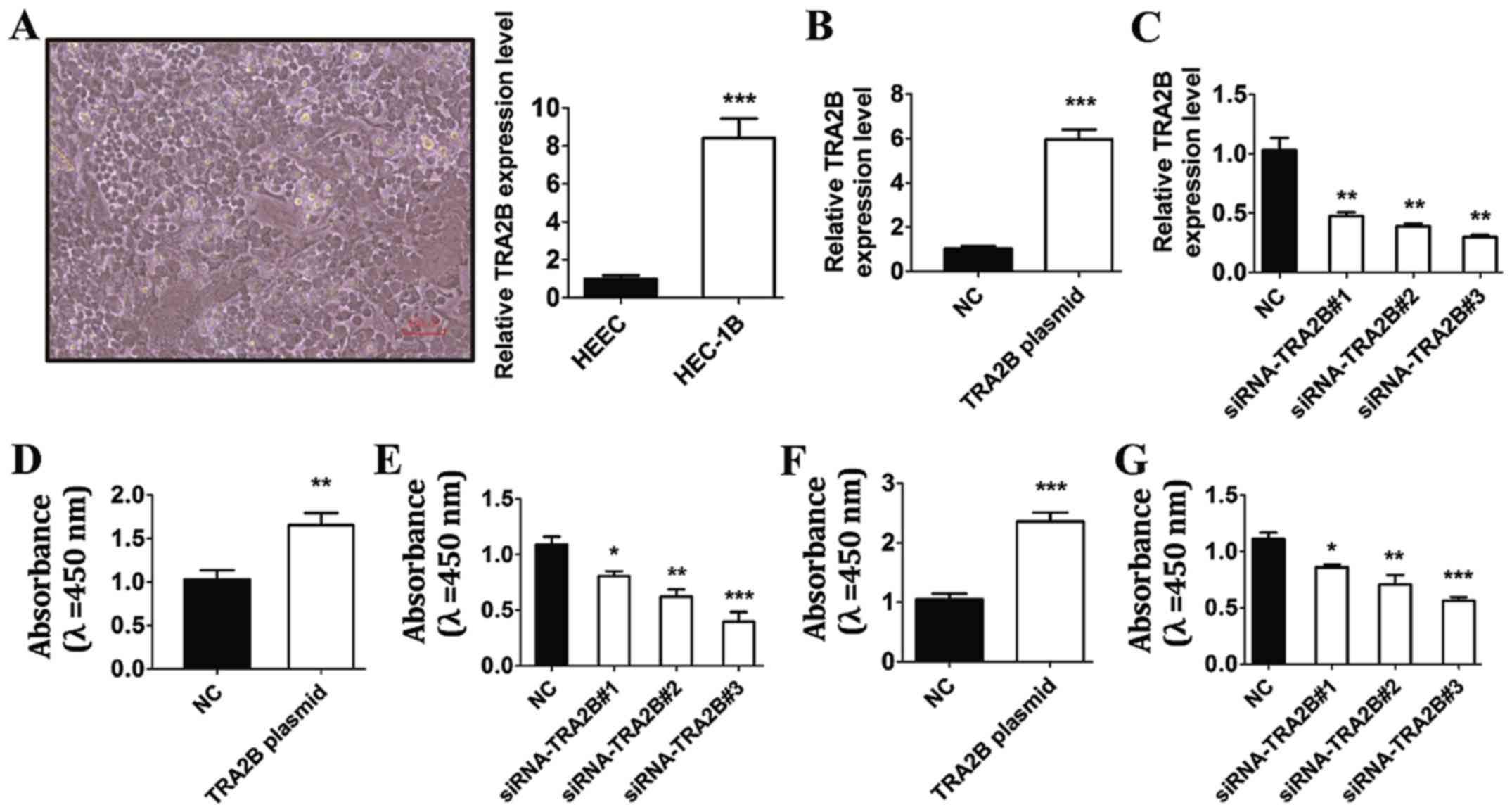
Initially, to explore the effect of TRA2B on the
viability of EC cells, the EC cells were cultured and used for
analysis on the expression of TRA2B. The morphological images of EC
cells are shown in Fig. 3A, and the
expression of TRA2B was significantly increased in EC cells
compared with human endometrial epithelial cells (HEEC) (Fig. 3A). We treated EC cells with TRA2B
plasmid, siRNA-TRA2B and its corresponding NC. RT-qPCR results
showed that EC cells exhibited high expression of TRA2B after
transfection of TRA2B plasmid (Fig.
3B). As shown in Fig. 3B,
RT-qPCR analysis indicated that the expression of TRA2B was low in
EC cells after transfection of siRNA-TRA2B#1, siRNA-TRA2B#2,
siRNA-TRA2B#3 compared with the NC group for 24 h (Fig. 3C).
Next, we treated EC cells with TRA2B plasmid,
siRNA-TRA2B#1, siRNA-TRA2B#2, siRNA-TRA2B#3 and NC for 24 h and
detected the role of TRA2B in the cell viability of HEC-1B cells by
cell counting kit-8 assay. As can be seen in Fig. 3D and E, the viability of EC cells was
extremely augmented by TRA2B plasmid. In contrast, transfection
with three siRNA-TRA2B for 24 h led to marked reduction of
viability in EC cells compared with the corresponding controls,
suggesting that inhibition of TRA2B attenuates the cell viability
of EC cells (Fig. 3D and E).
The effect of TRA2B was probed on the proliferation
capacity of EC cells by MTT assay. As Fig. 3F shows, the proliferation of EC cells
increased after transfection of TRA2B. Whereas, BMSCs transfected
with siRNA-TRA2B exhibited significantly lower absorbance values in
comparison with the control group, which indicated
siRNA-TRA2B-induced decline in the proliferation of EC cells
(Fig. 3G). These results indicated
that TRA2B silencing caused a suppression on the viability and
proliferation of EC cells.
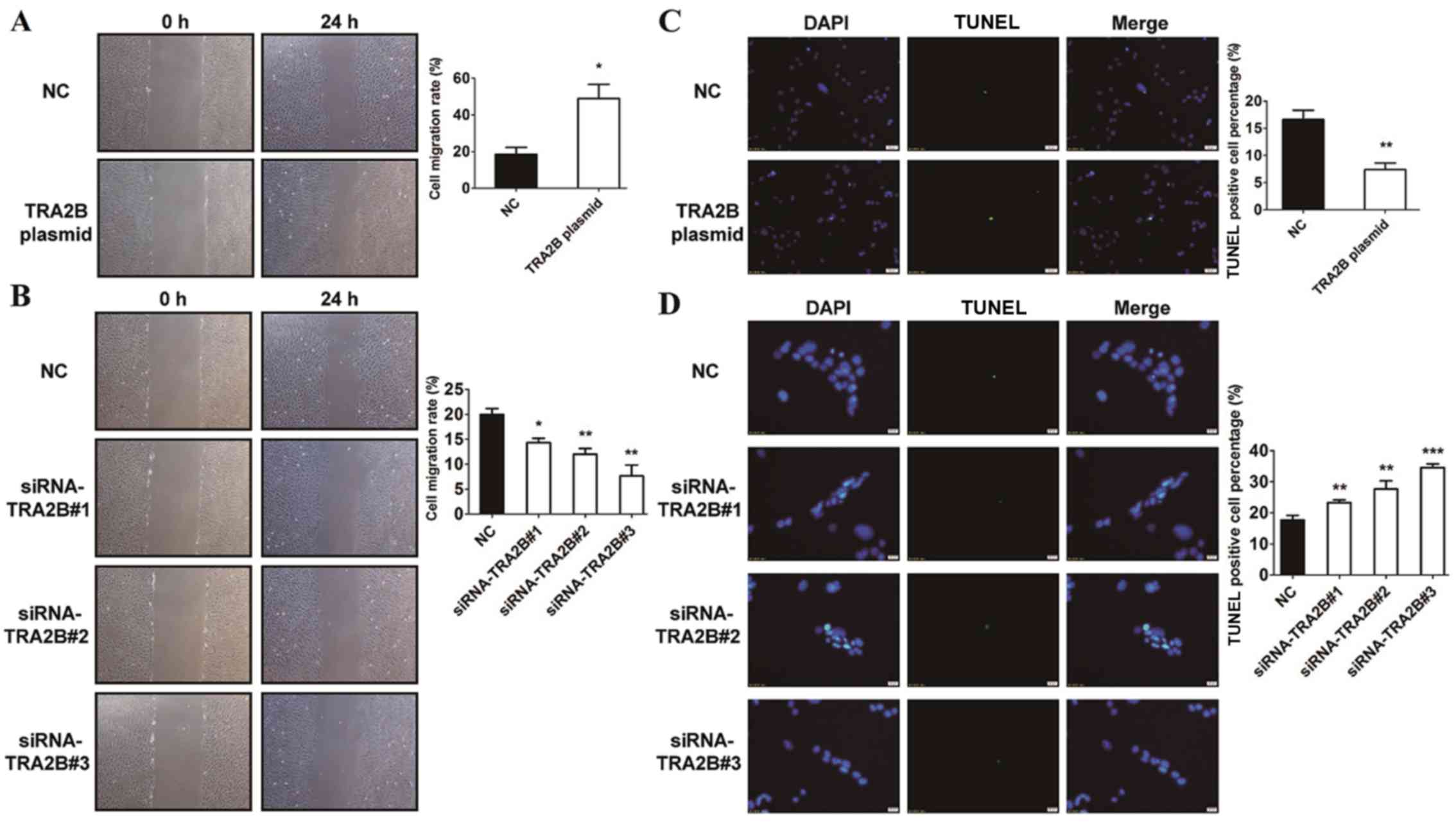
Suppression of TRA2B reduces invasion
and facilitates apoptosis of EC cells
The results above suggest that TRA2B was positively
associated with the cell viability and proliferation of EC cells.
To prove out hypothesis, we treated HEC-1B cells with TRA2B plasmid
and siRNA-TRA2B to investigate whether it can affect the
invasiveness of EC cells by Transwell invasion assay. The results
suggested that cells transfected with TRA2B plasmid exhibited
stronger ability to invade in comparison with NC (Fig. 4A). Furthermore, the Transwell results
displayed that the number of cells which passed through the
basement membrane was significantly reduced after interference of
the TRA2B gene with siRNA-TRA2B, indicating that suppression of
TRA2B significantly reduced the invasion ability of EC cells
(Fig. 4B). These data demonstrated
an inhibitory role for siRNA-TRA2B in the invasiveness of EC
cells.
To examine whether TRA2B could affect the apoptosis
of EC cells, we treated EC cells with TRA2B and siRNA-TRA2B and
stained the cells with TUNEL staining. The nucleus was stained with
DAPI solution for counting. As demonstrated in Fig. 4C, overexpression of TRA2B attenuated
apoptotic cells compared with NC group. Moreover, TRA2B silencing
significantly increased apoptosis of EC cells relative to the
corresponding control group (Fig.
4D). Consistently, TRA2B decreased apoptosis of the EC cells.
The data validated that silencing of TRA2B promotes apoptosis of EC
cells.
Discussion
Several genes have been recently investigated and
found to be differentially expressed in tumors and involved in the
progression and metastasis of human cancers (39–41). For
example, it is reported that ENO1 was confirmed as an upregulated
protein in uterine aspirate samples of patients with EC (42). Increasing evidence has also reported
on the relationship between TRA2B expression and several diseases
and cancers, and high expression of TRA2B has been linked to
aggressive disease and poor survival in cancer (43–46).
However, little is known of the effects of TRA2B on the progression
and invasion of EC.
In this study, we first analysed the expression of
TRA2B in the tissue of patients with EC. We only identified the
expression of the genes which have been discovered to be connected
with cancer. Our data indicated that among them, the expression
level of TRA2B in the tissue of patients with EC was greatly higher
than that in the normal tissue. It would be interesting to further
explore whether high TRA2B expression could be used to select
patients with EC. We hypothesized that TRA2B may be associated with
EC and even affect the cell viability, proliferation, invasion and
apoptosis of EC cells. In this study, CCK-8 assay showed that the
knockdown of TRA2B by the siRNA in EC cells was observed to reduce
cell viability. Downregulation of TRA2B inhibits the proliferation
of EC cells. Furthermore, the invasion capacity of EC cells was
decreased after treatment with TRA2B silencing in contrast to
control group. Loss of the TRA2B in EC cells enhances cell
apoptosis which was detected by TUNEL staining. Our data validation
declared that knockdown of TRA2B reduces cell viability,
proliferation, invasion of EC cells, but induces apoptosis of EC
cells. The present study provided an independent prognostic marker
in endometrial carcinoma.
In the present study, our data manifested high
expression of TRA2B in 68 EC patients. However, it is difficult to
study novel biomarkers in serum or plasma due to dynamic range and
the low concentration of biomarkers. This research focused on the
tissues and serum of patients with EC and healthy volunteers. The
strength of this study is the large cohort, including 68 evaluable
patients with EC. This gene has the potential to be a critical
biomarker and target for all patients with EC.
Taken together, our results present molecular
biological evidence of the association between TRA2B and EC and
reveal the role of TRA2B in EC cells. The results of this study
suggest that TRA2B can promote cell viability, proliferation and
invasion in EC. It is important to study the association of TRA2B
with EC. The clinical application of TRA2B as a potential molecular
biomarker for EC can increase the diagnostic process, reduce
medical expenses, and improve the quality of life.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
DP, XT, YO wrote the manuscript. DP and YO helped
with cell culture and transfection. QH and WZ were responsible for
RNA transfection. JW and MEP performed RT-qPCR. BD and XT
contributed to immunohistochemistry and MTT assay. All the authors
read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of Tongji Hospital Affiliated to Tongji University
(Shanghai, China). Patients who participated in this research had
complete clinical data. The signed informed consents were obtained
from the patients or the guardians.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Garg K, Karnezis AN and Rabban JT:
Uncommon hereditary gynaecological tumour syndromes: Pathological
features in tumours that may predict risk for a germline mutation.
Pathology. 50:238–256. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Seagle BL, Alexander AL, Lantsman T and
Shahabi S: Prognosis and treatment of positive peritoneal cytology
in early endometrial cancer: Matched cohort analyses from the
National Cancer Database. Am J Obstet Gynecol. 218:329.e1–329.e15.
2018. View Article : Google Scholar
|
|
3
|
Pal N, Broaddus RR, Urbauer DL,
Balakrishnan N, Milbourne A, Schmeler KM, Meyer LA, Soliman PT, Lu
KH, Ramirez PT, et al: Treatment of low-risk endometrial cancer and
complex atypical hyperplasia with the levonorgestrel-releasing
intrauterine device. Obstet Gynecol. 131:109–116. 2018.PubMed/NCBI
|
|
4
|
Troisi J, Sarno L, Landolfi A, Scala G,
Martinelli P, Venturella R, Di Cello A, Zullo F and Guida M:
Metabolomic signature of endometrial cancer. J Proteome Res.
17:804–812. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Smith D, Stewart CJR, Clarke EM, Lose F,
Davies C, Armes J, Obermair A, Brennan D, Webb PM, Nagle CM, et al:
ER and PR expression and survival after endometrial cancer. Gynecol
Oncol. 148:258–266. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Gibson DA, Collins F, Cousins FL, Esnal
Zufiaurre A and Saunders PT: The impact of 27-hydroxycholesterol on
endometrial cancer proliferation. Endocr Relat Cancer. 25:381–391.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Cho K, Havelock JC, Gilks B and Dunne C:
Case report: An identical twin with Sertoli-Leydig cell tumor.
Gynecol Endocrinol. 34:563–566. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Deguine C and Pulec JL: Attic
cholesteatoma and tympanosclerosis. Ear Nose Throat J. 76:3641997.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wen KC, Sung PL, Chou YT, Pan CM, Wang PH,
Lee OK and Wu CW: The role of EpCAM in tumor progression and the
clinical prognosis of endometrial carcinoma. Gynecol Oncol.
148:383–392. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hou R, Yao SS, Liu J, Wang LL, Wu L and
Jiang L: Dietary n-3 polyunsaturated fatty acids, fish consumption,
and endometrial cancer risk: A meta-analysis of epidemiological
studies. Oncotarget. 8:91684–91693. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Qiao Q and Li H: LncRNA FER1L4 suppresses
cancer cell proliferation and cycle by regulating PTEN expression
in endometrial carcinoma. Biochem Biophys Res Commun. 478:507–512.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Williams E, Villar-Prados A, Bowser J,
Broaddus R and Gladden AB: Loss of polarity alters proliferation
and differentiation in low-grade endometrial cancers by disrupting
Notch signaling. PLoS One. 12:e01890812017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Maybin JA, Murray AA, Saunders PT, Hirani
N, Carmeliet P and Critchley HO: Hypoxia and hypoxia inducible
factor-1α are required for normal endometrial repair during
menstruation. Nat Commun. 9:2952018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Knific T, Vouk K, Smrkolj Š, Prehn C,
Adamski J and Rižner TL: Models including plasma levels of
sphingomyelins and phosphatidylcholines as diagnostic and
prognostic biomarkers of endometrial cancer. J Steroid Biochem Mol
Biol. 178:312–321. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Beck TL, Schiff MA, Goff BA and Urban RR:
Robotic, laparoscopic, or open hysterectomy: Surgical outcomes by
approach in endometrial cancer. J Minim Invasive Gynecol.
25:986–993. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ebeid K, Meng X, Thiel KW, Do AV, Geary
SM, Morris AS, Pham EL, Wongrakpanich A, Chhonker YS, Murry DJ, et
al: Synthetically lethal nanoparticles for treatment of endometrial
cancer. Nat Nanotechnol. 13:72–81. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Pihlajamäki J, Lerin C, Itkonen P, Boes T,
Floss T, Schroeder J, Dearie F, Crunkhorn S, Burak F,
Jimenez-Chillaron JC, et al: Expression of the splicing factor gene
SFRS10 is reduced in human obesity and contributes to enhanced
lipogenesis. Cell Metab. 14:208–218. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Grellscheid SN, Dalgliesh C, Rozanska A,
Grellscheid D, Bourgeois CF, Stévenin J and Elliott DJ: Molecular
design of a splicing switch responsive to the RNA binding protein
Tra2β. Nucleic Acids Res. 39:8092–8104. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hirschfeld M, Jaeger M, Buratti E, Stuani
C, Grueneisen J, Gitsch G and Stickeler E: Expression of
tumor-promoting Cyr61 is regulated by hTRA2-β1 and acidosis. Hum
Mol Genet. 20:2356–2365. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Elks CE, Perry JR, Sulem P, Chasman DI,
Franceschini N, He C, Lunetta KL, Visser JA, Byrne EM, Cousminer
DL, et al GIANT Consortium, : Thirty new loci for age at menarche
identified by a meta-analysis of genome-wide association studies.
Nat Genet. 42:1077–1085. 2010. View
Article : Google Scholar : PubMed/NCBI
|
|
21
|
Mende Y, Jakubik M, Riessland M, Schoenen
F, Rossbach K, Kleinridders A, Köhler C, Buch T and Wirth B:
Deficiency of the splicing factor Sfrs10 results in early embryonic
lethality in mice and has no impact on full-length SMN/Smn
splicing. Hum Mol Genet. 19:2154–2167. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Benderska N, Becker K, Girault JA, Becker
CM, Andreadis A and Stamm S: DARPP-32 binds to tra2-beta1 and
influences alternative splicing. Biochim Biophys Acta.
1799:448–453. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Grellscheid S, Dalgliesh C, Storbeck M,
Best A, Liu Y, Jakubik M, Mende Y, Ehrmann I, Curk T, Rossbach K,
et al: Identification of evolutionarily conserved exons as
regulated targets for the splicing activator tra2β in development.
PLoS Genet. 7:e10023902011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Gabriel B, Zur Hausen A, Bouda J, Boudova
L, Koprivova M, Hirschfeld M, Jager M and Stickeler E: Significance
of nuclear hTra2-beta1 expression in cervical cancer. Acta Obstet
Gynecol Scand. 88:216–221. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Watermann DO, Tang Y, Zur Hausen A, Jäger
M, Stamm S and Stickeler E: Splicing factor Tra2-beta1 is
specifically induced in breast cancer and regulates alternative
splicing of the CD44 gene. Cancer Res. 66:4774–4780. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yang M, Li L, Wang J, Gao T, Sun Y, Li H,
Tong X and Ouyang Y: Heterogeneous nuclear ribonucleoproteins
(hnRNPs) and human transformer-2-beta1 (hTra2-beta1)-regulated
estrogen receptor-alpha improves prognosis of endometrial cancer.
Eur J Gynaecol Oncol. 35:701–707. 2014.PubMed/NCBI
|
|
27
|
Akaike Y, Masuda K, Kuwano Y, Nishida K,
Kajita K, Kurokawa K, Satake Y, Shoda K, Imoto I and Rokutan K: HuR
regulates alternative splicing of the TRA2β gene in human colon
cancer cells under oxidative stress. Mol Cell Biol. 34:2857–2873.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kajita K, Kuwano Y, Satake Y, Kano S,
Kurokawa K, Akaike Y, Masuda K, Nishida K and Rokutan K:
Ultraconserved region-containing Transformer 2β4 controls
senescence of colon cancer cells. Oncogenesis. 5:e2132016.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Munkley J, Livermore K, Rajan P and
Elliott DJ: RNA splicing and splicing regulator changes in prostate
cancer pathology. Hum Genet. 136:1143–1154. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Petru E, Lück HJ, Stuart G, Gaffney D,
Millan D and Vergote I; Gynecologic Cancer Intergroup (GCIG), :
Gynecologic Cancer Intergroup (GCIG) proposals for changes of the
current FIGO staging system. Eur J Obstet Gynecol Reprod Biol.
143:69–74. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wang QW, Su Y, Sheng JT, Gu LM, Zhao Y,
Chen XX, Chen C, Li WZ, Li KS and Dai JP: Anti-influenza A virus
activity of rhein through regulating oxidative stress, TLR4, Akt,
MAPK, and NF-kappaB signal pathways. PLoS One. 13:e01917932018.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Gu Y, Zhang J and Guan H: Expression of
EZH2 in endometrial carcinoma and its effects on proliferation and
invasion of endometrial carcinoma cells. Oncol Lett. 14:7191–7196.
2017.PubMed/NCBI
|
|
34
|
Zhang J, Pi J, Liu Y, Yu J and Feng T:
Knockdown of YTH N6-methyladenosine RNA binding protein 2 (YTHDF2)
inhibits proliferation and promotes apoptosis in MGC-803 gastric
cancer cells. Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi. 33:1628–1634.
2017.(In Chinese). PubMed/NCBI
|
|
35
|
Liu J, Qu X, Shao L, Hu Y, Yu X, Lan P,
Guo Q, Han Q, Zhang J and Zhang C: Pim-3 enhances melanoma cell
migration and invasion by promoting STAT3 phosphorylation. Cancer
Biol Ther. 19:160–168. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Grilz E, Marosi C, Königsbrügge O, Riedl
J, Posch F, Lamm W, Lang IM, Pabinger I and Ay C: Association of
complete blood count parameters, d-Dimer and soluble P-selectin
with risk of arterial thromboembolism in patients with cancer. J
Thromb Haemost. May 17–2019.(Epub ahead of print). View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Song D, Powles T, Shi L, Zhang L,
Ingersoll MA and Lu YJ: Bladder cancer, a unique model to
understand cancer immunity and develop immunotherapy approaches. J
Pathol. May 18–2019.(Epub ahead of print). View Article : Google Scholar
|
|
38
|
Meerson A, Eliraz Y, Yehuda H, Knight B,
Crundwell M, Ferguson D, Lee BP and Harries LW: Obesity impacts the
regulation of miR-10b and its targets in primary breast tumors. BMC
Cancer. 19:862019. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zhang S, Wang J, Ghoshal T, Wilkins D, Mo
YY, Chen Y and Zhou Y: lncRNA gene signatures for prediction of
breast cancer intrinsic subtypes and prognosis. Genes (Basel).
9:92018. View Article : Google Scholar
|
|
40
|
Fialkova V, Vidomanova E, Balharek T,
Marcinek J, Kudela E, Hanysova S, Visnovsky J, Dobrota D and Hatok
J: DNA methylation as mechanism of apoptotic resistance development
in endometrial cancer patients. Gen Physiol Biophys. 36:521–529.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Ma R, Feng N, Yu X, Lin H, Zhang X, Shi O,
Zhang H, Zhang S, Li L, Zheng M, et al: Promoter methylation of
Wnt/β-Catenin signal inhibitor TMEM88 is associated with
unfavorable prognosis of non-small cell lung cancer. Cancer Biol
Med. 14:377–386. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Martinez-Garcia E, Lesur A, Devis L,
Campos A, Cabrera S, van Oostrum J, Matias-Guiu X, Gil-Moreno A,
Reventos J, Colas E, et al: Development of a sequential workflow
based on LC-PRM for the verification of endometrial cancer protein
biomarkers in uterine aspirate samples. Oncotarget. 7:53102–53115.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Chen X, Huang J, Li J, Han Y, Wu K and Xu
P: Tra2betal regulates P19 neuronal differentiation and the
splicing of FGF-2R and GluR-B minigenes. Cell Biol Int. 28:791–799.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Shiraishi E, Imazato H, Yamamoto T, Yokoi
H, Abe S and Kitano T: Identification of two teleost homologs of
the Drosophila sex determination factor, transformer-2 in
medaka (Oryzias latipes). Mech Dev. 121:991–996. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Stoilov P, Daoud R, Nayler O and Stamm S:
Human tra2-beta1 autoregulates its protein concentration by
influencing alternative splicing of its pre-mRNA. Hum Mol Genet.
13:509–524. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Hofmann Y and Wirth B: hnRNP-G promotes
exon 7 inclusion of survival motor neuron (SMN) via direct
interaction with Htra2-beta1. Hum Mol Genet. 11:2037–2049. 2002.
View Article : Google Scholar : PubMed/NCBI
|