Introduction
Lobular endocervical glandular hyperplasia (LEGH)
was first described by Nucci et al (1) in 1999, as a benign lesion characterized
by a prominent lobular architecture lined by tall, mucin-rich
columnar cells located at the inner endocervix and surrounded by
larger cysts. These glands mimic gastric pyloric glands and produce
Periodic Acid-Schiff (PAS)-positive and HIK1083-positive
intracytoplasmic neutral mucin (2).
The incidence of LEGH was reported to be 0.7% in consecutive
hysterectomy specimens in a single institution (8/1169) with a mean
age range of 45–49 years (1,3). Clinically, patients present with
symptoms such as an increased watery discharge and cervical mass
(1). Gastric-type mucinous carcinoma
(GAS) is defined as adenocarcinoma with a voluminous clear and pale
eosinophilic cytoplasm and distinct cell borders (4). Minimal deviation adenocarcinoma (MDA),
also known as adenoma malignum, is very rare and defined as the
extremely well-differentiated form of GAS that often abundantly
produces pyloric mucin (5). Since
MDA/GAS exhibits an aggressive clinical behavior (6), its 5 year disease-specific survival
rate is significantly worse than that of non-gastric-type
adenocarcinoma (30% vs. 77%; P<0.0001) (5). Although LEGH was initially regarded as
a benign lesion, it was subsequently reported as a potential
precursor of MDA/GAS based on specific molecular findings as well
as clinicopathological features (7,8). We
previously demonstrated using a clonality analysis with the
inactivation pattern of the human androgen receptor gene (HUMARA
assay) that a subset of LEGH exhibited monoclonal growth and may be
a precursor of MDA (9). Moreover,
various findings on gene abnormalities common to MDA/GAS and
co-existing LEGH, such as the gain/loss of chromosomes and GNAS,
STK11, and KRAS mutations, have been reported (9–12). These
findings support the nature of LEGH as a precursor of MDA/GAS.
However, the molecular mechanisms involved in the carcinogenesis of
LEGH as well as its comprehensive genetic profiles have not yet
been elucidated. On the other hand, we often encounter clinical
LEGH cases showing stable imaging and cytology findings for many
years, which support a benign nature of LEGH. Therefore, to
elucidate the molecular characteristics of LEGH is of
importance.
In the present study, we performed whole-exome
sequencing (WES) using LEGH and normal tissues precisely collected
with laser microdissection (LM) to analyze the somatic variant
profiles of LEGH.
Patients and methods
Patients and tissue samples
The three LEGH cases described herein were
clinically diagnosed according to our management protocol (11,13). The
present study was approved by the Ethics Committee of Shinshu
University (approval no. 547) and patients provided written
consent. The 3 LEGH patients underwent simple hysterectomy. Of the
3 cases, one had adenomyosis and 2 exhibited increases in the size
of LEGH lesions. Several tissue specimens were collected from the
removed uteri. They were placed in O.C.T Compound (Sakura Finetek,
Tokyo, Japan), immediately frozen in liquid nitrogen, and stored at
−80°C until LM.
The remaining uterine tissues were fixed in 10%
buffered formalin and embedded in paraffin wax for a pathological
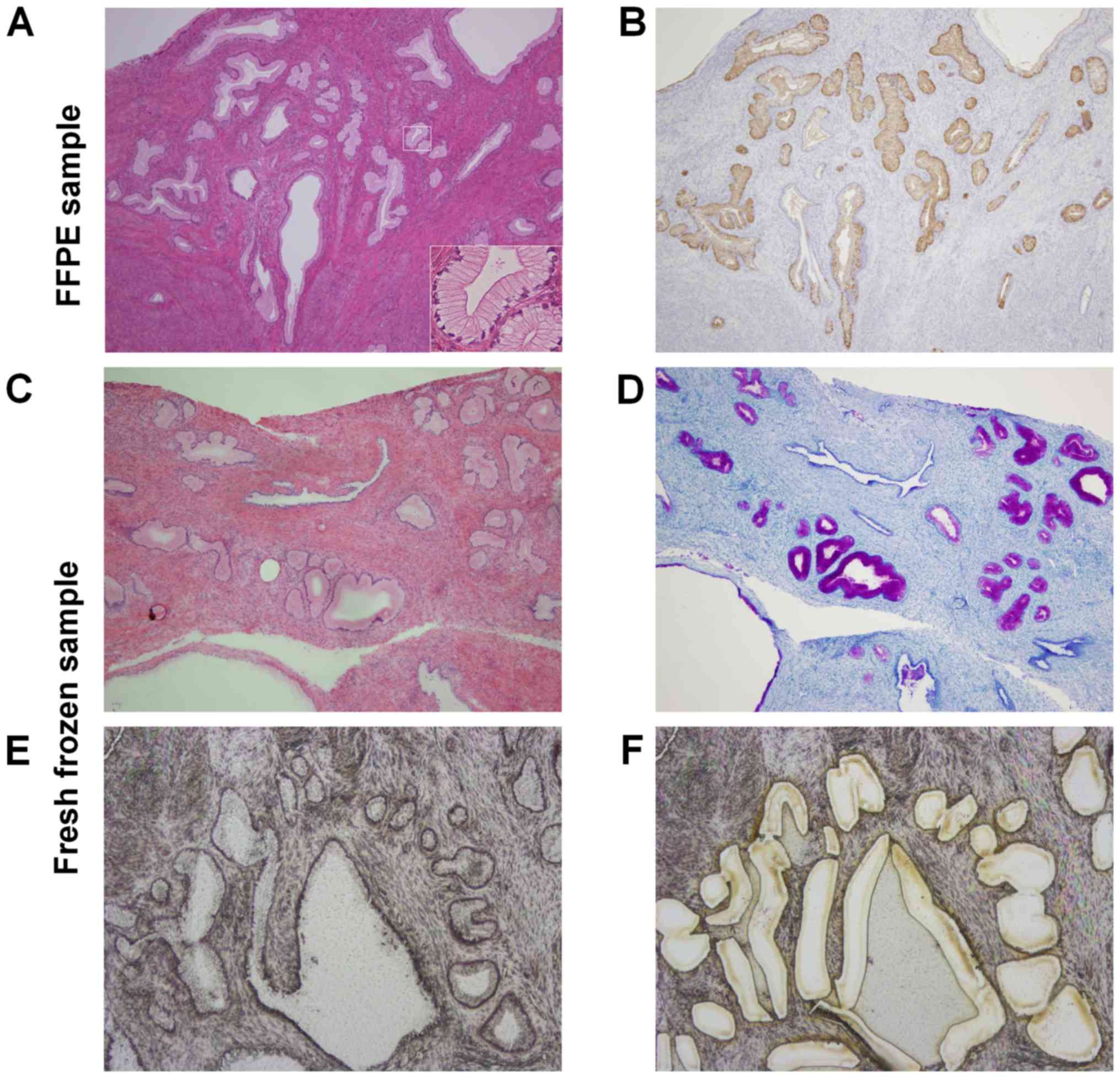
diagnosis. All three cases were histologically diagnosed with LEGH
(Fig. 1A). In order to exclude
intrinsic germline variants in WES, matched blood samples from all
cases were collected and used as an internal reference. There were
no patients with Peutz-Jeghers syndrome, which has been associated
with LEGH (14). Table I shows the characteristics of the 3
patients.
 | Table I.Characteristics of three patients with
LEGH. |
Table I.
Characteristics of three patients with
LEGH.
| Case | Age | WD | GM | Cytology | MRI findings | Surgical
indication | Surgery | Period before surgery
(months) |
|---|
| 1 | 39 | + | + | AGC-NOS | Cosmos | Lesion size-up | TAH | 68 |
| 2 | 41 | + | + | AGC-NOS | Cosmos | Lesion size-up | TLH | 62 |
| 3 | 25 | + | + | AGC-NOS | Cosmos | Adenomyosis | TLH | 81 |
Immunohistochemistry
Immunohistochemical staining for pyloric mucin was
performed (Fig. 1B) using an
anti-αGlcNAc antibody (clone HIK1083, 1:15 dilution, Kantokagaku,
Tokyo, Japan) as the primary antibody. The staining procedure was
performed according to a previous study (15). Negative controls were established by
omitting primary antibodies from the procedure (15).
LM
Fifty to sixty serial sections (thickness of 10 µm
for LM and 4 µm for H&E) were cut from fresh frozen samples
using a cryostat (Leica CM1950; Leica BIOSYSTEMS, Nussloch,
Germany).
One in every 10 sections was stained by H&E and
Alcian Blue/Periodic Acid-Schiff (AB/PAS) to confirm LEGH, while
the others were stained with hematoxylin only (Fig. 1C and D). In order to enrich LEGH
cells more than 80%, LEGH lesions were precisely collected using
the LM system (LMD6500; Leica MICROSYSTEMS, Germany) (Fig. 1E and F).
DNA extraction, WES
The genomic DNAs of LEGH and blood samples were
purified using QIAamp DNA Micro kits and QIAamp DNA Blood Mini kits
(Qiagen, Hilden, Germany) following the manufacturer's protocols.
Double-stranded DNA concentrations were quantified using the Qubit
dsDNA HS Assay and the quality of DNA was assessed by the 2200
TapeStation system (Agilent Technologies, Santa Clara, CA, USA).
Three micrograms of genomic DNA, except for the sample from case 1
(0.3 µg), was used for WES. WES was performed using SureSelectXT
Human All Exon V6 kits, except for case 1 for which we selected the
low input protocol of the same kits according to the manufacturer's
instructions (Agilent Technologies). Data were generated by
100-base paired-end reads on an HiSeq 2500 platform (Illumina Inc.,
San Diego, CA, USA). FASTQ sequences were aligned to the human
reference genome (GCRh37/hg19) with BWA-MEM software version 0.7.11
(16) and Picard (http://broadinstitute.github.io/picard)
was applied for post-alignment procedures as sorting, indexing, and
marking duplicates.
Variant calling
Since our major objective was to detect somatic gene
alterations in LEGH, germline alterations were not analyzed in the
present study. We used SAMtools 1.2 (17) and VarScan 2.3.9 to call somatic
mutations, copy number alterations (CNAs), and loss of
heterozygosity (LOH) in tumor-normal pairs (18). Single nucleotide variants (SNVs) and
indels (insertion-deletions) for somatic and germline sites were
called using VarScan v2.3.9 with almost all default settings
(19), except for a parameter of
Fisher's exact test; somatic P-value 0.001. The somatic P-value
threshold to call a somatic site was set as low as possible in
order to avoid false positives due to the low coverage depth as
recently reported (20). The effects
of all non-synonymous somatic mutations on gene function were
predicted using the annotation tool SnpEff v4.2 (21) to extract and select protein
structure-altering variants showing the functional impact of
Moderate or High. Furthermore, we browsed the Catalogue Of Somatic
Mutations In Cancer (COSMIC) v78 (https://cancer.sanger.ac.uk/cosmic), the Cancer Gene
Census (https://cancer.sanger.ac.uk/census), and ClinVar
(https://www.ncbi.nlm.nih.gov/clinvar/) to clarify
whether the mutations detected had previously been reported as
being associated with diseases. We also detected CNAs with Samtools
and VarScan in the exome data of LEGH and matched normal samples by
comparing their log2 ratios of read depths within contiguous
coverage regions separated by 100 base-pairs and by visually
inspecting them for deviations from normal (thresholds in default
settings: 0.20 for focal amplifications, −0.10 for deletions)
(18).
Results
Patient characteristics
The mean age of the three patients was 35 years old
and the mean follow-up period was 70 months (Table I). All three cases showed positive
immunostaining for HIK1083 (Fig.
1B).
WES
WES was performed on DNA isolated from LEGH tissues
and matched blood lymphocytes. The mean coverages of the target
regions of LEGH and blood samples were 32.8× and 33.6×,
respectively. Fifty somatic variants were identified. After the
variants detected in blood samples were filtered out, we identified
13 functional variants, including 12 missense and 1
insertion-deletion variants (Table
II). The mean mutation rate (± SD) of synonymous and
non-synonymous mutations of the 3 patients was 0.37 (±0.37)/Mb
(case 1; 0.79/Mb, case 2; 0.11/Mb, case 3; 0.21/Mb) (Table II).
 | Table II.Number of somatic mutations and
mutation rate in each case. |
Table II.
Number of somatic mutations and
mutation rate in each case.
|
| Non-synonymous
variant |
|
|---|
|
|
|
|
|---|
| Case | Subtotal | Missense | Insertion-deletion
(Indel) | Mutation rate
Synonymous + non-synonymous (per Mb) |
|---|
| 1 | 8 | 7 | 1 | 0.79 |
| 2 | 1 | 1 | 0 | 0.11 |
| 3 | 4 | 4 | 0 | 0.21 |
| Total | 13 | 12 | 1 |
|
Signature
The spectra of SNVs in the three cases were
characterized by a predominance of C>T/G>A transitions,
particularly at CpCpG nucleotides, and T>C/A>G transitions
followed by C>G/C>G and C>A/G>T transversions (data not
shown). These spectra did not match the seven validated mutational
signatures of cervical cancer displayed in COSMIC (https://cancer.sanger.ac.uk/cosmic/signatures)
(22).
Commonly mutated genes
We identified protein-altering somatic variants in
13 genes, including 12 non-synonymous mutations, and one
frame-shifting deletion. However, we did not detect frequent
mutations or mutations relevant to carcinogenesis in any of the 13
somatic variants (Table III). The
mutations in KPRP, OR2M4, and ZNF645 were found in COSMIC but were
not related to carcinogenic diseases. We also did not identify any
functional mutations in ClinVar except p.L141P in COL4A3 indicating
‘Benign.’ Therefore, none of the mutations identified appeared to
contribute functionally to the progression of LEGH.
 | Table III.Somatic mutations identified by a
whole-exome analysis of three patients with LEGH. |
Table III.
Somatic mutations identified by a
whole-exome analysis of three patients with LEGH.
| A, Patient 1 |
|---|
|
|---|
|
| Mutation position
(GRCh37/hg 19) |
|
| SnpEff |
|
|
|---|
|
|
|
|
|
|
|
|
|---|
| Gene | Nucleotide
(genomic) | Amino acid
(protein) | Variant allele
frequencya | Mutation type | Annotation | Impact | COSMIC ID | ClinVar allele
ID |
|---|
| KPRP |
chr1:152733724C>T | p.R554W | 0.39 (12:31) | Substitution | Missense | Moderate | COSM1334231 | N/A |
| OR2M4 |
chr1:248402643C>T | p.P138L | 0.30 (13:44) | Substitution | Missense | Moderate | COSM322280 | N/A |
| TUBA3D |
chr2:132237643C>T | p.A126V | 0.24 (14:59) | Substitution | Missense &
splice variant | Moderate | N/A | N/A |
| SST |
chr3:187386990C>A | p.D72Y | 0.35 (8:23) | Substitution | Missense | Moderate | N/A | N/A |
| CASC5 |
chr15:40915190A>G | p.R936G | 0.61 (19:31) | Substitution | Missense | Moderate | N/A | N/A |
| UNK |
chr17:7380988G>T | p.W266L | 0.21 (6:29) | Substitution | Missense | Moderate | N/A | N/A |
| ZNF645 |
chrX:22292037G>A | p.R310H | 0.43 (24:56) | Substitution | Missense | Moderate | COSM4589382 | N/A |
| SHROOM2 |
chrX:9900287delC | p.P989fs | 0.24 (21:88) | Deletion | Frameshift | High/LOF | N/A | N/A |
|
| B, Patient
2 |
|
|
| Mutation
position (GRCh37/hg 19) |
|
| SnpEff |
|
|
|
|
|
|
|
|
|
|
| Gene | Nucleotide
(genomic) | Amino acid
(protein) | Variant allele
frequencya | Mutation
type |
Annotation | Impact | COSMIC
ID | ClinVar allele
ID |
|
| COL4A3 |
chr2:228111435T>C | p.L141P | 0.66 (19:29) | Substitution | Missense | Moderate | N/A | N/A |
|
| C, Patient
3 |
|
|
| Mutation
position (GRCh37/hg 19) |
|
| SnpEff |
|
|
|
|
|
|
|
|
|
|
| Gene | Nucleotide
(genomic) | Amino acid
(protein) | Variant allele
frequencya | Mutation
type |
Annotation | Impact | COSMIC
ID | ClinVar allele
ID |
|
| CAMTA1 |
chr1:7723593A>G | p.K329R | 0.49 (34:69) | Substitution | Missense | Moderate | N/A | N/A |
| NBPF1 |
chr1:16901668T>C | p.K726E | 0.30 (51:168) | Substitution | Missense | Moderate | N/A | N/A |
| VWA5B1 |
chr1:20662938A>G | p.K634R | 0.65 (22:34) | Substitution | Missense | Moderate | N/A | N/A |
| ESRRA |
chr11:64083320T>C | p.L385P | 0.42 (13:31) | Substitution | Missense | Moderate | N/A | N/A |
CNAs in LEGH
We did not detect any copy number amplification or
deletion in case 1 or 2. In case 3, one focal amplification, with
an estimated copy number value of 2.9, was found between the range
p35.3 to p36.31 in chromosome 1. However, cancer-related genes from
the Cancer Gene Census in this range were all tumor suppressor
genes, such as ARID1A and ID3, which suggested that functional copy
number loss did not occur in this case. As a result, we did not
find any significant CNAs related to the progression of LEGH.
Discussion
LEGH was regarded as a putative MDA/GAS precursor in
the 2014 WHO classification, and an analysis of its genetic nature
was expected to clarify the neoplastic nature of LEGH. Although the
present study detected 13 variants in 3 LEGH cases, they did not
appear to be oncogenic. There has been one report of aberrant
COL4A3 expression being related to the pathogenesis of gastric
carcinoma (23); however, the
mutation of COL4A3 in the present results was ‘Benign’ in ClinVar.
Therefore, the neoplastic or pre-cancerous nature of LEGH has yet
to be elucidated in detail.
All three cases showed a low mutation rate,
non-specific mutational signatures, and lower malignant potential
missense or frame shift mutations. Of these mutations, KRPR, OR2M4,
and ZNF645 were already reported in COSMIC. KPRP is an epidermal
marker of differentiating keratinocytes, and its expression is
elevated in psoriatic lesions (24).
Regarding OR2M4, this olfactory receptor protein is one of the
members of a large family of G-protein-coupled receptors, the
signaling pathways of which are important for autism spectrum
disorder (25). Furthermore, ZNF645,
a human RING finger protein, possesses the domain of E3 ubiquitin
ligase activity (26). These
functions of all three COSMIC genes do not appear to be related to
carcinogenesis.
The strengths of the present study are the LCM
selection of samples to sequence, and comprehensive evaluation by
WES analysis, which is the first report in this pathology. Also,
limitations include the sample size, and the inherent shortcomings
of current databases regarding this pathology, calling for future
studies in the subject.
As previously reported, the GNAS, KRAS, and STK11
mutations were all exclusively detected in 11 (58%) LEGH cases
(12). We also detected activating
GNAS mutations in two ‘LEGH with atypia’ cases (14%) out of a total
of 14 cases (9 LEGH and 5 ‘LEGH with atypia’), and a STK11 mutation
was identified in only one MDA case (20%) (9,11).
Collectively, the present results imply that somatic driver
variants are not generated in the early stage of ‘pure’ or
‘non-atypical’ LEGH, but may occur in the late stage, i.e., LEGH
already associated with atypia or MDA. Differences between the
present results and previous findings may be attributed to their
LEGH specimens having already acquired atypia.
In conclusion, although the present results indicate
the metaplastic nature of LEGH, further research, including WES on
atypical LEGH/LEGH with MDA, is needed to clarify the neoplastic
characteristics of LEGH.
Acknowledgements
The authors would like to thank Ms Fumi Tsunoda and
Mr Eiji Uchida (Shinshu University School of Medicine) for their
technical assistance.
Funding
The present study was supported by JSPS KAKENHI
(grant no. 15K10712).
Availability of data and materials
The datasets generated and/or analyzed during the
present study are available from the corresponding author on
reasonable request.
Authors' contributions
KI designed the study, contributed to all
experiments, data analysis and manuscript drafting. TM contributed
to sample collection, data analysis, manuscript drafting and
revising. AT and HA participated in the study design and data
analysis of the whole-exome sequencing. RA, HT, SY and MO
contributed to the recruitment of patients, collection of the LEGH
tissues by laser microdissection, and DNA extraction used in the
whole-exome sequencing. SA contributed to the pathological
diagnosis and study design, and collected the LEGH tissues by laser
microdissection. TS was the supervisor of this study, contributed
to study design, data analysis, and drafting and revising the
manuscript. All authors were given the opportunity to revise the
manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of Shinshu University (approval no. 547) and patients
provided written consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Nucci MR, Clement PB and Young RH: Lobular
endocervical glandular hyperplasia, not otherwise specified: A
clinicopathologic analysis of thirteen cases of a distinctive
pseudoneoplastic lesion and comparison with fourteen cases of
adenoma malignum. Am J Surg Pathol. 23:886–891. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Mikami Y, Hata S, Fujiwara K, Imajo Y,
Kohno I and Manabe T: Florid endocervical glandular hyperplasia
with intestinal and pyloric gland metaplasia: Worrisome benign
mimic of ‘adenoma malignum’. Gynecol Oncol. 74:504–511. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Mikami Y, Hata S, Melamed J, Fujiwara K
and Manabe T: Lobular endocervical glandular hyperplasia is a
metaplastic process with a pyloric gland phenotype. Histopathology.
39:364–372. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kojima A, Mikami Y, Sudo T, Yamaguchi S,
Kusanagi Y, Ito M and Nishimura R: Gastric morphology and
immunophenotype predict poor outcome in mucinous adenocarcinoma of
the uterine cervix. Am J Surg Pathol. 31:664–672. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Silverberg SG and Hurt WG: Minimal
deviation adenocarcinoma (‘adenoma malignum’) of the cervix: A
reappraisal. Am J Obstet Gynecol. 121:971–975. 1975. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Karamurzin YS, Kiyokawa T, Parkash V,
Jotwani AR, Patel P, Pike MC, Soslow RA and Park KJ: Gastric-type
endocervical adenocarcinoma: An aggressive tumor with unusual
metastatic patterns and poor prognosis. Am J Surg Pathol.
39:1449–1457. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Nara M, Hashi A, Murata S, Kondo T,
Yuminamochi T, Nakazawa K, Katoh R and Hoshi K: Lobular
endocervical glandular hyperplasia as a presumed precursor of
cervical adenocarcinoma independent of human papillomavirus
infection. Gynecol Oncol. 106:289–298. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Mikami Y and McCluggage WG: Endocervical
glandular lesions exhibiting gastric differentiation: An emerging
spectrum of benign, premalignant, and malignant lesions. Adv Anat
Pathol. 20:227–237. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Takatsu A, Miyamoto T, Fuseya C, Suzuki A,
Kashima H, Horiuchi A, Ishii K and Shiozawa T: Clonality analysis
suggests that STK11 gene mutations are involved in progression of
lobular endocervical glandular hyperplasia (LEGH) to minimal
deviation adenocarcinoma (MDA). Virchows Arch. 462:645–651. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kawauchi S, Kusuda T, Liu XP, Suehiro Y,
Kaku T, Mikami Y, Takeshita M, Nakao M, Chochi Y and Sasaki K: Is
lobular endocervical glandular hyperplasia a cancerous precursor of
minimal deviation adenocarcinoma? A comparative molecular-genetic
and immunohistochemical study. Am J Surg Pathol. 32:1807–1815.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ando H, Miyamoto T, Kashima H, Takatsu A,
Ishii K, Fujinaga Y and Shiozawa T: Usefulness of a management
protocol for patients with cervical multicystic lesions: A
retrospective analysis of 94 cases and the significance of GNAS
mutation. J Obstet Gynaecol Res. 42:1588–1598. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Matsubara A, Sekine S, Ogawa R, Yoshida M,
Kasamatsu T, Tsuda H and Kanai Y: Lobular endocervical glandular
hyperplasia is a neoplastic entity with frequent activating GNAS
mutations. Am J Surg Pathol. 38:370–376. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Takatsu A, Shiozawa T, Miyamoto T,
Kurosawa K, Kashima H, Yamada T, Kaku T, Mikami Y, Kiyokawa T,
Tsuda H, et al: Preoperative differential diagnosis of minimal
deviation adenocarcinoma and lobular endocervical glandular
hyperplasia of the uterine cervix: A multicenter study of
clinicopathology and magnetic resonance imaging findings. Int J
Gynecol Cancer. 21:1287–1296. 2011.PubMed/NCBI
|
|
14
|
Hirasawa A, Akahane T, Tsuruta T,
Kobayashi Y, Masuda K, Banno K, Fujii T, Susumu N, Itsubo T,
Kameyama K, et al: Lobular endocervical glandular hyperplasia and
peritoneal pigmentation associated with peutz-jeghers syndrome due
to a germline mutation of STK11. Ann Oncol. 23:2990–2992. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ohya A, Yamanoi K, Shimojo H, Fujii C and
Nakayama J: Gastric gland mucin-specific O-glycan expression
decreases with tumor progression from precursor lesions to
pancreatic cancer. Cancer Sci. 108:1897–1902. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Li H and Durbin R: Fast and accurate short
read alignment with Burrows-Wheeler transform. Bioinformatics.
25:1754–1760. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Li H: A statistical framework for SNP
calling, mutation discovery, association mapping and population
genetical parameter estimation from sequencing data.
Bioinformatics. 27:2987–2993. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Koboldt DC, Zhang Q, Larson DE, Shen D,
McLellan MD, Lin L, Miller CA, Mardis ER, Ding L and Wilson RK:
VarScan 2: Somatic mutation and copy number alteration discovery in
cancer by exome sequencing. Genome Res. 22:568–576. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Koboldt DC, Larson DE and Wilson RK: Using
varscan 2 for germline variant calling and somatic mutation
detection. Curr Protoc Bioinformatics. 44:15.4.1–17. 2013.
|
|
20
|
Mandai M, Watanabe A, Kurimoto Y, Hirami
Y, Morinaga C, Daimon T, Fujihara M, Akimaru H, Sakai N, Shibata Y,
et al: Autologous induced stem-cell-derived retinal cells for
macular degeneration. N Engl J Med. 376:1038–1046. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Cingolani P, Platts A, Wang le L, Coon M,
Nguyen T, Wang L, Land SJ, Lu X and Ruden DM: A program for
annotating and predicting the effects of single nucleotide
polymorphisms, SnpEff: SNPs in the genome of drosophila
melanogaster strain w1118; iso-2; iso-3. Fly (Austin). 6:80–92.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Alexandrov LB, Nik-Zainal S, Wedge DC,
Aparicio SA, Behjati S, Biankin AV, Bignell GR, Bolli N, Borg A,
Børresen-Dale AL, et al: Signatures of mutational processes in
human cancer. Nature. 500:415–421. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Nie XC, Wang JP, Zhu W, Xu XY, Xing YN, Yu
M, Liu YP, Takano Y and Zheng HC: COL4A3 expression correlates with
pathogenesis, pathologic behaviors, and prognosis of gastric
carcinomas. Hum Pathol. 44:77–86. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lee WH, Jang S, Lee JS, Lee Y, Seo EY, You
KH, Lee SC, Nam KI, Kim JM, Kee SH, et al: Molecular cloning and
expression of human keratinocyte proline-rich protein (hKPRP), an
epidermal marker isolated from calcium-induced differentiating
keratinocytes. J Invest Dermatol. 125:995–1000. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kuo PH, Chuang LC, Su MH, Chen CH, Chen
CH, Wu JY, Yen CJ, Wu YY, Liu SK, Chou MC, et al: Genome-wide
association study for autism spectrum disorder in taiwanese han
population. PLoS One. 23:e01386952015. View Article : Google Scholar
|
|
26
|
Liu YQ, Bai G, Zhang H, Su D, Tao DC, Yang
Y, Ma YX and Zhang SZ: Human RING finger protein ZNF645 is a novel
testis-specific E3 ubiquitin ligase. Asian J Androl. 12:658–666.
2010. View Article : Google Scholar : PubMed/NCBI
|