|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2018. CA Cancer J Clin. 68:7–30. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Sato H, Kotake K, Sugihara K, Takahashi H,
Maeda K and Uyama I: Study Group for Peritoneal Metastasis from
Colorectal Cancer By the Japanese Society for Cancer of the Colon
and Rectum: Clinicopathological factors associated with recurrence
and prognosis after R0 resection for stage IV colorectal cancer
with peritoneal metastasis. Dig Surg. 33:382–391. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Corte H, Manceau G, Blons H and
Laurent-Puig P: MicroRNA and colorectal cancer. Dig Liver Dis.
44:195–200. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Gonzalez-Pons M and Cruz-Correa M:
Colorectal cancer biomarkers: Where are we now? Biomed Res Int.
2015:1490142015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Xie X, Zheng X, Han Z, Chen Y, Zheng Z,
Zheng B, He X, Wang Y, Kaplan DL, Li Y, et al: A biodegradable
stent with surface functionalization of combined-therapy drugs for
colorectal cancer. Adv Healthc Mater. 7:e18012132018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Valastyan S: Roles of microRNAs and other
non-coding RNAs in breast cancer metastasis. J Mammary Gland Biol
Neoplasia. 17:23–32. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhang N, Wang X, Huo Q, Sun M, Cai C, Liu
Z, Hu G and Yang Q: MicroRNA-30a suppresses breast tumor growth and
metastasis by targeting metadherin. Oncogene. 33:3119–3128. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Gregory PA, Bert AG, Paterson EL, Barry
SC, Tsykin A, Farshid G, Vadas MA, Khew-Goodall Y and Goodall GJ:
The miR-200 family and miR-205 regulate epithelial to mesenchymal
transition by targeting ZEB1 and SIP1. Nat Cell Biol. 10:593–601.
2008. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Seitz H: Redefining microRNA targets. Curr
Biol. 19:870–873. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Xia T, Chen S, Jiang Z, Shao Y, Jiang X,
Li P, Xiao B and Guo J: Long noncoding RNA FER1L4 suppresses cancer
cell growth by acting as a competing endogenous RNA and regulating
PTEN expression. Sci Rep. 5:134452015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yue B, Sun B, Liu C, Zhao S, Zhang D, Yu F
and Yan D: Long non-coding RNA Fer-1-like protein 4 suppresses
oncogenesis and exhibits prognostic value by associating with
miR-106a-5p in colon cancer. Cancer Sci. 106:1323–1332. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Salmena L, Poliseno L, Tay Y, Kats L and
Pandolfi PP: A ceRNA hypothesis: The Rosetta Stone of a hidden RNA
language? Cell. 146:353–358. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Karreth FA, Reschke M, Ruocco A, Ng C,
Chapuy B, Léopold V, Sjoberg M, Keane TM, Verma A, Ala U, et al:
The BRAF pseudogene functions as a competitive endogenous RNA and
induces lymphoma in vivo. Cell. 161:319–332. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Poliseno L, Salmena L, Jiangwen Z, Carver
B, Haveman WJ and Pandolfi PP: A coding-independent function of
gene and pseudogene mRNAs regulates tumour biology. Nature.
465:1033–1038. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Qu J, Li M, Zhong W and Hu C: Competing
endogenous RNA in cancer: A new pattern of gene expression
regulation. Int J Clin Exp Med. 8:17110–17116. 2015.PubMed/NCBI
|
|
16
|
Lo PK, Zhang Y, Wolfson B, Gernapudi R,
Yao Y, Duru N and Zhou Q: Dysregulation of the BRCA1/long
non-coding RNA NEAT1 signaling axis contributes to breast
tumorigenesis. Oncotarget. 7:65067–65089. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Liang WC, Fu WM, Wong CW, Wang Y, Wang WM,
Hu GX, Zhang L, Xiao LJ, Wan DC, Zhang JF and Waye MM: The lncRNA
H19 promotes epithelial to mesenchymal transition by functioning as
miRNA sponges in colorectal cancer. Oncotarget. 6:22513–22525.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li LJ, Zhao W, Tao SS, Leng RX, Fan YG,
Pan HF and Ye DQ: Competitive endogenous RNA network: Potential
implication for systemic lupus erythematosus. Expert Opin Ther
Targets. 21:639–648. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Xu J, Zhang R and Zhao J: The novel long
noncoding RNA TUSC7 inhibits proliferation by sponging MiR-211 in
colorectal cancer. Cell Physiol Biochem. 41:635–644. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
He F, Song Z, Chen H, Chen Z, Yang P, Li
W, Yang Z, Zhang T, Wang F, Wei J, et al: Long noncoding RNA
PVT1-214 promotes proliferation and invasion of colorectal cancer
by stabilizing Lin28 and interacting with miR-128. Oncogene.
38:164–179. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Bosma FT: WHO classification of tumours of
the digestive system. 2010.
|
|
22
|
Rice TW: 7th Edition AJCC/UICC Staging,
Esophagus and Esophagogastric Junction. 2017. View Article : Google Scholar
|
|
23
|
Diboun I, Wernisch L, Orengo CA and
Koltzenburg M: Microarray analysis after RNA amplification can
detect pronounced differences in gene expression using limma. BMC
Genomics. 7:2522006. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
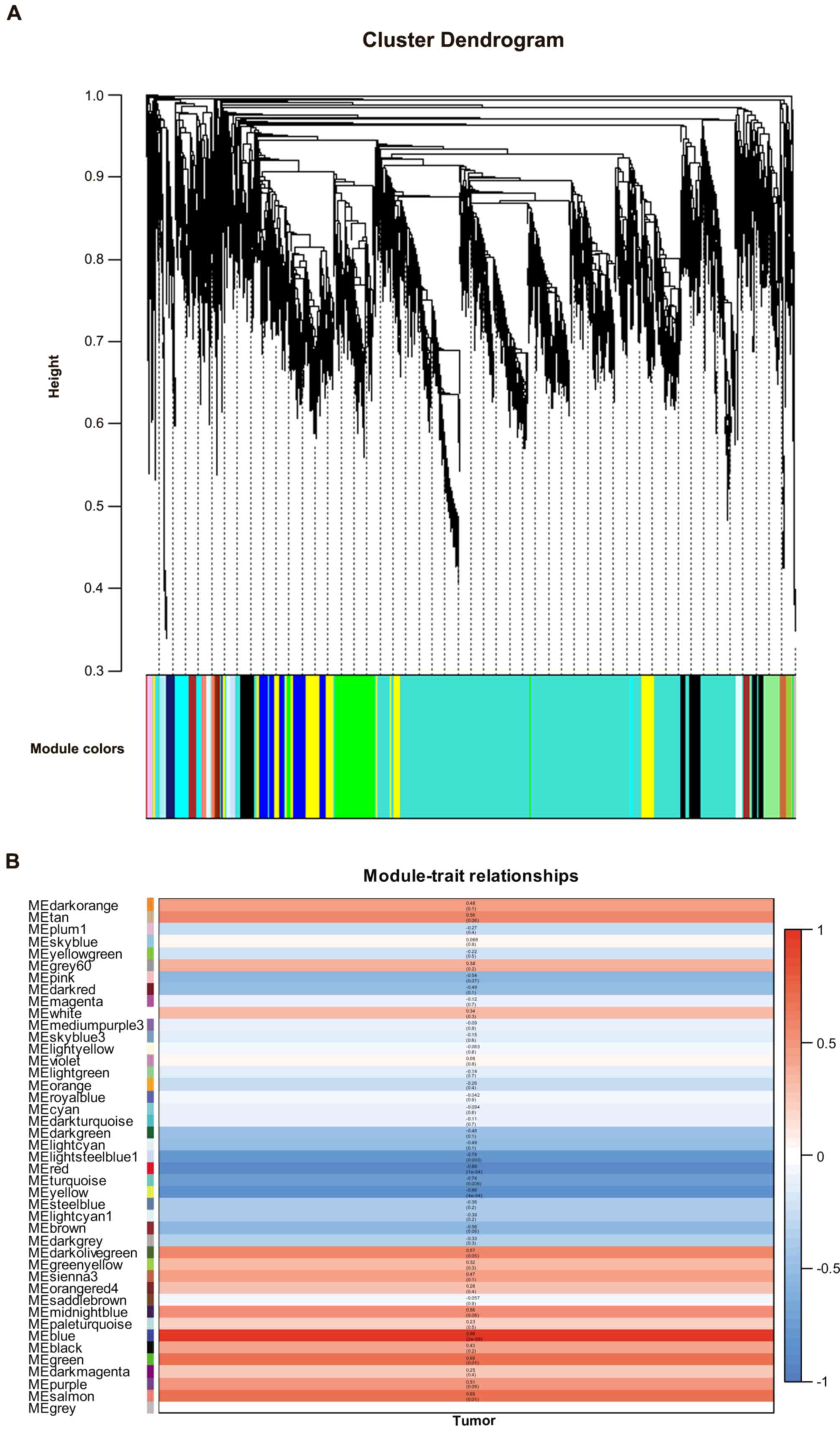
Zhang B and Horvath S: A general framework
for weighted gene co-expression network analysis. Stat Appl Genet
Mol Biol. 4:Article172005. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Gao B, Shao Q, Choudhry H, Marcus V, Dong
K, Ragoussis J and Gao ZH: Weighted gene co-expression network
analysis of colorectal cancer liver metastasis genome sequencing
data and screening of anti-metastasis drugs. Int J Oncol.
49:1108–1118. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Langfelder P and Horvath S: WGCNA: An R
package for weighted correlation network analysis. BMC
Bioinformatics. 9:5592008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ashburner M, Ball CA, Blake JA, Botstein
D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT,
et al: Gene ontology: Tool for the unification of biology. The Gene
Ontology Consortium. Nat Genet. 25:25–29. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wong N and Wang X: miRDB: An online
resource for microRNA target prediction and functional annotations.
Nucleic Acids Res. 43:(Database Issue). D146–D152. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lewis BP, Burge CB and Bartel DP:
Conserved seed pairing, often flanked by adenosines, indicates that
thousands of human genes are microRNA targets. Cell. 120:15–20.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Paraskevopoulou MD, Vlachos IS, Karagkouni
D, Georgakilas G, Kanellos I, Vergoulis T, Zagganas K, Tsanakas P,
Floros E, Dalamagas T and Hatzigeorgiou AG: DIANA-LncBase v2:
Indexing microRNA targets on non-coding transcripts. Nucleic Acids
Res. 44:D231–D238. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Tomczak K, Czerwińska P and Wiznerowicz M:
The cancer genome atlas (TCGA): An immeasurable source of
knowledge. Contemp Oncol (Pozn). 19:A68–A77. 2015.PubMed/NCBI
|
|
32
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Cao H, Mok A, Miskie B and Hegele RA:
Single-nucleotide polymorphisms of the proprotein convertase
subtilisin/kexin type 5 (PCSK5) gene. J Hum Genet. 46:730–732.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Bajikar SS, Wang CC, Borten MA, Pereira
EJ, Atkins KA and Janes KA: Tumor-auppressor inactivation of GDF11
occurs by precursor sequestration in triple-negative breast cancer.
Dev Cell. 43:418–435.e13. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zhang DY, Zou XJ, Cao CH, Zhang T, Lei L,
Qi XL, Liu L and Wu DH: Identification and functional
characterization of long non-coding RNA MIR22HG as a tumor
suppressor for hepatocellular carcinoma. Theranostics. 8:3751–3765.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Cui Z, An X, Li J, Liu Q and Liu W: LncRNA
MIR22HG negatively regulates miR-141-3p to enhance DAPK1 expression
and inhibits endometrial carcinoma cells proliferation. Biomed
Pharmacother. 104:223–228. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Althubit M, Rada M, Samuel J, Escorsa JM,
Najeeb H, Lee KG, Lam KP, Jones GD, Barlev NA and Macip S: BTK
modulates p53 activity to enhance apoptotic and senescent
responses. Cancer Res. 76:5405–5414. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Feng L, Xie Y, Zhang H and Wu Y:
Down-regulation of NDRG2 gene expression in human colorectal cancer
involves promoter methylation and microRNA-650. Biochem Biophys Res
Commun. 406:534–538. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Xie BH, He X, Hua RX, Zhang B, Tan GS,
Xiong SQ, Liu LS, Chen W, Yang JY, Wang XN and Li HP: Mir-765
promotes cell proliferation by downregulating INPP4B expression in
human hepatocellular carcinoma. Cancer Biomark. 16:405–413. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Zheng Z, Luan X, Zha J, Li Z, Wu L, Yan Y,
Wang H, Hou D, Huang L, Huang F, et al: TNF-α inhibits the
migration of oral squamous cancer cells mediated by
miR-765-EMP3-p66Shc axis. Cell Signal. 34:102–109. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Godoy P, Cadenas C, Hellwig B, Marchan R,
Stewart J, Reif R, Lohr M, Gehrmann M, Rahnenführer J, Schmidt M
and Hengstler JG: Interferon-inducible guanylate binding protein
(GBP2) is associated with better prognosis in breast cancer and
indicates an efficient T cell response. Breast Cancer. 21:491–499.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Wang X, Wang SS, Zhou L, Yu L and Zhang
LM: A network-pathway based module identification for predicting
the prognosis of ovarian cancer patients. J Ovarian Res. 9:732016.
View Article : Google Scholar : PubMed/NCBI
|