Introduction
Prostate cancer (PCa), which exhibits complicated
pathogenesis and treatment difficulties, is the most common
malignancy of the male reproductive system worldwide, accounting
for ~29,430 deaths in the USA in 2018 (1). PCa is a global public issue that
threatens human health and life, with increasing morbidity and
mortality rates each year (2).
Metastatic PCa is commonly treated with androgen deprivation
therapy; however, resistance can still develop quickly, which leads
to castration-resistant PCa (CRPC) (3). Docetaxel is widely used as the standard
first-line chemotherapy treatment for patients with CRPC (3). The majority of patients with CRPC who
receive docetaxel chemotherapy develop resistance to this
treatment. Additionally, with increasing treatment times and doses,
complications may occur (4).
Therefore, further investigation regarding the mechanism of
docetaxel-resistant PCa may improve the prognosis of patients with
PCa.
Although the mechanism underlying PCa drug
resistance has been extensively studied, its cause and pathogenesis
remain poorly understood. The current consensus is that the
mechanism of docetaxel-resistant PCa is associated with multiple
factors, including androgen receptor splice variant expression
(5), changes in the expression of
β-tubulin (6), multidrug resistance
induced by abnormal expression of the ATP binding cassette (ABC)
transporter family and abnormal expression of signaling pathway
factors (7), including the
PI3K/AKT/mTOR (8), Wnt (9) and NF-κB/interleukin (IL)6 pathways
(10). Additionally, abnormal
expression levels of EMT and stem-like cell markers have been
detected in PCa cells resistant to docetaxel, which lead to a
downregulation of cadherin 1 (CDH1) and an upregulation of
vimentin, zinc finger E-box binding homeobox 1 and the stem-like
cell marker CD44 molecule (CD44) (11). Taken together, these studies suggest
that docetaxel resistance in PCa occurs due to alterations in
numerous factors and/or genetic changes, rather than a single
factor. Although these basic and clinical studies have investigated
the resistance of docetaxel in PCa in the past few decades with the
aim of revealing the potential underlying mechanisms, the effect of
treatment remains unsatisfactory (5–11).
Therefore, understanding the precise molecular mechanisms
associated with the development of docetaxel resistance in PCa is
essential for the improvement of effective diagnosis and treatment
strategies. Microarray technologies, which have widely been used to
investigate large scale gene expression simultaneously, presents an
effective method to investigate the expression of tens of thousands
of genes and identify the mechanisms of numerous diseases,
particularly cancers. The integration and analysis of microarray
data provide valuable information for the study of docetaxel
resistance (12).
In this present study, the GSE33455 microarray
dataset (13), which includes two
docetaxel-sensitive and two docetaxel-resistant cell lines, was
downloaded from the Gene Expression Omnibus (GEO) database, and
differentially expressed genes (DEGs) between the two types of PCa
cell lines were identified. Functional enrichment analyses and
functional annotation were performed and a protein-protein
interaction (PPI) network was constructed and analyzed to screen
potential therapeutic targets for docetaxel resistance using the
Search Tool for the Retrieval of Interacting Genes/Proteins
(STRING) database and Cytoscape software. Reverse
transcription-quantitative PCR analysis was performed to
investigate the expression of the hub genes in docetaxel-sensitive
and docetaxel-resistant PCa cell lines and to further validate
their potential roles in docetaxel resistance in PCa.
Materials and methods
Data collection
The microarray expression profile dataset GSE33455
(13) was downloaded from the GEO
database (http://www.ncbi.nlm.nih.gov/geo), which is based on
the GPL570 [HG-U133_Plus_2] Affymetrix Human Genome U133 Plus 2.0
Array. The dataset contained 12 sets of data from four cell lines,
including PCa cell lines DU-145 (docetaxel-sensitive), PC-3
(docetaxel-sensitive), DU-145R (docetaxel-resistant) and PC-3R
(docetaxel-resistant).
Analysis of DEGs
The original expression data underwent background
correction and quartile data normalization and was converted into
gene expression measures using the robust multiarray average
(14) in the R Affy package (release
3.9; http://www.bioconductor.org/packages/release/bioc/html/affy.html).
The DEGs between docetaxel-sensitive and docetaxel-resistant
samples were analyzed using the limma package (15) in Bioconductor (http://www.bioconductor.org), and a DEG was considered
to be significant according to the following criteria:
|log[fold-change (FC)]|>2 and false discovery rate (FDR)
<0.05. Subsequently, a heatmap was constructed and the DEGs were
identified using the pheatmap package of R software (16).
Gene ontology (GO) and Kyoto
Encyclopedia of Genes and Genomes (KEGG) pathway analyses
GO is a tool used to annotate genes, collect and
analyze information according to cellular component (CC),
biological process (BP) and molecular function (MF) terms following
the criteria P<0.05 (17). KEGG
is an online database and analysis tool for integrating and
interpreting large molecular datasets (18). Database for Annotation, Visualization
and Integrated Discovery (DAVID; http://david.ncifcrf.gov) is a website composed of a
comprehensive biological database and analysis tools that assist
with the understanding of the biological meaning of gene lists
(19). In the present study, DAVID
and Metascape (https://metascape.org) were used for
GO and KEGG pathway enrichment analyses of the DEGs. FDR<0.05
was considered to indicate a statistically significant result.
Analysis of the PPI network
The STRING database is an online biological database
that collects comprehensive information on proteins to evaluate the
PPI information (20). In the
current study, significant gene pairs of the PPI network were
visually represented using Cytoscape; a combined score >0.4 was
considered as significant and the strength of an interaction was
modelled by the number of lines (21). Cytoscape is a bioinformatics software
used to perform computational analysis of cellular networks and
merge experimental omics datasets together (22). The hub genes were selected using the
CytoHubba network analyzer plug-in (23). In addition, analysis of the most
important module was performed using the MCODE plug-in for
Cytoscape. Subsequently, Metascape (http://metascape.org/gp/index.html) software was used
for functional enrichment analysis of the module genes.
Analysis of hub gene expression
levels
RNA-sequencing data of 497 PCa and 52 adjacent
normal tissue samples were downloaded from The Cancer Genome Atlas
(TCGA) database (https://cancergenome.nih.gov/) to examine the
expression levels of the hub genes. Gene Expression Profiling
Interactive Analysis (GEPIA; http://gepia2.cancer-pku.cn/#index, accessed on May
4th, 2019) is a newly developed interactive web server for
analyzing the RNA-sequencing expression data of 9,736 tumor samples
and 8,587 normal samples from TCGA and GTEx projects using a
standard processing pipeline. GEPIA provides customizable
functions, including tumor/normal differential expression analysis,
profiling according to cancer types or pathological stages, patient
survival analysis, similar gene detection and correlation analysis
(24). The present study used GEPIA
to analyze the associations between the identified hub genes. GEPIA
uses the non-log scale for calculation and uses the log-scale axis
for visualization. This function of GEPIA performs pairwise gene
expression correlation analysis for given sets of TCGA and/or GTEx
expression data using a variety of methods, including Pearson,
Spearman and Kendall analyses.
Cell lines
The human PCa cell lines DU-145 and PC-3, purchased
from the Type Culture Collection of the Chinese Academy of Sciences
were maintained in MEM (Gibco; Thermo Fisher Scientific, Inc.) or
F12K (Gibco; Thermo Fisher Scientific, Inc.), respectively. The
media were supplemented with 10% fetal calf serum (Gibco; Thermo
Fisher Scientific, Inc.). The DU-145R and PC-3R cell lines were
developed by docetaxel dose escalation, as previously described
(25). Cells were cultured at 37°C
in a 5% CO2 incubator.
RNA extraction and reverse
transcription-quantitative PCR
Total RNA was extracted from cells using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.) and reverse transcribed into cDNA templates using
PrimeScript® RT Reagent kit (Takara Biotechnology Co.,
Ltd.) according to the manufacturer's instructions. The mRNA
expression levels were evaluated using SYBR® Green
Master Mix (Takara Biotechnology Co., Ltd.) and a CFX96 PCR machine
(Bio-Rad Laboratories, Inc.). The thermocycling conditions were as
follows: 10 min at 95°C, 40 cycles of 15 sec at 95°C and 60 sec at
60°C, followed by 1 h at 4°C. ß-actin was used as an internal
reference for normalization. Compared with the control, the fold
change in mRNA levels was calculated using the 2−ΔΔCq
method (26). The specific PCR
primers for the hub genes and β-actin as the housekeeping gene were
designed with Primer Express version 2.0 (Applied Biosystems,
Carlsbad, CA, USA) and are presented in the Table SI.
Statistical analysis
The PCR data were presented as the mean ± standard
deviation and analyzed using SPSS 19.0 statistical software (IBM
Corp.). Differences between the two types of PCa cell lines were
analyzed using Student's t-test. P<0.05 was considered to
indicate a statistically significant difference.
Results
Identification of DEGs
The gene expression dataset GSE33455 was downloaded
from the GEO database and included data for two docetaxel-sensitive
PCa cell lines and two docetaxel-resistant PCa cell lines.
Following differential expression analysis, 756 DEGs were
identified between docetaxel-sensitive and docetaxel-resistant PCa,
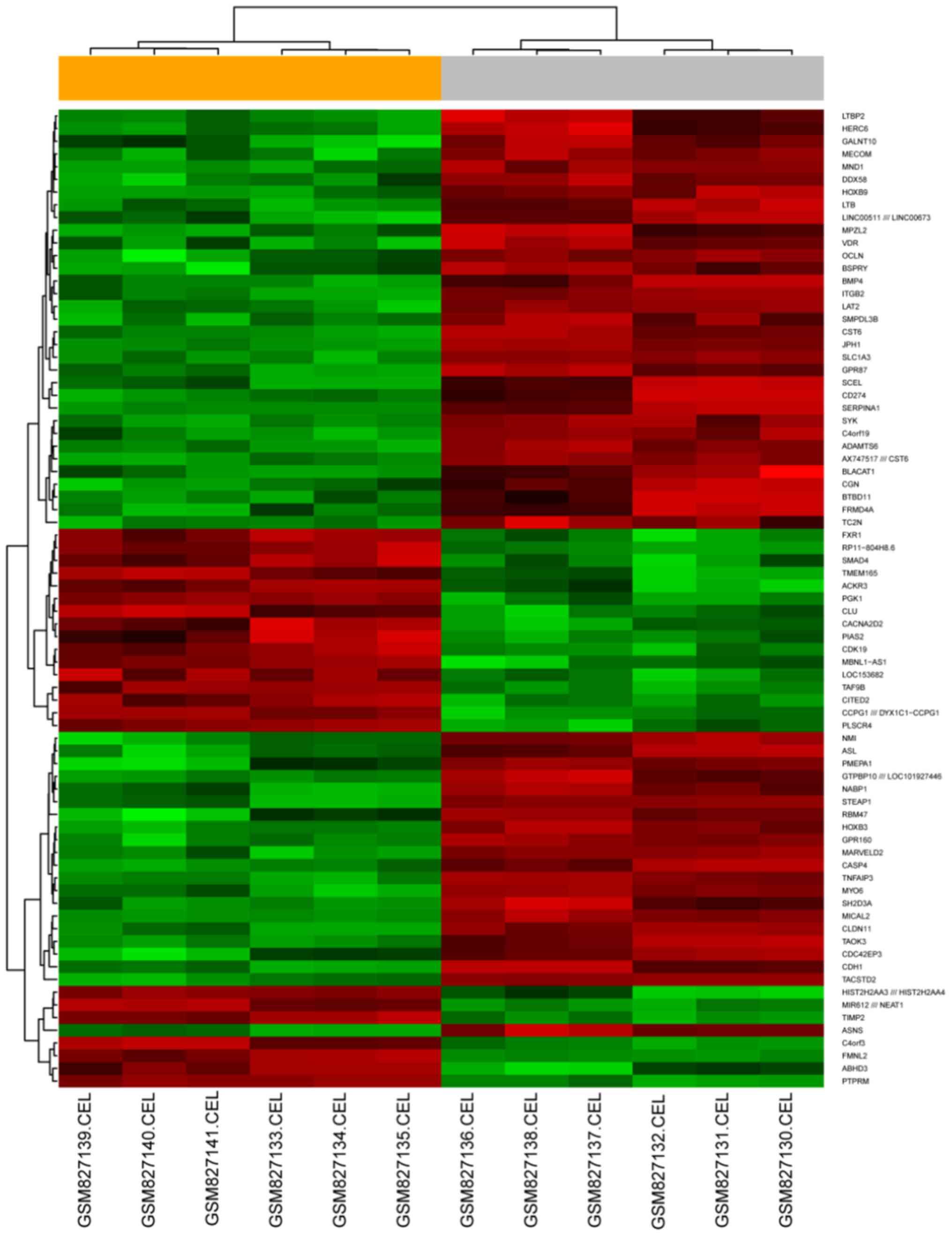
including 247 upregulated and 509 downregulated genes. A heatmap
was constructed using the top 100 DEGs based on their FC (Fig. 1).
GO and pathway enrichment
analyses
The biological functions of all identified DEGs were
evaluated by GO and pathway enrichment analyses, which were
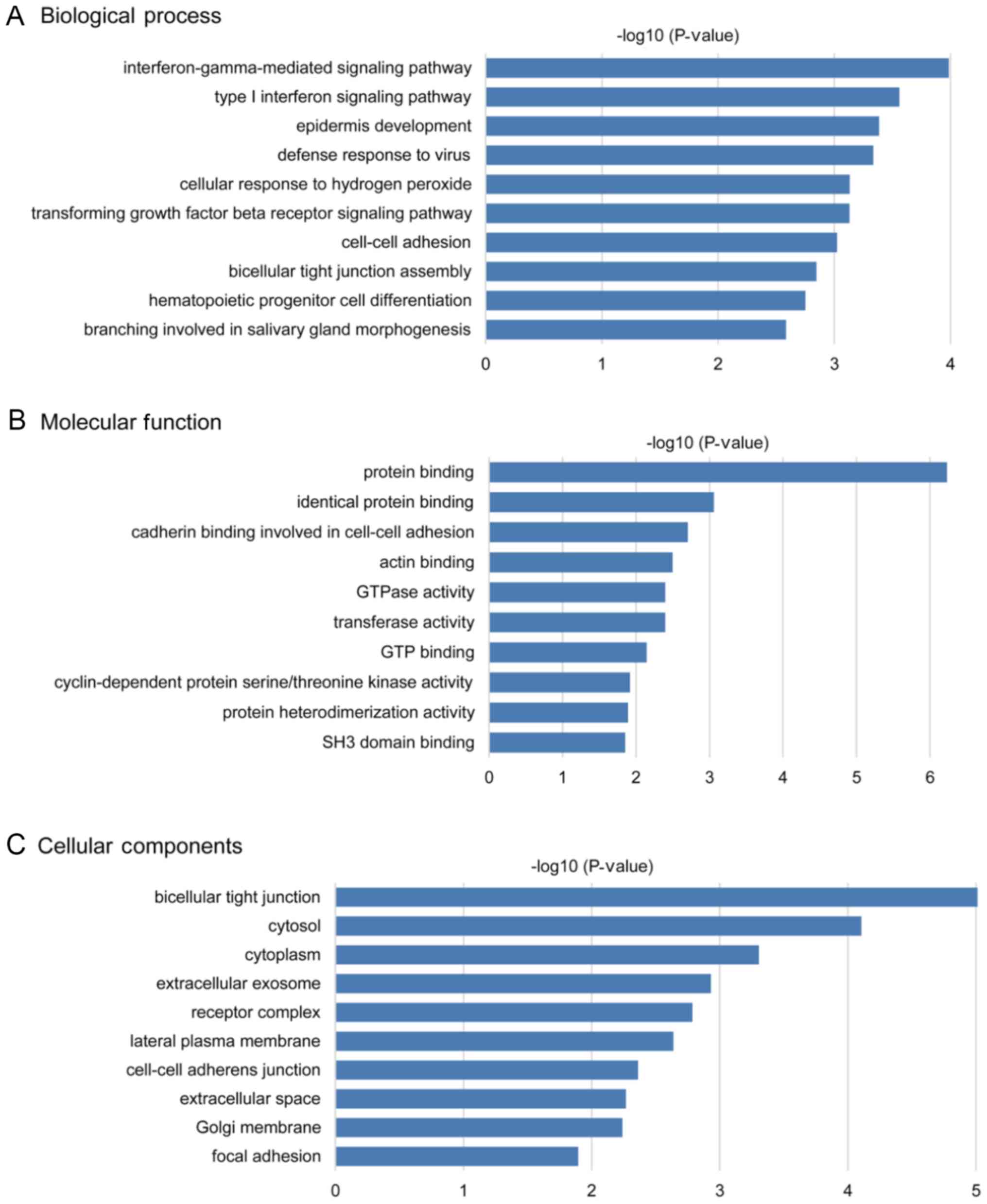
performed using DAVID and Metascape. In the enrichment analysis of
BPs, the DEGs were significantly enriched in ‘type I interferon
signaling pathway’, ‘interferon-gamma-mediated signaling pathway’,
‘epidermis development’, ‘defense response to virus’ and
‘transforming growth factor beta receptor signaling pathway’
(Fig. 2A). In the MF analysis, the
DEGs were significantly enriched in ‘protein binding’, ‘identical
protein binding’, ‘cadherin binding involved in cell-cell adhesion’
and ‘actin binding’ (Fig. 2B). In
the CC analysis, the DEGs were predominantly enriched in
‘bicellular tight junction’, ‘cytosol’, ‘extracellular exosome’ and
‘receptor complex’ (Fig. 2C). The
top five GO terms of the DEGs are presented in Tables SII and SIII.
KEGG pathway analysis indicated that the DEGs were
significantly enriched in ‘metabolic pathways’, ‘pathways in
cancer’, ‘PI3K-Akt signaling pathway’ and other significant
signaling pathways with the highest gene numbers (P<0.05;
Table SIV; Fig. 3). The majority of these pathways are
closely associated with the occurrence and progression of
tumors.
PPI network analysis and module
selection
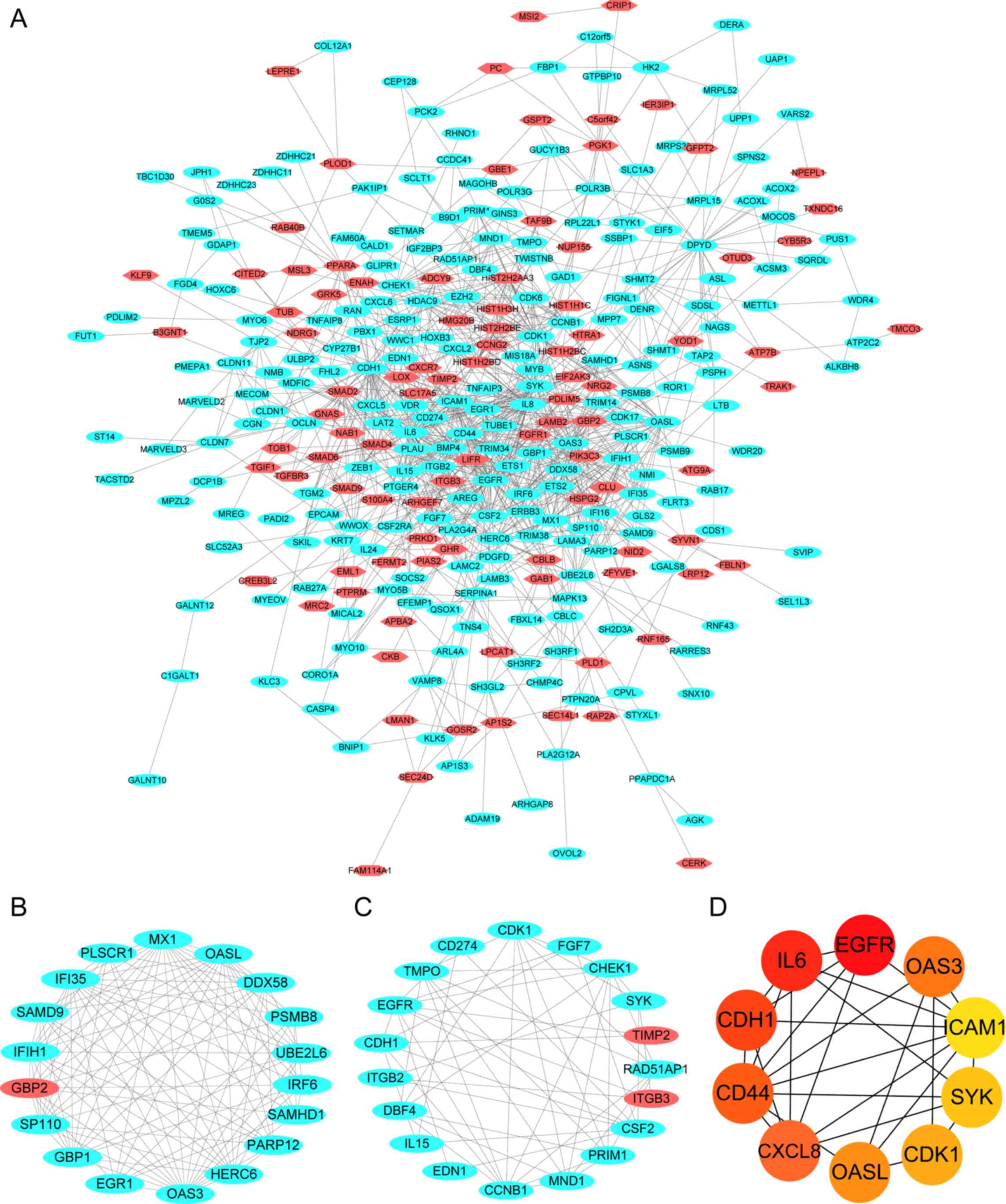
The PPI network of the DEGs was constructed using
the STRING online database and consisted of 324 nodes and 1,087
edges (Fig. 4A). The results were
then transferred to Cytoscape software to analyze the interactions
between the candidate DEGs in PCa. The Cytoscape cytoHubba Network
Analyzer plug-in selected ten hub genes from the PPI network by
identifying the top ten nodes ranked by degree. To investigate the
significant modules in this PPI network, two significant modules
were obtained by Cytotype MCODE, with enrichment scores of 11.053
and 5.3, respectively, and all of the MCODE scores for the two
significant modules were >5. The results of functional
enrichment analysis indicated that module one consisted of 18 nodes
and 105 edges (Fig. 4B), which were
predominantly enriched in the ‘defense response to virus’,
‘interferon-gamma-mediated signaling pathway’ and ‘negative
regulators of DDX58/IFIH1 signaling’ (Table I). Module two consisted of 19 nodes
and 53 edges (Fig. 4C), which were
predominantly associated with the ‘regulation of peptidyl-tyrosine
phosphorylation’, ‘DNA replication’, ‘cell-cell adhesion’ and the
‘Rap1 signaling pathway’ (Table
II).
 | Table I.GO and KEGG pathway enrichment
analysis of module 1. |
Table I.
GO and KEGG pathway enrichment
analysis of module 1.
| Term | Description |
-log10(P-value) | Genes |
|---|
| R-HSA-913531 | Interferon
signaling | 20.627 | EGR1, GBP1, GBP2,
IFI35, IRF6, MX1, OAS3, PSMB8, OASL, UBE2L6, DDX58, SAMHD1 |
| GO:0051607 | Defense response to
virus | 11.488 | GBP1, MX1, OAS3,
PLSCR1, OASL, DDX58, SAMHD1, IFIH1, SP110, PSMB8 |
| GO:0060333 |
Interferon-gamma-mediated signaling
pathway | 8.152 | GBP1, GBP2, IRF6,
OAS3, OASL |
| R-HSA-936440 | Negative regulators
of DDX58/IFIH1 signaling | 5.599 | UBE2L6, DDX58,
IFIH1, MX1, OAS3, PLSCR1, PSMB8, SAMHD1, GBP1, EGR1 |
| R-HSA-1169408 | ISG15 antiviral
mechanism | 4.627 | MX1, UBE2L6,
DDX58 |
| R-HSA-983168 | Antigen processing:
Ubiquitination & Proteasome degradation | 2.744 | PSMB8, UBE2L6,
HERC6 |
| GO:0002429 | Immune
response-activating cell surface receptor signaling pathway | 2.362 | GBP1, PLSCR1,
PSMB8 |
 | Table II.GO and KEGG pathway enrichment
analysis of module 2. |
Table II.
GO and KEGG pathway enrichment
analysis of module 2.
| Term | Description |
-log10(P-value) | Genes |
|---|
| GO:0050730 | Regulation of
peptidyl-tyrosine phosphorylation | 9.269 | CSF2, EGFR, FGF7,
IL15, ITGB2, ITGB3, SYK, CD274, CDH1, EDN1, TIMP2 |
| GO:0042554 | Superoxide anion
generation | 8.073 | EDN1, EGFR, ITGB2,
SYK, CCNB1, CDK1, FGF7, ITGB3, CD274, CSF2, TIMP2, IL15 |
| GO:0006260 | DNA
replication | 6.949 | CDK1, CHEK1, CSF2,
EGFR, PRIM1, DBF4, CCNB1, TMPO, MND1, EDN1, IL15, TIMP2, FGF7, SYK,
ITGB2, CDH1 |
| GO:0007568 | Ageing | 6.941 | CDK1, CHEK1, EDN1,
IL15, ITGB2, TIMP2 |
| hsa04015 | Rap1 signaling
pathway | 6.155 | CDH1, EGFR, FGF7,
ITGB2, ITGB3, IL15, SYK, CDK1, CSF2, EDN1, CD274, PRIM1, CHEK1 |
| GO:0098609 | Cell-cell
adhesion | 5.708 | CDH1, EGFR, IL15,
ITGB2, ITGB3, SYK, CD274, TIMP2 |
| GO:0010035 | Response to
inorganic substance | 5.550 | CCNB1, CDK1, CDH1,
CSF2, EDN1, EGFR, ITGB3, FGF7, SYK, CD274, IL15, CHEK1, ITGB2 |
| GO:0051052 | Regulation of DNA
metabolic process | 4.842 | CDK1, CHEK1, CSF2,
EGFR, RAD51AP1, MND1, EDN1, SYK |
| hsa05166 | HTLV–I
infection | 4.259 | CHEK1, CSF2, IL15,
ITGB2, EDN1, FGF7, TIMP2 |
| hsa05203 | Viral
carcinogenesis | 3.215 | CDK1, CHEK1,
SYK |
The top ten DEGs with high degrees of connectivity
were considered as the hub genes of resistant PCa, and a degree
>28 was identified as the central node degree used to determine
hub genes as the degree of the tenth gene was 29. These hub genes,
ranked by node degree, including intercellular adhesion molecule 1
(ICAM1), Spleen-associated tyrosine kinase SYK, Cyclin-dependent
kinase 1 (CDK1), 2′-5′-oligoadenylate synthetase-like (OASL),
2′-5′-oligoadenylate synthetase 3 (OAS3), CXC motif chemokine
ligand 8 (CXCL8), CD44, CDH1, epidermal growth factor receptor
(EGFR) and IL6, were identified as the key candidate genes, which
may serve crucial roles in cancer drug resistance (Fig. 4D). The degrees and functions of the
top ten hub genes in the PPI network are presented in Table III, and these genes/proteins may be
associated with the docetaxel resistance of PCa.
 | Table III.Degrees and functions of the top 10
hub genes in the protein-protein interaction network. |
Table III.
Degrees and functions of the top 10
hub genes in the protein-protein interaction network.
| Gene | Full name | Degree | Function |
|---|
| ICAM1 | Intercellular
Adhesion Molecule 1 | 29 | Overexpressed in
various cancers and may be involved in the progression of
cancer |
| SYK | Spleen-associated
tyrosine kinase | 30 | Immune cell
signaling pathways, including proliferation, differentiation and
phagocytosis |
| CDK1 | Cyclin-dependent
kinase 1 | 34 | Regulates cell
viability, cell cycle progression, apoptosis and DNA damage repair
of tumor cells |
| OASL |
2′-5′-oligoadenylate synthetase-like | 35 | Negative role in
the anti-tumor immune response |
| OAS3 |
2′-5′-oligoadenylate synthetase 3 | 36 | Cellular innate
antiviral response, apoptosis, cell growth, differentiation and
gene regulation |
| CXCL8 | CXC motif chemokine
ligand 8 | 38 | Overexpressed in
multiple cancer types; promotes the acquisition of mesenchymal
features, stemness, resistance to therapy |
| CD44 | CD44 molecule | 38 | Cell-surface
glycoprotein involved in cell-cell interactions, cell adhesion and
migration |
| CDH1 | Cadherin 1 | 43 | Loss of CDH1 is
associated with migration, invasion and poor prognosis of multiple
cancers |
| IL6 | Interleukin 6 | 57 | Immune response to
cancer and inflammatory diseases |
| EGFR | Epidermal growth
factor receptor | 59 | Promotes the
proliferation of multiple cancer types |
Hub gene validation using TCGA
database
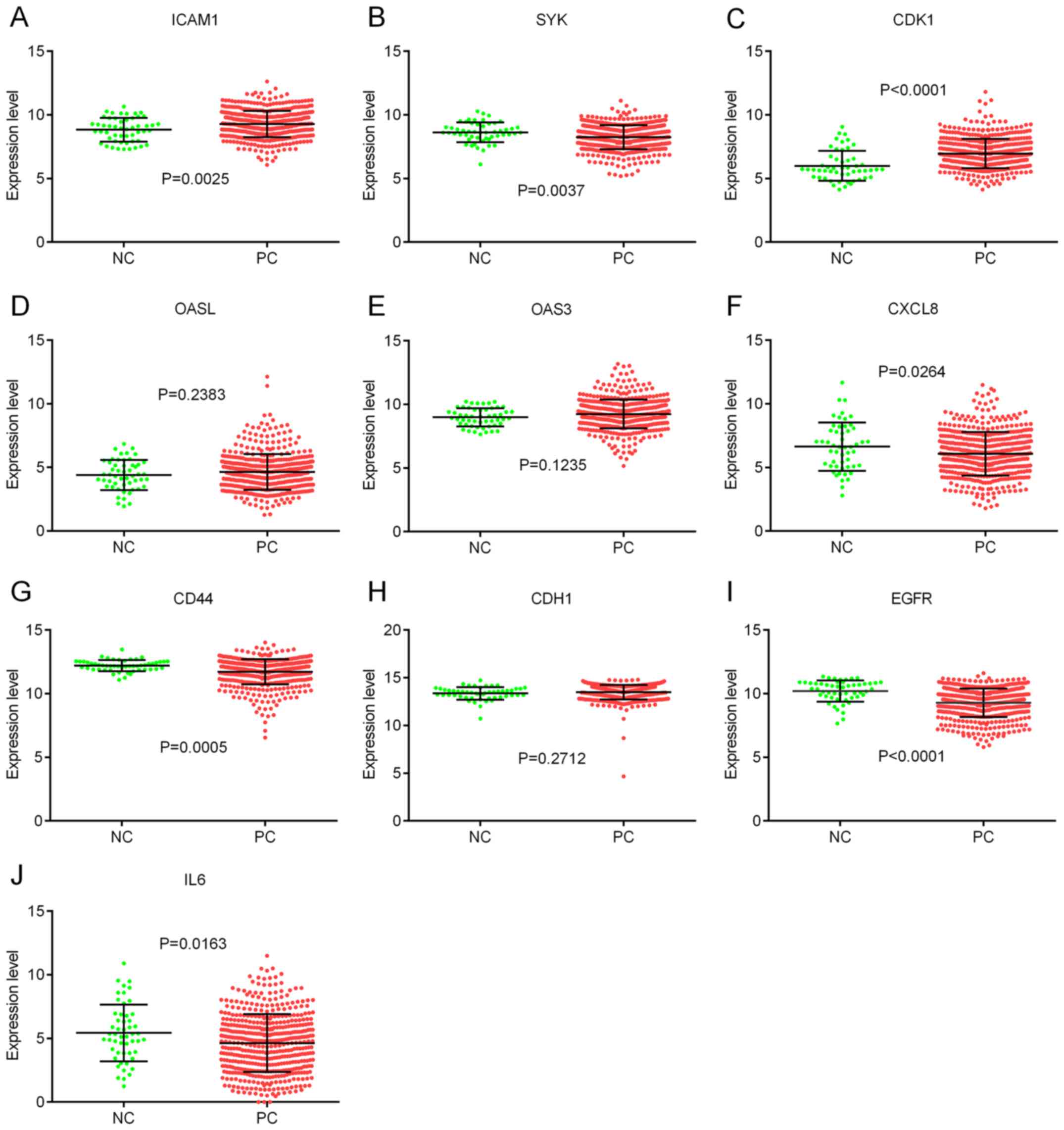
To validate the hub genes, the expression levels of
the hub genes were analyzed using data from TCGA database. The
results indicated that the expression levels of ICAM1 and CDK1 were
significantly higher in PCa tissues compared with normal tissues
(Fig. 5A and C), whereas the
expression levels of SYK, CXCL8, CD44, EGFR and IL6 were lower in
PCa tissues compared with normal tissues (Fig. 5B, F, G, I and J). However, there was
no significant difference in the expression levels of OASL, OAS3
and CDH1 between PCa tissues and normal tissues (Fig. 5D, E and H).
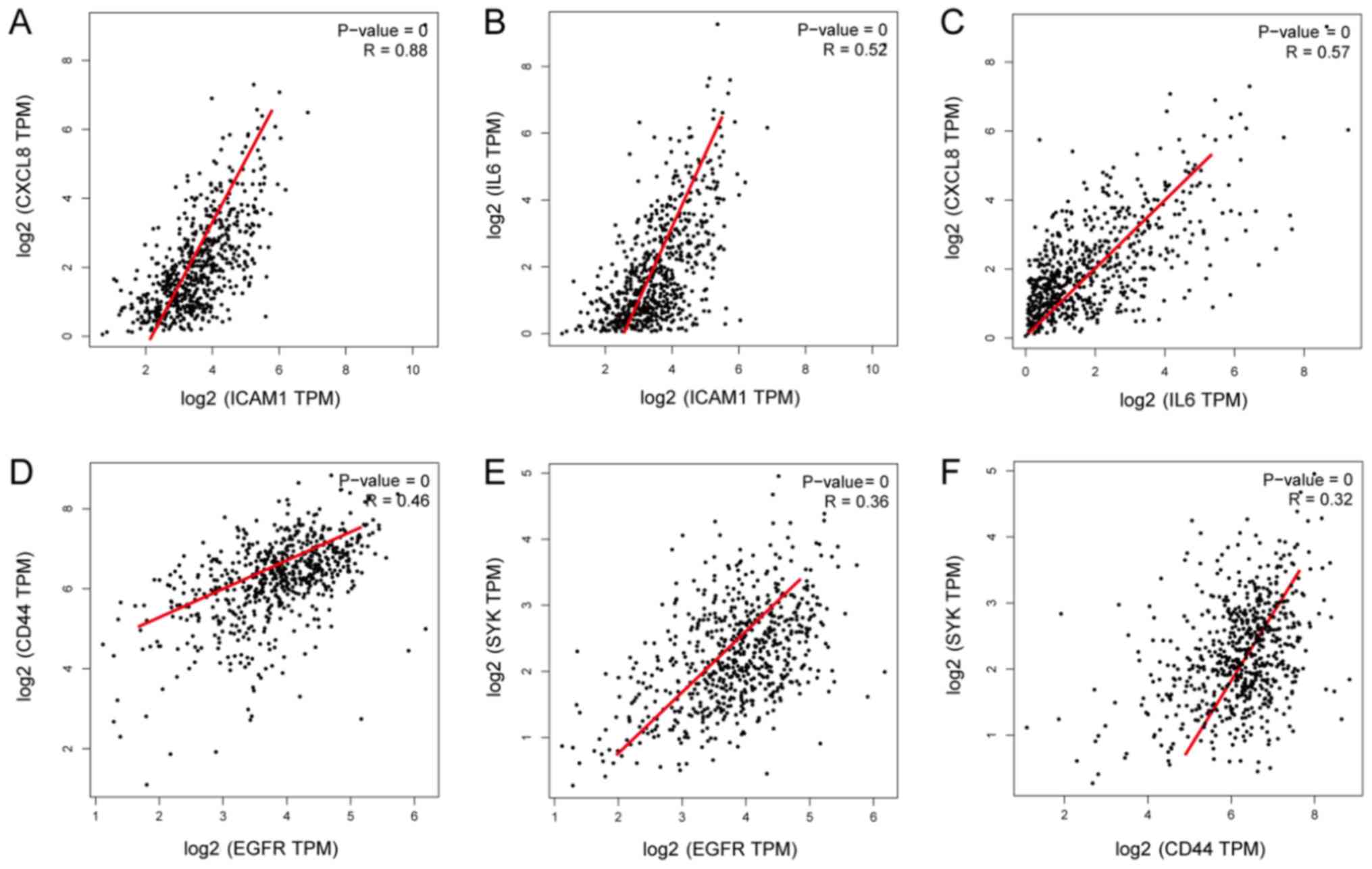
Correlations among the expression of
the ten hub genes
As significant differences were identified in the
expression levels of EGFR, SYK, ICAM1 and CD44 in PCa compared with
adjacent normal tissues, the GEPIA database was used in the present
study to analyze the correlations among these genes. The results
revealed that ICAM1 may be associated with IL6 and CXCL8. ICAM1 and
CXCL8 were positively correlated (P<0.001; R=0.88), and ICAM1
and IL6 were positively correlated (P<0.001; R=0.52) (Fig. 6A and B). EGFR and CD44 were
positively correlated (P<0.001; R=0.46), and EGFR and SYK were
positively correlated (P<0.001; R=0.36) (Fig. 6D and E).
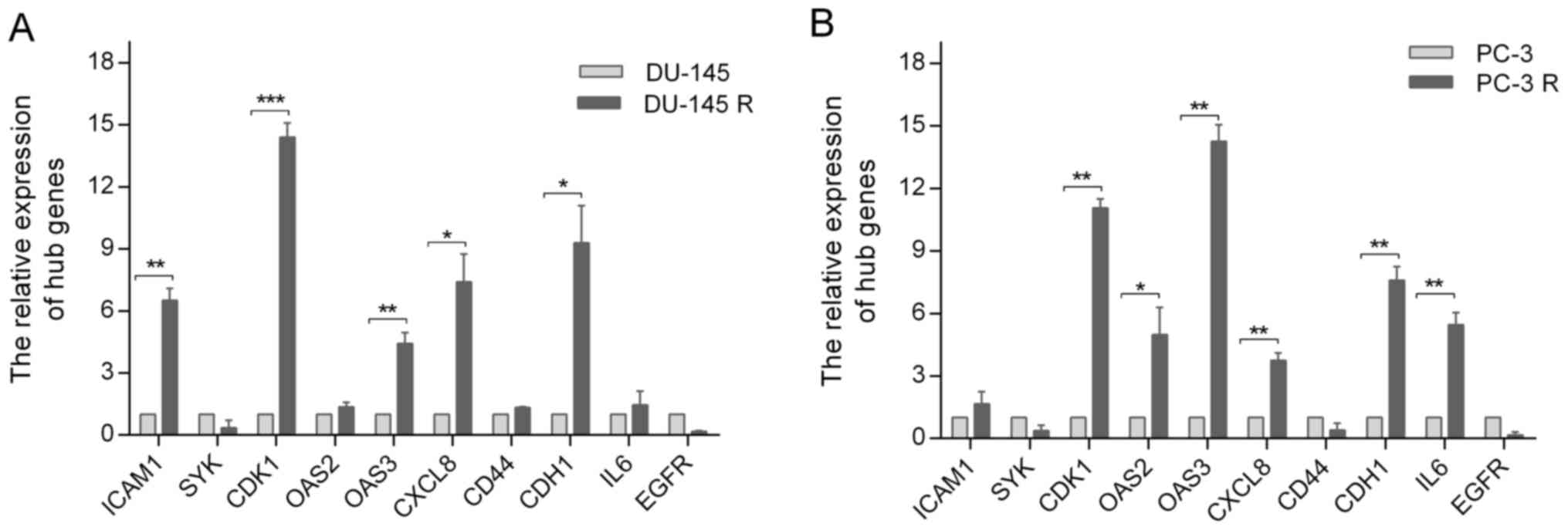
Expression of the hub genes in
docetaxel-sensitive and docetaxel-resistant cell lines
To further validate the potential role of the hub
genes in docetaxel-resistant PCa, their expressions in
docetaxel-sensitive and docetaxel-resistant PCa cell lines were
investigated by RT-qPCR, which demonstrated that the relative
expression levels of ICAM1, CDK1, OAS3, CXCL8 and CDH1 in DU-145
cells were significantly lower compared with those in DU-145R cells
(Fig. 7A), whereas CDK1, OAS2, OAS3,
CXCL8, CDH1 and IL6 expression levels in PC-3 cells were lower
compared with those in PC-3R cells (Fig.
7B).
Discussion
In the present study, 756 DEGs were identified
between two docetaxel-sensitive prostate cell lines and two
docetaxel-resistant prostate cell lines by analysis of the GSE33455
dataset. The DEGs included 247 upregulated genes and 509
downregulated genes. The interactions among these DEGs were
investigated with KEGG and GO enrichment analyses; the DEGs were
predominantly enriched in the ‘interferon-gamma-mediated signaling
pathway’ in the BP category. In addition, other notable enriched
terms included ‘bicellular tight junction assembly’, ‘cell-cell
adhesion’ and the ‘transforming growth factor beta receptor
signaling pathway’, all of which are closely associated with tumor
metastasis and drug resistance. In the MF category, the DEGs were
associated with ‘protein binding’, ‘identical protein binding’,
‘cadherin binding involved in cell-cell adhesion’, ‘actin binding’
and ‘GTPase activity’; these data suggested that the DEGs may
affect the binding of proteins, cadherin, actin and GTPase
activity. In the CC category, the DEGs were mainly enriched in the
‘bicellular tight junction’, ‘cytosol’, ‘extracellular exosome’ and
‘receptor complex’; these data indicated that the DEGs were mainly
involved in substance transfer and transport in the cytoplasm of
cells.
According to the KEGG analysis, the DEGs were mainly
enriched in ‘pathways in cancer’, ‘metabolic pathways’, the
‘PI3K-Akt signaling pathway’, the ‘Jak-STAT signaling pathway’,
‘proteoglycans in cancer’ and the ‘NF-κB signaling pathway’. Chen
et al (8) have demonstrated
that upregulated inositol polyphosphate-4-phosphatase type II B
induces apoptosis and enhances sensitivity to docetaxel via the
PI3K/Akt signaling pathway in PC3-DR and DU-145-DR cells. NF-κB
signaling serves a crucial role in regulating invasion, metastasis,
proliferation, angiogenesis and drug resistance in tumor cells. A
previous study reported that the NF-κB pathway may be a potential
target for combination therapy during the advanced stages of
thyroid cancer (27). In addition,
NF-κB, pAkt, macrophage inhibitory cytokine-1 and EGFR, which are
significantly overexpressed in PCa samples, induce
caspase-dependent apoptosis and increase the sensitivity of
cytotoxic effects caused by docetaxel in chemo-resistant SP
WPE1-NB26 cells (28). This
indicates the crucial roles of the PI3K/Akt and NF-κB signaling
pathways in PCa resistance to docetaxel.
In the present study, a PPI network was constructed
using the DEGs with 324 nodes and 1,087 edges, and two notable
modules termed module one and module two were obtained. Module one
comprised 18 genes, including interferon induced protein 35,
phospholipid scramblase 1, guanylate binding protein 1 and
ubiquitin conjugating enzyme E2 L6, which were enriched in ‘defense
response to virus’, ‘interferon-gamma-mediated signaling pathway’
and ‘negative regulators of DDX58/IFIH1 signaling’. These
enrichments result in the interaction of multiple signal
transduction pathways in effector cells and the expression of
associated stimulatory genes, which have many biological functions,
including antivirus, antitumor and immune regulatory functions
(29). Module two consisted of 19
genes, including CDK1, cyclin B1, meiotic nuclear divisions 1, DNA
primase subunit 1, checkpoint kinase 1, colony stimulating factor 2
and CDH1, which were mainly associated with ‘regulation of
peptidyl-tyrosine phosphorylation’, ‘DNA replication’, ‘cell-cell
adhesion’ and the ‘Rap1 signaling pathway’. It has been reported
that these enrichment results are mainly involved in the processes
of tumor adhesion, invasion, metastasis and drug resistance
(30). The top ten DEGs with a
degree of connectivity >28 were considered as the hub genes,
including ICAM1, SYK, CDK1, OASL, OAS3, CXCL8, CD44, CDH1, EGFR and
IL6, which may serve critical roles in docetaxel-resistance in PCa.
Analysis using data from TCGA database demonstrated that the
expression levels of ICAM1 and CDK1 were significantly higher in
PCa tissues compared with normal tissues, whereas the expression
levels of SYK, CXCL8, CD44, EGFR and IL6 were significantly lower.
Tumor cells promote growth by avoiding or preventing the immune
response (31). Therefore, it can be
hypothesized that ICAM-1, a co-stimulatory molecule, may promote
tumor survival by signaling to natural killer cells and cytotoxic T
lymphocytes (32). Several studies
have demonstrated that CXCL8 is associated with the migration and
proliferation of various types of cancer cells, including PCa cells
(33,34). CXCL8 promotes the proliferation and
progression of cancer cells and increases the resistance to
cytotoxic drugs in androgen-independent PCa by upregulating the
expression of survival factors, which promotes the growth and
development of tumors (35).
Correlation analysis of the hub genes identified in
the present study revealed that the expression levels of ICAM1 and
CXCL8 were positively correlated (P<0.01, R=0.88), suggesting
that ICAM1 may serve an important role in docetaxel-resistance in
PCa. Ghotra et al (36)
demonstrated that the protein tyrosine kinase SYK may be a new
therapeutic target for advanced PCa as it stimulates the growth and
migration of PCa cells. CDK1 is essential for cell viability as it
serves important roles in numerous biological events, including
activating checkpoint proteins, repairing DNA damage and regulating
the cell cycle (37). A previous
study has indicated that abnormal activation of CDK1 promotes the
proliferation and survival of PCa cells by phosphorylating and
suppressing FOXO1 (38). Previous
studies have demonstrated that 2′-5′-oligoadenylate synthetase
(OAS) is induced by interferons when infected by viruses, as
OAS-like (OASL) has a regulatory function in antiviral innate
immunity via interferon signaling; the genetic variation of OAS may
increase the risk of chronic lymphocytic leukemia (39,40).
CD44 performs versatile functions as a cell membrane receptor,
including cell adhesion, invasion and metastasis in tumor cells
(41). CD44 has also been identified
on cancer-initiating cells and stem cells (42). CD44 performs a tumor-promoter
function by mediating the invasion, proliferation and migration of
PCa PC-3 cells; inhibition of CD44 decreases the glucose
consumption and increase the sensitivity to docetaxel of PC-3
cells. This suggests that CD44 exhibits a regulatory effect on the
progression and drug resistance of PCa cells (43). Furthermore, Jiang et al
(44) reported that a mutation of
the CDH1 gene is associated with metastasis and invasion in
numerous types of cancer, as it changes the transcriptional
activity of epithelial cells. Epigenetic loss of CDH1 is associated
with multidrug resistance in human hepatocellular carcinoma cells
(44). Additionally, EGFR has been
demonstrated to be a driver of tumorigenesis by promoting the
proliferation and development of a number of different cancer types
(45). Hour et al (46) have demonstrated a positive
correlation between EGFR expression and resistance to docetaxel,
which was mediated by EGFR via the Akt/ABCB1 pathway in PCa cells,
and an increased susceptibility to docetaxel-based treatment while
dealt with EGFR inhibition. These findings indicate that EGFR
serves a crucial role in docetaxel-resistant PCa. IL6, an
inflammatory factor associated with inflammation-driven cancer,
performs an important role in the resistance to EGFR drugs. A
recent study suggested that co-targeting EGFR and IL6 may exhibit
potential as a new cancer treatment, as crosstalk between the EGFR
and IL6 signaling pathways contributes to drug resistance (47). The present study demonstrated
positive correlations between EGFR and CD44 and between EGFR and
SYK expression levels.
In summary, a total of 756 DEGs and ten hub genes
were identified in the current study. Bioinformatics analysis
demonstrated that ICAM1, CXCL8, CD44, SYK, EGFR and IL6 were
upregulated in the docetaxel-resistant PCa cell lines, and RT-qPCR
analysis confirmed that a number of the hub genes, including CHK1,
OAS3, CXCL8 and CDH1, were highly expressed in the
docetaxel-resistant cell lines; these data suggested that these
genes may be the core genes involved in the mechanism of docetaxel
resistance in PCa. The data from the present study suggested that
these genes may be closely associated the carcinogenesis,
progression, prognosis and drug resistance of PCa.
In conclusion, the present preliminary study
revealed several hub genes associated with docetaxel resistance by
comprehensive bioinformatics analysis, which may assist with
improving the understanding of the underlying molecular mechanisms
of docetaxel resistance. Combined targeted therapy of multiple
genes and pathways is of great significance to investigate the
mechanism of docetaxel-resistance. However, a limitation of the
present study is that only a single platform based on docetaxel
sensitivity and docetaxel resistance in PCa was analyzed.
Furthermore, the current study was focused on bioinformatics and
the results only verified by RT-qPCR in cell lines; therefore, the
conclusion remains to be confirmed by in vivo experiments.
Studies involving experiments and larger sample sizes are required
to further confirm the present results in the future.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was funded by the Project of Youth
Science Foundation of Jiangxi Science and Technology Office (grant
no. 20171BAB215015).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
LD and XG contributed to the study design, data
acquisition and analysis and drafted the manuscript. HC
participated in the study design, data acquisition and revision of
the manuscript. TZ, FX, ZD and CL assisted in the performance of
the statistical analysis. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2019. CA Cancer J Clin. 69:7–34. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wong MC, Goggins WB, Wang HH, Fung FD,
Leung C, Wong SY, Ng CF and Sung JJ: Global incidence and mortality
for prostate cancer: Analysis of temporal patterns and trends in 36
countries. Eur Urol. 70:862–874. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Tsao CK, Galsky MD and Oh WK: Docetaxel
for metastatic Hormone-sensitive prostate cancer: Urgent need to
minimize the risk of neutropenic fever. Eur Urol. 70:707–708. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Fizazi K, Ulys A, Sengeløv L, Moe M,
Ladoire S, Thiery- Vuillemin A, Flechon A, Guida A, Bellmunt J,
Climent MA, et al: A randomized, double-blind, placebo-controlled
phase II study of maintenance therapy with tasquinimod in patients
with metastatic castration-resistant prostate cancer responsive to
or stabilized during first-line docetaxel chemotherapy. Ann Oncol.
28:2741–2746. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Thadani-Mulero M, Portella L, Sun S, Sung
M, Matov A, Vessella RL, Corey E, Nanus DM, Plymate SR and
Giannakakou P: Androgen receptor splice variants determine taxane
sensitivity in prostate cancer. Cancer Res. 74:2270–2282. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ploussard G, Terry S, Maillé P, Allory Y,
Sirab N, Kheuang L, Soyeux P, Nicolaiew N, Coppolani E, Paule B, et
al: Class III beta-tubulin expression predicts prostate tumor
aggressiveness and patient response to docetaxel-based
chemotherapy. Cancer Res. 70:9253–9264. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhu Y, Liu C, Nadiminty N, Lou W, Tummala
R, Evans CP and Gao AC: Inhibition of ABCB1 expression overcomes
acquired docetaxel resistance in prostate cancer. Mol Cancer Ther.
12:1829–1836. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Chen H, Li H and Chen Q: INPP4B reverses
docetaxel resistance and epithelial-to-mesenchymal transition via
the PI3K/Akt signaling pathway in prostate cancer. Biochem Biophys
Res Commun. 477:467–4672. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
De Bessa Garcia SA, Pavanelli AC, Cruz E
Melo N and Nagai MA: Prostate apoptosis response 4 (PAR4)
expression modulates WNT signaling pathways in MCF7 breast cancer
cells: A possible mechanism underlying PAR4-mediated docetaxel
chemosensitivity. Int J Mol Med. 39:809–818. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Codony-Servat J, Marín-Aguilera M, Visa L,
García-Albéniz X, Pineda E, Fernández PL, Filella X, Gascón P and
Mellado B: Nuclear factor-kappa B and interleukin-6 related
docetaxel resistance in castration-resistant prostate cancer.
Prostate. 73:512–521. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Marín-Aguilera M, Codony-Servat J, Reig Ò,
Lozano JJ, Fernández PL, Pereira MV, Jiménez N, Donovan M, Puig P
and Mengual L: Epithelial-to-mesenchymal transition mediates
docetaxel resistance and high risk of relapse in prostate cancer.
Mol Cancer Ther. 13:1270–1284. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Sotiriou C and Piccart MJ: Taking
gene-expression profiling to the clinic: When will molecular
signatures become relevant to patient care? Nat Rev Cancer.
7:545–553. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
13
|
Marín-Aguilera M, Codony-Servat J, Kalko
SG, Fernández PL, Bermudo R, Buxo E, Ribal MJ, Gascón P and Mellado
B: Identification of docetaxel resistance genes in
castration-resistant prostate cancer. Mol Cancer Ther. 11:329–339.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Irizarry RA, Hobbs B, Collin F,
Beazer-Barclay YD, Antonellis KJ, Scherf U and Speed TP:
Exploration, normalization, and summaries of high density
oligonucleotide array probe level data. Biostatistics. 4:249–264.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW,
Shi W and Smyth GK: Limma powers differential expression analyses
for RNA-sequencing and microarray studies. Nucleic Acids Res.
43:e472015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wang L, Cao C, Ma Q, Zeng Q, Wang H, Cheng
Z, Zhu G, Qi J, Ma H, Nian H and Wang Y: RNA-seq analyses of
multiple meristems of soybean: Novel and alternative transcripts,
evolutionary and functional implications. BMC Plant Biol.
14:1692014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Tian Z, Wang C, Guo M, Liu X and Teng Z:
An improved method for functional similarity analysis of genes
based on Gene Ontology. BMC Syst Biol. 10 (Suppl 4):S1192016.
View Article : Google Scholar
|
|
18
|
Kanehisa M, Goto S, Sato Y, Furumichi M
and Tanabe M: KEGG for integration and interpretation of
large-scale molecular data sets. Nucleic Acids Res 40 (Database
Issue). D109–D114. 2012. View Article : Google Scholar
|
|
19
|
Huang da W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Franceschini A, Szklarczyk D, Frankild S,
Kuhn M, Simonovic M, Roth A, Lin J, Minguez P, Bork P, von Mering C
and Jensen LJ: STRING v9.1: Protein-protein interaction networks,
with increased coverage and integration. Nucleic Acids Res 41
(Database Issue). D808–D815. 2013.
|
|
21
|
Smoot ME, Ono K, Ruscheinski J, Wang PL
and Ideker T: Cytoscape 2.8: New features for data integration and
network visualization. Bioinformatics. 27:431–432. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Almeida D, Azevedo V, Silva A and Baumbach
J: PetriScape-A plugin for discrete Petri net simulations in
Cytoscape. J Integr Bioinform. 13:2842016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Chin CH, Chen SH, Wu HH, Ho CW, Ko MT and
Lin CY: cytoHubba: Identifying hub objects and sub-networks from
complex interactome. BMC Syst Biol. 8 (Suppl 4):S112014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Tang Z, Li C, Kang B, Gao G, Li C and
Zhang Z: GEPIA: A web server for cancer and normal gene expression
profiling and interactive analyses. Nucleic Acids Res. 45:W98–W102.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lin JZ, Wang ZJ, De W, Zheng M, Xu WZ, Wu
HF, Armstrong A and Zhu JG: Targeting AXL overcomes resistance to
docetaxel therapy in advanced prostate cancer. Oncotarget.
8:41064–41077. 2017.PubMed/NCBI
|
|
26
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Pozdeyev N, Berlinberg A, Zhou Q, Wuensch
K, Shibata H, Wood WM and Haugen BR: Targeting the NF-κB pathway as
a combination therapy for advanced thyroid cancer. PLoS One.
10:e01349012015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Mimeault M, Johansson SL and Batra SK:
Pathobiological implications of the expression of EGFR, pAkt, NF-κB
and MIC-1 in prostate cancer stem cells and their progenies. PLoS
One. 7:e319192012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Haralambieva IH, Ovsyannikova IG, Umlauf
BJ, Vierkant RA, Shane Pankratz V, Jacobson RM and Poland GA:
Genetic polymorphisms in host antiviral genes: Associations with
humoral and cellular immunity to measles vaccine. Vaccine.
29:8988–8997. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Xue Y, Rushton MD and Maringele L: A novel
checkpoint and RPA inhibitory pathway regulated by Rif1. PLoS
Genet. 7:e10024172011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Croci DO and Salatino M: Tumor immune
escape mechanisms that operate during metastasis. Curr Pharm
Biotechnol. 12:1923–1936. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Slavin-Chiorini DC, Catalfamo M,
Kudo-Saito C, Hodge JW, Schlom J and Sabzevari H: Amplification of
the lytic potential of effector/memory CD8+ cells by vector-based
enhancement of ICAM-1 (CD54) in target cells: Implications for
intratumoral vaccine therapy. Cancer Gene Ther. 11:665–680. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Brat DJ, Bellail AC and Van Meir EG: The
role of interleukin-8 and its receptors in gliomagenesis and
tumoral angiogenesis. Neuro Oncol. 7:122–133. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Roumeguère T, Legrand F, Rassy EE,
Kaitouni MI, Albisinni S, Rousseau A, Vanhaeverbeek M, Rorive S,
Decaestecker C, Debeir O, et al: A prospective clinical study of
the implications of IL-8 in the diagnosis, aggressiveness and
prognosis of prostate cancer. Future Scie OA. 4:FSO2662017.
View Article : Google Scholar
|
|
35
|
Araki S, Omori Y, Lyn D, Singh RK,
Meinbach DM, Sandman Y, Lokeshwar VB and Lokeshwar BL:
Interleukin-8 is a molecular determinant of androgen independence
and progression in prostate cancer. Cancer Res. 67:6854–6862. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Ghotra VP, He S, van der Horst G, Nijhoff
S, de Bont H, Lekkerkerker A, Janssen R, Jenster G, van Leenders
GJ, Hoogland AM, et al: SYK is a candidate kinase target for the
treatment of advanced prostate cancer. Cancer Res. 75:230–240.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Prevo R, Pirovano G, Puliyadi R, Herbert
KJ, Rodriguez- Berriguete G, O'Docherty A, Greaves W, McKenna WG
and Higgins GS: CDK1 inhibition sensitizes normal cells to DNA
damage in a cell cycle dependent manner. Cell Cycle. 17:1513–1523.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Liu P, Kao TP and Huang H: CDK1 promotes
cell proliferation and survival via phosphorylation and inhibition
of FOXO1 transcription factor. Oncogene. 27:4733–4744. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Dunn GP, Old LJ and Schreiber RD: The
immunobiology of cancer immunosurveillance and immunoediting.
Immunity. 21:137–148. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Sava GP, Speedy HE, Di Bernardo MC, Dyer
MJ, Holroyd A, Sunter NJ, Marr H, Mansouri L, Deaglio S, Karabon L,
et al: Common variation at 12q24.13 (OAS3) influences chronic
lymphocytic leukemia risk. Leukemia. 29:748–751. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Suarez JS, Gurler Main H, Muralidhar GG,
Elfituri O, Xu HL, Kajdacsy-Balla AA and Barbolina MV: CD44
regulates formation of spheroids and controls organ-specific
metastatic colonization in epithelial ovarian carcinoma. Mol Cancer
Res. May 30–2019.(Epub ahead of print). doi:
10.1158/1541-7786.MCR-18-1205.
|
|
42
|
Miletti-González KE, Murphy K, Kumaran MN,
Ravindranath AK, Wernyj RP, Kaur S, Miles GD, Lim E, Chan R,
Chekmareva M, et al: Identification of function for CD44
intracytoplasmic domain (CD44-ICD): Modulation of matrix
metalloproteinase 9 (MMP-9) transcription via novel promoter
response element. J Biol Chem. 287:18995–19007. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Li W, Qian L, Lin J, Huang G, Hao N, Wei
X, Wang W and Liang J: CD44 regulates prostate cancer
proliferation, invasion and migration via PDK1 and PFKFB4.
Oncotarget. 8:65143–65151. 2017.PubMed/NCBI
|
|
44
|
Jiang L, Chan JY and Fung KP: Epigenetic
loss of CDH1 correlates with multidrug resistance in human
hepatocellular carcinoma cells. Biochem Biophys Res Commun.
422:739–744. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Sigismund S, Avanzato D and Lanzetti L:
Emerging functions of the EGFR in cancer. Mol Oncol. 12:3–20. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Hour TC, Chung SD, Kang WY, Lin YC, Chuang
SJ, Huang AM, Wu WJ, Huang SP, Huang CY and Pu YS: EGFR mediates
docetaxel resistance in human castration-resistant prostate cancer
through the Akt-dependent expression of ABCB1 (MDR1). Arch Toxicol.
89:591–605. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Ray K, Ujvari B, Ramana V and Donald J:
Cross-talk between EGFR and IL-6 drives oncogenic signaling and
offers therapeutic opportunities in cancer. Cytokine Growth Factor
Rev. 41:18–27. 2018. View Article : Google Scholar : PubMed/NCBI
|