Introduction
The Epstein-Barr virus (EBV) is a ubiquitous
lymphotropic herpesvirus within the human gamma herpesvirus group.
This virus infects >95% of individuals in a population during
childhood and early adolescence in an asymptomatic process. In
adolescent individuals, it may cause infectious mononucleosis,
which manifests as a fever, pharyngitis and lymphadenopathy
(1,2).
EBV establishes its infection mainly in two types of
cells, lymphocytes and epithelial cells, and this infection
persists in the host, establishing latency. EBV infection is
associated with the occurrence of numerous diseases, including:
Nasopharyngeal carcinoma (NPC) and EBV-associated gastric cancer
(EBVaGC) of epithelial origin; Burkitt lymphoma (BL), acquired
immune deficiency syndrome-related lymphoma (ARL) and
post-transplantation lymphoproliferative disorder (PTLD) of B-cell
origin; natural killer (NK)/T-cell lymphoma of T-cell origin;
lymphoid tumors; and Hodgkin's disease. The viruses that replicate
in cells with genetic lesions may contribute to oncogenesis.
Previous data suggested that EBV infection is responsible for the
clonal expansion of premalignant nasopharyngeal epithelial cells
(3).
In EBV-associated lymphomas, the virus adopts
different types of latent infection, ranging from type I-restricted
expression patterns of the viral genome to a wider spectrum of type
II and type III, under which the full spectrum of viral genomic
products is expressed. Lymphomas with latency type I EBV infection,
which is exemplified by BL, certain ARLs and PTLD, carry
cytogenetic or genetic mutations leading to oncogene activation or
inactivation of tumor suppressor genes (4). In diffuse large B-cell lymphoma
(DLBCL), B-cell lymphoma 6 (BCL-6) is frequently translocated,
while other genetic alterations are also present, including cMyc
and BCL-2 translocations, as well as p53, N-RAS and tumor necrosis
factor (TNF)-α-induced protein 3 mutations (4). All cases of BL possess chromosomal
translocation resulting in Myc immunoglobulin gene juxtaposition,
promoting the transformation potential of the Myc oncogene.
In lymphomas with latency type III EBV infection,
transforming viral proteins are expressed. Activation of the
Notch-1 signaling pathway by EBV-determined nuclear antigen 2
(EBNA-2) is responsible for the transformation of this type of
lymphoma (5). The membrane integral
proteins, latent membrane protein 1 (LMP1), LMP2A and LMP2B,
possess transforming potential. Among these, LMP1 is the viral
homolog of mammalian cluster of differentiation 40, which is
present as an active TNF receptor. LMP1 aggregates on the cell
surface, and integrates signals of growth and proliferation by
activating the nuclear factor (NF)-κB and c-Jun amino-terminal
kinase signaling pathways when the plasmic carboxyl-terminal tail
of LMP1 associates co-factors such as TNF receptor-associated
factors (TRAFs) and TNF receptor type 1-associated DEATH domain
protein (6). By contrast, LMP2A,
which is also expressed in human tumors with latency II and III EBV
infections, is homologous to the mammalian B-cell receptor. It also
contributes to NF-κB signaling by controlling TRAF2 expression,
which in turn is important for LMP1 signaling (7).
MicroRNAs (miRNAs or miRs) are a group of newly
emerging non-coding small RNAs that bind to the 3′-untranslated
region (3′-UTR) of mRNAs of specific genes and inhibit the
translation of proteins. A study examining the EBV-infected GC cell
line AGS revealed that miRs encoded by EBV bind to the 3′-UTRs of
transcripts of max-interacting protein 1, activating transcription
factor 5 (ATF-5) and ATF-6 (8). By
abrogating the inhibition of Myc, EBV encoded BART miRs contribute
to the activation of proto-oncogene Myc. As members of the unfolded
protein group, ATF-5, ATF-6 and X-box-binding protein 1, induced by
BART miRs, subsequently inhibit the expression of the EBV-encoded
lytic protein transactivator protein BZLF1 to maintain the latency
of the EBV infection in host cells. The decreased expression of
ATF-5 and ATF-6 facilitates entry into the viral lytic cycle
(8). These findings suggested that,
in addition to coding for protein products, EBV also generates
non-coding RNAs to contribute in host-virus interaction to mediate
malignant transformation.
miRs consist of 19–25 nucleotides (nt) in length and
are generated from mRNA catalyzed by type II DNA polymerases, which
initially forms primary RNA and then matures into miRNA. It is
known that miRs possess the ability to form a stem-loop,
imperfectly complementary, hairpin structure with a 33-base pair
(bp) stem and a terminal loop on mRNAs (9).
Drosha, a type III RNAse enzyme, initially forms a
microprocessor complex; next, together with helicases, it
recognizes and cleaves the folded miRs located 11 bp away from the
junction between the single- and double-stranded RNA at the base of
the hairpin stem (10,11). A pre-miRNA that is 60–100 nt long is
then generated and retains the hairpin of its precursor.
Furthermore, in the cytoplasm, the RNA-induced signaling complex
containing Dicer (an RNAse type III enzyme) cleaves off the
terminal loop from the pre-miRNA to generate a mature
double-stranded miR (10,11).
In addition to targeting the 3′-UTRs of mRNAs of
specific genes, the binding sites of miRs in other regions on mRNA
strands have been described (9).
miRs accelerate the degradation of mRNAs, as well as to inhibit the
translation of mRNAs. It has also been reported that the expression
of miRs affects multiple biological processes in the human body, as
well as viral parasitism and replication.
The present review aims to summarize the latest
advances in the study of EBV encoded miRs in regards with their
biogenesis, cell type dependent expression profile, activities in
regulating host immunity, apoptosis, cell cycle and tumor
progression, to explore their possible application in diagnosis and
prognosis of EBV associated human tumors.
Cell type-dependent expression spectrum of
EBV-encoded miRs
miRs are a group of non-coding small RNAs that do
not encode any protein products. In EBV-infected cells, the viral
genome encodes multiple proteins and miRs to maintain the malignant
phenotype.
Five viral miRNAs were first identified in 2004 in
EBV-infected cells [13]; over the last 10 years, additional EBV
miRNAs were identified by sequencing studies [15-17]. At least 44
mature miRNAs arise from the 25 EBV precursor miRNAs (pre-miRNAs):
Three miRNAs flank the BHRF1 ORF encoding a viral Bcl2 homolog,
including miR-BHRF1-1, miR-BHRF1-2, miR-BHRF1-3, and two large
clusters of miRNAs arise from introns within the BART region. The
three BHRF1 miRs are expressed in EBV latency III infections
(12–14). miR-BHRF1-1 is located at the 5′ end
of the BHRF1 lytic mRNA transcription start site and overlaps the
TATA box of the EBV replication-activated BHRF1 promoter, whereas
miR-BHRF1-2 and miR-BHRF1-3 are located in the 3′-UTR of the BHRF1
(13).
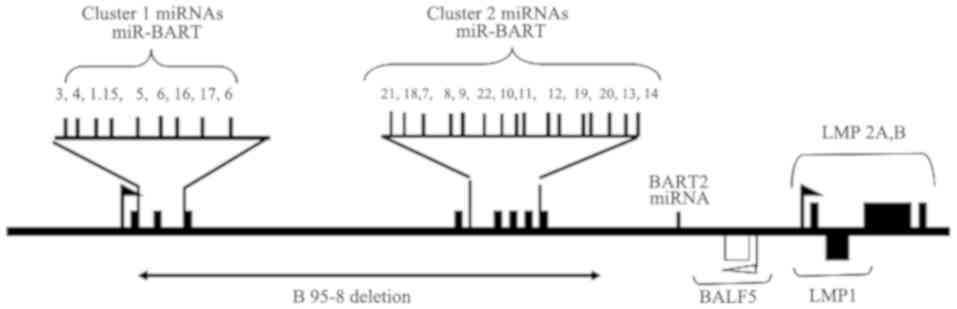
Among 25 miRs encoded by the EBV genome, three miRs
arise in the BHRF 1 locus, and the remaining 22 miR-BARTs are
located in three clusters (15,16). The
BART miRs are divided into two subclusters: Subcluster 1, which
includes miR-BART1, 3–6 and 15–17, and subcluster 2 that consists
of miR-BART7-14 and 18–21 (17). As
indicated in Fig. 1, subtypes of the
BART cluster 2 members, including miR-BART18-3p, 7–5p, 10-3p,
10–5p, 11–3p, 13-5p and 14-5p, were expressed in latency type III
infections (18,19). In addition, miR-BART2-5p and
miR-BART2-3p have been reported to be the downstream members of
these two subclusters (20). The
EBV-encoded BART miRs are expressed in virtually all EBV
infection-associated human tumors, ranging from BL (latency I),
NK/T-cell lymphoma (latency I) and EBV aGC (latency I), to Hodgkin
disease (latency II), NPC (latency II), and EBV-associated B
lymphoma and PTLD (latency III).
Expression of EBV-encoded miRs in
B-cell lymphomas
Despite the full transformation potential in B
lymphocytes, the prototype EBV strain B95-8 contains a deletion in
the BART region, which encodes part of the BART clusters of miRs
(20). When B cells were
immortalized to form the LCL by infecting them with the B95-8
strain of EBV, the miRs from the affected region were not expressed
(21). BHRF1 miRNAs are only found
in cells that harbor the virus in type III latency, such as PTLD
arising during immunosuppression (22), but not for instance in GC or NPC
(17,23–25).
Expression profile of EBV-encoded miRs
in NK/T cells
An analysis of the EBV antigenic expression profile
of EBV-positive NK/T-cell lymphomas arising in the nasal cavity of
immunocompetent patients revealed that EBV adopts type I latency,
as LMP1 or EBNA2 are not expressed (26). In addition, miRs derived from the
BHRF1 cluster were not found in the tumor cells. In NPC, the EBV
miRs accounted for 5–19% of the total miRs, whereas the viral
miRNAs in NK/T-cell lymphoma represented only 2.3% of the total
miRs. The five most expressed EBV-miRs, BART7, 5, 11-5p, 1-5p and
19-3p, accounted for ~50% of the viral-encoded miRs and ~1% of the
total miRs (15,26). Similar to the expression profile of
EBV-encoded miRs in NPC tumor biopsies, all the analyzed BART miRs
were detected in peripheral T-cell lymphoma samples (26,27).
Expression profile of EBV-encoded miRs
in gastric cancer
Non-invasive gastric carcinoma tissues are
associated with latent infections of EBV (28). In EBVaGC, the tumor cells exhibit EBV
latency I infections and express the EBV-encoded small RNA EBNA1.
In certain human GC cells, LMP2A could also be detected, whereas
LMP1 is often absent (29). As type
I latency cells, EBVaGC cells express different BART miRs. A
comprehensive profiling of EBV-miRNAs was analyzed in patients with
gastric cancer (17). The prevalence
of EBVa GC was 5.0% (52 out of 1039) in our series. The most
abundant EBV-miRNAs of EBVa GC were Bart4, followed by Bart11,
Bart2, Bart6, Bart9, and Bart18, in the decreasing order. Of them,
Bart9 exhibited the same seed sequence as to hsa miR-200a and
miR-141. Expression of E-cadherin of EBV-positive SNU-719 was
increased after BART9 knockdown. In infected cells, the viral BART
miRs were expressed at high levels, and a small fraction of
cellular miRs exhibited consistently decreased expression with
specific downregulation of tumor suppressor miRs. Furthermore, the
AGS-EBV cell lines was reported to have an expression pattern
similar to that of the NPC cells and other EBV-associated
epithelial malignancies in which the BART miRs are highly expressed
and the BHRF1 miRs are barely detectable (17).
Expression profile of EBV-encoded miRs
in NPC
Two methods have been commonly used to determine the
abundance of BART miRs in NPC tissues and cell lines. One of these
methods is stem-loop polymerase chain reaction (PCR) for performing
quantitative (q)PCR with very small RNA templates, in which miRs
serve as such templates. Profiling the expression levels of BART
miRs from the EBV-harboring NPC cell line C666-1 and NPC biopsies
was performed using this method (14,24,25,30).
Furthermore, relative abundance of different BART miRs can also be
determined by direct sequencing of miR species into small RNA
libraries. Several BART miRs were detected in terms of relative
abundance in C666-1 cells, including miR-BART15, 10 and 19-3p. The
relative abundance of the miRs varies considerably when detected
with these two different methods, namely qPCR and direct sequencing
(14,25,31,32).
In a previous study, the profiling of 39 of the 40
known mature EBV miRs was assayed with a multiplex reverse
transcription-PCR (33). With this
approach, a comprehensive profile of EBV miRs in primary NPC
tumors, including estimates of miRNA copy number per tumor cell,
was obtained. It was reported that BART-derived miRs are present in
a wide range of copy numbers from <103 per cell in
both primary tumors and the widely used NPC-derived C666-1 cell
line (33).
miR-BART7 is an miRNA that is encoded by cluster 2
in the EBV genome and is highly expressed in undifferentiated
NPC-derived cells infected with EBV (11,18).
Mature EBV-miR-BART7 (MIMAT0003416) is a single-stranded molecule
containing 22 nt (5′-caucauaguccaguguccaggg-3′). Further
investigation confirmed that EBV-miR-BART7 is also highly expressed
in NPC biopsies, suggesting that it may have an oncogenic role
(33).
EBV-encoded miRs target cellular genes to
regulate the biological activities of the host
As discussed earlier, EBV-encoded miRs are expressed
in a cell type-dependent manner. The BHRF1 3′-UTR in the BHRF1
cluster with miRs 1–1, 1–2, 1–3 and the third immediately 5′ to the
BHRF1 lytic mRNA transcription start site (34) are mainly expressed by the EBV strain
harbored in latency type III cells, including EBV-transformed cells
in vitro, PTLD and B-cell lymphomas in complication with
PTLD. BART miRs are expressed in latency types I and II, adopted by
EBV-positive cell lines including BL and NPC, respectively
(24).
In a previous study, a virus mutant D123 that lacks
all three members of cluster BHRF1 miRs was constructed (35). The B-cell transforming capacity of
the D123 EBV mutant was decreased by >20-fold relative to the
wild-type or mutant viruses. However, the B cells harboring EBV
deletion with BHRF1 miRs displayed higher latent gene expression
levels and latent protein production when compared with their
wild-type counterparts. The growth of B cells infected with the
deletion-mutant virus was markedly decreased, and the percentage of
cells entering the S phase of the cell cycle was two-fold reduced.
Therefore, BHRF1 miRs accelerate B-cell expansion at lower latent
gene expression levels. The data of this previous study suggested
that expressed miRs expand the viral reservoir and reduce the viral
antigenic load, hence facilitating the persistence of EBV in the
host (36). EBV-encoded miRs also
contribute to carcinogenesis by apoptosis inhibition and immune
evasion.
EBV has been reported to be tightly associated with
NPC (36–38). The virus adopts the latency II
characteristic of restricted viral genomic expression profiles due
to the immune competence of the hosts. The expression of viral
proteins has been demonstrated to be downregulated, whereas
EBV-encoded miRs in the BART cluster were highly expressed
(14,19). EBV-miR-BART5-3p was reported to be
upregulated and to promote the growth of NPC and gastric cancer
cells. BART5-3p directly binds to the 3′-UTR of mRNA of the p53
coding gene, and downregulates cyclin dependent kinase inhibitor 1A
(CDKN1A), BAX and FAS expression, which has been reported to lead
to acceleration of the cell cycle progression and inhibition of
cell apoptosis. It has also been observed that BART5-3p facilitated
the degradation of p53 (37,39). The findings suggest a mechanism
underlying the strategies utilized by EBV to maintain latent
infection and promote the development of carcinomas associated with
EBV infection (36).
The targets of EBV-encoded multiple cancer-related
proteins have been described. These target molecules include
BCL-2-interacting mediator of cell death (Bim) (40), p53 upregulated modulator of apoptosis
(PUMA) (41), determination of
interleukin-4 commitment 1(DICE1) (42), the tumor suppressor phosphatase and
tensin homolog (PTEN) (43,44) and E-cadherin (45). BART miRs also target a number of
genes associated with host immune regulation, including importin 7
(43,46), Dicer (44,47) and
major histocompatibility complex class I-related chain B (48), as well as EBV encoded transforming
proteins, LMP1 and LMP2 (49,50), as
listed in Table I.
 | Table I.Molecular targets of EBV-encoded miRs
and their functions. |
Table I.
Molecular targets of EBV-encoded miRs
and their functions.
| EBV miR
species | Molecular
targets | Function of target
molecules | (Refs.) |
|---|
| miR-BART5 | PUMA | Pro-apoptotic
effector of p53 | (41) |
| miR-BART3-5p | DICE1 | TSG in NPC | (42) |
| miR-BART9 | E-cadherin | Promotes invasion
and metastatic properties of NPC cells | (45) |
| miR-BART17-5p,
miR-BART17-1-5p and miR-BART17-16 | LMP1 | Oncogenic
potential | (19,49) |
| miR-BART22 | LMP2 | Involvement in
cellular transformation | (19,50) |
Impact of EBV-encoded miRs on the
anti-EBV immune response
Viruses have evolved numerous strategies to evade
host immune responses to support viral replication (51). It has been reported that growth of
cells infected with mutant EBV with miR deletions is markedly
slowed down (36). Evidence has
suggested that EBV-encoded miRs promote the proliferation and
survival of infected B cells and tumor cells, modulate immune
evasion and reduce immune recognition against EBV (52–56).
It was demonstrated that the pre-miR-BHRF1-2 and 1–3
stem-loops are present in the 3′-UTR of transcripts encoding
EBNA-leader protein (EBNA-LP; also known as EBNA5), an EBV antigen
expressed early in viral infections (57). Excision of pre-miR-BHRF1-2 and 1–3 by
Drosha to generate mature miRs destabilizes these mRNAs and reduces
the expression of the encoded protein. Experiments using mutant EBV
strains with inactivated pre-miR-BHRF1-2 and 1–3 revealed the
upregulated expression of EBNA-LP, and EBNA-LP-regulated miRs and
proteins, including LMP1 (49). The
data suggested that the expression of BHRF1 miRs serves a role in
immune evasion by reducing the levels of viral proteins that evoke
immune response (56).
miRs have been known to modulate innate and adaptive
immunity through transcriptionally regulating multiple genes. miRs
encoded by EBV and other human herpesviruses are less immunogenic
due to their small size; these miRs regulate the expression of both
viral and host proteins (52,55), and
work efficiently to evade the anti-viral immune defense. It has
been reported that herpes virus-encoded miRs affect host immunity
by targeting the expression of ligands of NK cell receptors
(48), chemokines (58) and inflammasome components (59,60), and
components of innate immune signaling pathways (61–64).
In EBV-transformed B cells and GC cells, miR-BART16
directly targets and downregulates CREB-binding protein, which is a
key transcriptional coactivator in interferon (IFN) signaling
(65). It also abrogates the
production of IFN responsive genes in response to IFN-α stimulation
and inhibits the antiproliferative effect of IFN-α in latently
infected BL cells. By obstructing the type I IFN-induced antiviral
response, miR-BART16 facilitates the establishment of a latent EBV
infection and promotes viral replication (65).
Signaling of interleukin-1 (IL-1) serves an
important role in inflammation and early activation of the
antiviral innate immune response. EBV-encoded miRs have been
identified to target the 3′-UTR of IL-1 receptor 1 mRNA, as
confirmed by 3′-UTR luciferase reporter assays (66). This viral miR activity disrupts IL-1
autocrine and paracrine signaling loops and provides a survival
advantage by dampening excessive inflammation that is detrimental
to the infected cell.
Impact of EBV-encoded miRs on host
cell apoptosis
As a form of programmed cell death, apoptosis is a
host mechanism to eliminate unwanted cells in order to maintain
homeostasis. It also functions to combat viral infection by
triggering the death of virus-infected cells. Apoptosis is
initiated and executed in complicated cascades (67); the apoptotic pathways in which
mitochondria and caspases (CASPs) serve central roles are modulated
by different factors, notably pro- and anti-apoptotic members of
the BCL-2 family (68).
It has been reported that viruses have evolved
different strategies to allow viral genomic products to inhibit
apoptosis to counteract antiviral immunity (69). The viral genome-encoded products
include proteins and miRs to modulate host apoptosis. Whether BART
miRs, which are frequently expressed in EBV-associated carcinomas,
are required in the transformation of human epithelial cells
remains unclear due to the lack of an experimental model for
investigation. However, the data from the gastric carcinoma-derived
cell line AGS support this speculation. Upon ectopic expression of
BART miRs, the anchorage-independent growth of AGS cells, which are
negative for the EBV genome but readily infected with EBV, was
significantly enhanced and apoptosis was inhibited (37,70,71).
During the process, pro-apoptotic genes are downregulated by
individual BART miRs (38,39,53,72,73). A
previous study reported that five of the 22 miR-BART pre-miRs were
anti-apoptotic, as well as the seven identified pro-apoptotic
cellular mRNA targets (six of which were novel) that significantly
contributed to the observed anti-apoptotic phenotype (72).
An additional pro-apoptotic member of the BCL-2
family, the BH3-interacting domain death agonist protein (also
known as BID), has been demonstrated to be downregulated in EBVaGC
tissue samples and gastric carcinoma cell lines infected with EBV,
resulting in inhibition of apoptosis (69). The expression of BAD, another BCL-2
family pro-apoptotic protein, has also been reported to be
significantly lower in EBV-infected AGS-EBV cells when compared
with that in EBV-negative AGS cells. In addition, five BART miRs
that showed seed matching with the 3′-UTR of the BAD mRNA were
identified. Of these identified miRs, only miR-BART20-5p reduced
BAD expression when individually transfected into AGS cells. The
EBV-encoded miRs were also demonstrated to reduce apoptosis and
enhance cell growth (70,71). miR-BART20-5p has been reported to
increase chemoresistance to 5-fluorouracil and docetaxel (53,73).
A total of 12 EBV miRs were found to have one or
more seed binding sites in the 3′-UTR of the caspase 3 (CASP3)
mRNA, while nine induced significant repression of a full-length
CASP3 reporter. Furthermore, three EBV miRs, including BART22,
repressed the endogenous CASP3 protein (74). With regards to the crucial role of
CASP3 in executing apoptosis, the data supports the notion that
EBV-encoded miRs exert apoptosis inhibition to facilitate viral
replication. It has also been reported that EBV-miR-BART15-3p
sensitized GC cells to an anticancer drug, fluorouracil, through
downregulation of Tax1-binding protein 1 (75).
EBV-encoded miRs target the cell cycle
regulation
EBV-encoded miRs regulate diverse biological
processes, including B-cell activation, cell proliferation,
apoptosis and cell cycle entry through targeting multiple
intracellular signaling pathways. Upregulated expression of PR
domain zinc finger protein 1 (PRDM1), a master regulator of B-cell
terminal differentiation and a tumor suppressor gene in aggressive
lymphomas (such as in DLBCL), induced apoptosis and cell cycle
arrest in the LCL. It has also been reported that PRDM1 is
regulated by miR-BHRF1-2, which repressed the reporter activity by
specific interactions with the seed region within the PRDM1 3′-UTR
(76). The study also suggested that
inhibition of miR-BHRF1-2 negatively regulated the cell cycle and
decreases the expression of Small Cajal body-specific RNA 20, a
small nucleolar RNA that is also downregulated by PRDM1
overexpression. The interaction between miR-BHRF1-2 and PRDM1 may
thus be one of the mechanisms by which miR-BHRF1-2 promotes EBV
lymphomagenesis (76).
When viral mutants with deletions of BHRF1 miRs
infected target cells, the proliferation support of infected B
cells was evidently lost early after infection. The cells with EBV
lacking the BHRF1 miRs progressed through the cell cycle less
efficiently and died due to apoptosis more frequently than cells
infected with the parental EBV. According to phenotypic
characterizations, EBV-encoded BHRF1 miRs supported B-cell
activation mediated by EBV, but were not involved in maintaining
viral latency (74,76)
The role of EBV-encoded miRs on the transformation
of B cells and growth of LCLs was examined using a mutant BHRF1 miR
cluster. Deficient BHRF1 miRs still increased the number of
B-cell-derived lymphoblastoid cell lines (LCLs) in the S phase, and
this finding prompted a screening of the expression of cell cycle
regulators. Reduced expression of the negative cell cycle regulator
CDKN1B/p27 was identified after 30 days in culture, while
CDKN1B/p27 expression was eliminated after 2 weeks of infection
(75,77). The reduced expression of p27 suggests
that the effects of PTEN overexpression were counteracted in the
infected cells. Indeed, PTEN blocks the repressive effects of Akt
on p27 and thus increases p27 expression (76,78).
EBV miRs promote tumor
progression
Certain EBV-encoded miRs are present in high levels
in EBV-positive NPC tissues, as well as in the plasma of the
patients. This raises the possibility that, similar to the viral
proteins expressed, these miRs may contribute to the genesis and/or
progression of NPC, and serve as an indicator for diagnosis and
prognosis. Data have also suggested that EBV-encoded miRs promoted
tumor metastasis (79).
The cellular tumor suppressor PTEN was reported to
be directly targeted by EBV-miR-BART1 (43). The PTEN expression level has been
shown to be reduced, while downstream PTEN-regulated signaling
pathways are activated, including the PI3K-Akt, focal activating
kinase-p130 and SHC-transforming protein 1-MAPK/ERK1/2 signaling
pathways, and epithelial-mesenchymal transition is induced; hence,
the migration, invasion and metastasis of the tumor cells is
increased. Exogenous PTEN reversed all phenotypes generated by
BART1, highlighting the role of PTEN in suppressing
EBV-miR-BART-driven metastasis in NPC (43,77).
These data provided an insight into the EBV-mediated metastasis
regulation and proposed novel clinical intervention strategies of
NPC.
More than 20 BART miRs are detectable at abundant
levels in EBV-positive NPC. Their expression profiles were studied
in EBV latently infected Mutu I and Mutu III cell lines,
EBV-positive NPC cells and noncancerous nasopharynx cells (24,30). The
results revealed that the miRs BART3, 7 and 13 were highly
expressed and regularly secreted into the extracellular
environment. High extracellular levels of these two EBV-BART-miRs
were also identified in the plasma of patients with NPC, whereas
the expression was absent in the plasma of non-NPC and healthy
individual controls (79). However,
the implication of miR-BART7 and 13 in the occurrence of NPC
remains to be further elucidated.
miR-BART7 exists at significantly higher levels in
patients with NPC when compared with those in healthy individuals,
and this miR was detectable in all the patient plasma samples
examined, independent of the EBV DNA level (78). Furthermore, in vitro
expression of miR-BART7 enhanced the proliferation, migration and
invasion of NPC cells, while NPC cells expressing miR-BART7 were
found to be more resistant to cisplatin (35).
EBV-encoded miRs as biomarkers for the
diagnosis and prognosis of EBV-associated tumors
EBV is associated with various types of
non-Hodgkin's lymphoma, ranging from BL to opportunistic lymphomas,
ARL and tumors that occur in immunologically compromised patients
(80). Detection of genomic products
of EBV and their antibodies in the peripheral blood is important
for the diagnosis of these diseases. In hairy cell lymphoma, the
effects of elevated organochlorines and antibodies against EBV
proteins were determined (81).
Titers of antibodies to the EBV early antigens and EBNAs were
correlated to concentrations of organochlorines in order to
evaluate the impact of these factors in the pathogenesis of hairy
cell lymphoma (81).
The expression and their contribution of EBV miRs to
the disease progression of chronic lymphocytic leukemia were
further assessed in a previous study (80). It was reported that the expression
level of EBV BHRF1-1, which is located immediately upstream and
downstream of the BHRF1 open reading frame, and is frequently
expressed in EBV latency III infections, was high in patients with
chronic lymphocytic leukemia and was predictive of shorter overall
survival. This predictive power was retained even when common
prognostic factors were included in the multivariate analysis. The
ability of BHRF1-1 expression levels to define the outcome in a
validation group of patients was also confirmed (80).
miRs synthesized by different internal organs are
released into the extracellular microenvironment in different
forms, including exosomes, apoptotic bodies and microvesicles. They
are tissue specific and detectable in the blood (82–84), and
are stable in the serum or plasma with reproducible and consistent
levels in different individuals of the same species (83,84). Due
to their persistence in circulation and their stable nature during
diseases, miRs can serve as novel noninvasive diagnostic and
prognostic biomarkers for diseases, including cancer (84,85). The
miR expression profile was examined in a variety of cancer types,
and the expression signature of miRs can be used to distinguish
cancer cells from normal cells (86)
or one cancer type from another (85,87–89), as
well as to predict the response to a particular anticancer
therapeutic agent (90) or the
clinical outcome of patients (91–94).
miRs signatures are reported to be implicated in the
prognosis of EBV-associated human tumors. For instance, NPC is very
sensitive to radiation, and the majority of patients responded well
to radiotherapy (95). Despite the
efficacy of radiotherapy against this tumor, the problem of
recurrence in cervical lymph nodes and distal metastasis is still
encountered subsequent to treatment (96). Clinically, NPC has a high incidence
of lymph node metastasis and occurs in a majority of the patients
prior to their first visit to the clinic (97,98). The
demand to improve the efficacy of anti-NPC therapy has prompted the
identification of genes that are implicated in lymph node
involvement, which is an indicative of disease progression and
therapeutic failure (99–101). Thus, the investigation of the role
of miRs in NPC may provide a mechanistic explanation of lymph node
metastasis, as well as more a sensitive indicator for the prognosis
of NPC.
The discovery of molecular biomarkers that can be
incorporated into the cancer staging system could improve the
accuracy of prognostic prediction. Ideal biomarkers should be
easily accessible and noninvasive; therefore, circulating nucleic
acids are of great interest. Upregulation of EBV-encoded miRs
(including BART1-3p, 2-5p, 5, 6-5p, 6-3p, 7, 8, 9, 14, 17-5p, 18-5p
and 19-3p) was revealed in NPC biopsies using high throughput
screening. It was observed that tumor and serum miR copy numbers
from the patients and controls were correlated. The data further
suggested that viral miRs are generally more upregulated than miRs
of human origin and that their presence in the serum of patients
with NPC was positively correlated with the copy numbers of the
same miRs in tumor cells (23).
Furthermore, cellular target genes were predicted by gene
expression analysis using bioinformatics tools, revealing that the
tumor suppressor PTEN and Wnt signaling pathways involved in NPC
were targeted. High levels of the EBV-miRs BART2-5p, 6-5p and 17-5p
were revealed in the serum of patients with NPC, but not in that of
healthy individuals. Their levels in NPC tumors were significantly
correlated with the levels in serum samples, which may allow them
to be used as diagnostic or prognostic markers (95).
Several EBV-encoded miRs, including BART1, 9, 16, 17
and 22, have been found to be expressed in NPC biopsies, while they
were reported to have both cellular and viral targets, influencing
in turn multiple host properties, such as growth, proliferation,
survival and evasion of host immunity (32,102,103).
The biological function of serum-associated BART miRs remains
unclear, although available evidence has suggested that its
epigenetic regulation may target the host immune response (65,66).
Their expression and functional downregulation in circulating blood
could enhance to attack virus-infected NPC cells by host immunity
However, the downregulation of miRs expression in circulation and
their function has been proved to enhance the attack of EBV
infected NPC cells mediated by the host immunity (16). The BART miRs are possibly released
into the blood from NPC cells in exosomes, apoptotic bodies or
microvesicles.
The potential use of miR-BART3, 7 and 13 as NPC
biomarkers has been evaluated, as they are highly expressed and
regularly secreted into the extracellular environment of NPC cells
(79). This study reported that the
levels of miR-BART7 and miR-BART13, but not miR-BART3, were
distinctly present in patients with NPC, with elevated levels being
particularly apparent among patients in advanced stages of the
disease. These data suggested that the EBV-encoded miR-BART7 and
miR-BART13 may serve as novel serological biomarkers for the
diagnosis of NPC and prediction of the efficacy of treatment.
The oncogenic potential of EBV-miR-BART7 has been
suggested due to its high expression in NPC cells, as well as NPC
biopsies (16,21,30,33,34). It
has been reported that in vitro expression of miR-BART7
enhanced the proliferation, migration and invasion of NPC cells,
and enabled their high resistance to cisplatin (35). High-throughput gene expression
analysis further suggested that EBV-miR-BART7 affects multiple
cancer-related pathways (33,79). A
comparison was conducted between the circulating BART7 and EBV DNA
(34). These data supported the
usefulness of this viral miR as a biomarker of undifferentiated
NPC, which is associated with EBV infection.
Conclusions
As epigenetic regulators of gene expression, miRs
target numerous genes, accelerating their degradation to maintain
EBV replication and persistence through inhibiting apoptosis and
suppressing the host immune response. The effects significantly
contribute to the achievement of malignancy of EBV-infected cells.
Currently, it is accepted that EBV infection serves an important
role in the early stages of NPC occurrence, and that it is
implicated in the expansion of premalignant, dysplastic epithelial
cell clones. In addition, LMP1 is regarded as a transforming
protein that contributes to disease progression. Emerging evidence
also supported the role of EBV-encoded miRs in the genesis of NPC.
BART miRs are expressed in abundance in EBV-positive NPC, which may
be due to their small size enabling them to escape innate immune
defense. Finally, BART miRs are specific for EBV-infected NPC
cells, and may therefore serve as good candidates for therapeutic
targets of NPC. It is anticipated that a novel miR-based
therapeutic strategy will be designed, either alone or in
combination with pre-existing regimens, to treat EBV-infected
malignancies.
Acknowledgements
The authors would like to thank Professor Ingemar
Ernberg and Dr. Ljudmila Matskova (Karolinska Institutet, Sweden)
for their stimulating discussions on the role of microRNAs in the
regulation of gene expression.
Funding
The present study was supported by a research grant
from the Medical Research Fund, Commission of Health and Family
Plan of the Province of Guangdong (awarded to XZ; grant no.
A2018356).
Availability of data and materials
Not applicable.
Authors' contributions
XZ, QQ and ZH conceived, wrote and corrected the
paper; YY, MF and BZ wrote and revised the paper. All authors
approved the final version of the manuscript before submission.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
miR
|
microRNA
|
|
EBV
|
Epstein-Barr virus
|
|
EBNA
|
EBV-determined nuclear antigen
|
|
LMP
|
latent membrane protein
|
|
NPC
|
nasopharyngeal carcinoma
|
|
EBVaGC
|
EBV-associated gastric cancer
|
|
PTLD
|
post-transplantation
lymphoproliferative disorder
|
|
BHRF1
|
BamHI reading frame 1
|
|
BART
|
BamHI A region transcripts
|
References
|
1
|
Kutok JL and Wang F: Spectrum of Epstein
Barr virus associated diseases. Annu Rev Pathol. 1:375–404. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Williams H and Crawford DH: Epstein-Barr
virus: The impact of scientific advances on clinical practice.
Blood. 107:862–869. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lo KW, To KF and Huang DP: Focus on
nasopharyngeal carcinoma. Cancer Cell. 5:423–428. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Cesarman E: Gammaherpesvirus and
lymphoproliferative disorders in immunocompromised patients. Cancer
Lett. 305:163–174. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hsieh JJ and Hayward SD: Masking of the
CBF1/RBPJ kappa transcriptional repression domain by Epstein-Barr
virus EBNA2. Science. 268:560–563. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Yoshizaki T, Kondo S, Wakisaka N, Murono
S, Endo K, Sugimoto H, Nakanishi S, Tsuji A and Ito M: Pathogenic
role of Epstein-Barr virus latent membrane protein-1 in the
development of nasopharyngeal carcinoma. Cancer Lett. 337:1–7.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Guasparri I, Bubman D and Cesarman E: EBV
LMP2A affects LMP1-mediated NF-kappaB signaling and survival of
lymphoma cells by regulating TRAF2 expression. Blood.
111:3813–3820. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Marquitz AR, Mathur A, Edwards RH and
Raab-Traub N: Host gene expression is regulated by two types of
noncoding RNAs transcribed from the Epstein-Barr virus BamHI A
rightward transcript region. J Virol. 89:11256–11268. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Winter J, Jung S, Keller S, Gregory RI and
Diederichs S: Many roads to maturity: microRNA biogenesis pathways
and their regulation. Nat Cell Biol. 11:228–234. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Han J, Lee Y, Yeom KH, Nam JW, Heo I, Rhee
JK, Sohn SY, Cho Y, Zhang BT and Kim VN: Molecular basis for the
recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell.
125:887–901. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Busson P, Keryer C, Ooka T and Corbex M:
EBV-associated nasopharyngeal carcinomas: From epidemiology to
virus-targeting strategies. Trends Microbiol. 12:356–360. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Pfeffer S, Zavolan M, Grässer FA, Chien M,
Russo JJ, Ju J, John B, Enright AJ, Marks D, Sander C and Tuschl T:
Identification of virus-encoded microRNAs. Science. 304:734–736.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhu JY, Pfuhl T, Motsch N, Barth S,
Nicholls J, Grässer F and Meister G: Identification of novel
Epstein-Barr virus microRNA genes from nasopharyngeal carcinomas. J
Virol. 83:3333–3341. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Edwards RH, Marquitz AR and Raab-Traub N:
Epstein-Barr virus BART microRNAs are produced from a large intron
prior to splicing. J Virol. 82:9094–9106. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Qiu J, Cosmopoulos K, Pegtel M, Hopmans E,
Murray P, Middeldorp J, Shapiro M and Thorley-Lawson DA: A novel
persistence associated EBV miRNA expression profile is disrupted in
neoplasia. PLoS Pathog. 7:e10021932011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Tsai CY, Liu YY, Liu KH, Hsu JT, Chen TC,
Chiu CT and Yeh TS: Comprehensive profiling of virus microRNAs of
Epstein-Barr virus-associated gastric carcinoma: Highlighting the
interactions of ebv-Bart9 and host tumor cells. J Gastroenterol
Hepatol. 32:82–91. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Sakamoto K, Sekizuka T, Uehara T, Hishima
T, Mine S, Fukumoto H, Sato Y, Hasegawa H, Kuroda M and Katano H:
Next-generation sequencing of miRNAs in clinical samples of
Epstein-Barr virus-associated B-cell lymphomas. Cancer Med.
6:605–618. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lo AKF, Dawson CW, Jin DY and Lo KW: The
pathological roles of BART miRNAs in nasopharyngeal carcinoma. J
Pathol. 227:392–403. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Baer R, Bankier AT, Biggin MD, Deininger
PL, Farrell PJ, Gibson TJ, Hatfull G, Hudson GS, Satchwell SC,
Séguin C, et al: DNA sequence and expression of the B95-8
Epstein-Barr virus genome. Nature. 310:207–211. 1984. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Pratt ZL, Kuzembayeva M, Sengupta S and
Sugden B: The microRNAs of Epstein-Barr Virus are expressed at
dramatically differing levels among cell lines. Virology.
386:387–397. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Xia T, O'Hara A, Araujo I, Barreto J,
Carvalho E, Sapucaia JB, Ramos JC, Luz E, Pedroso C, Manrique M, et
al: EBV microRNAs in primary lymphomas and targeting of CXCL-11 by
ebv-mir-BHRF1-3. Cancer Res. 68:1436–1442. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kim DN, Chae HS, Oh ST, Kang JH, Park CH,
Park WS, Takada K, Lee JM, Lee WK and Lee SK: Expression of viral
microRNAs in Epstein-Barr virus-associated gastric carcinoma. J
Virol. 81:1033–1036. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Cosmopoulos K, Pegtel M, Hawkins J,
Moffett H, Novina C, Middeldorp J and Thorley-Lawson DA:
Comprehensive profiling of Epstein-Barr virus microRNAs in
nasopharyngeal carcinoma. J Virol. 83:2357–2367. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Chen SJ, Chen GH, Chen YH, Liu CY, Chang
KP, Chang YS and Chen HC: Characterization of Epstein-Barr virus
miRNAome in nasopharyngeal carcinoma by deep sequencing. PLoS One.
5(pii): e127452010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Motsch N, Alles J, Imig J, Zhu J, Barth S,
Reineke T, Tinguely M, Cogliatti S, Dueck A, Meister G, et al:
MicroRNA profiling of Epstein-Barr virus-associated NK/T-cell
lymphomas by deep sequencing. PLoS One. 7:e421932012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Jun SM, Hong YS, Seo JS, Ko YH, Yang CW
and Lee SK: Viral microRNA profile in Epstein-Barr virus-associated
peripheral T cell lymphoma. Brit J Haematol. 142:320–323. 2008.
View Article : Google Scholar
|
|
28
|
Arikawa J, Tokunaga M, Tashiro Y, Tanaka
S, Sato E, Haraguchi K, Yamamoto A, Toyohira O and Tsuchimochi A:
Epstein-Barr virus-positive multiple early gastric cancers and
dysplastic lesions: A case report. Pathol Int. 47:730–734. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Fukayama M: Epstein-Barr virus and gastric
carcinoma. Pathol Int. 60:337–350. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wong AM, Kong KL, Tsang JW, Kwong DL and
Guan XY: Profiling of Epstein-Barr virus-encoded microRNAs in
nasopharyngeal carcinoma reveals potential biomarkers and oncomirs.
Cancer. 118:698–710. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee
DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, et al:
Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic
Acids Res. 33:e1792005. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Amoroso R, Fitzsimmons L, Thomas WA, Kelly
GL, Rowe M and Bell AI: Quantitative studies of Epstein-Barr
virus-encoded microRNAs provide novel insights into their
regulation. J Virol. 85:996–1010. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Chan JY, Gao W, Ho WK, Wei WI and Wong TS:
Overexpression of Epstein-Barr virus-encoded microRNA-BART7 in
undifferentiated nasopharyngeal carcinoma. Anticancer Res.
32:3201–3210. 2012.PubMed/NCBI
|
|
34
|
Cai X, Schäfer A, Lu S, Bilello JP,
Desrosiers RC, Edwards R, Raab-Traub N and Cullen BR: Epstein-Barr
virus microRNAs are evolutionarily conserved and differentially
expressed. PLoS Pathog. 2:e232006. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Neuhierl B and Delecluse HJ: Molecular
genetics of DNA viruses: Recombinant virus technology. Methods Mol
Biol. 292:353–370. 2005.PubMed/NCBI
|
|
36
|
Feederle R, Linnstaedt SD, Bannert H, Lips
H, Bencun M, Cullen BR and Delecluse HJ: A viral microRNA cluster
strongly potentiates the transforming properties of a human
herpesvirus. PLoS Pathog. 7:e10012942011. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Tsao SW, Tsang CM and Lo KW: Epstein-Barr
virus infection and nasopharyngeal carcinoma. Philos Trans R Soc
Lond B Biol Sci. 372(pii): 201602702017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Young LS and Dawson CW: Epstein-Barr virus
and nasopharyngeal carcinoma. Chin J Cancer. 33:581–590.
2014.PubMed/NCBI
|
|
39
|
Zheng X, Wang J, Wei L, Peng Q, Gao Y, Fu
Y, Lu Y, Qin Z, Zhang X, Lu J, et al: Epstein-Barr virus MicroRNA
miR-BART5-3p inhibits p53 expression. J Virol. 92(pii): e01022–18.
2018.PubMed/NCBI
|
|
40
|
Marquitz AR, Mathur A, Nam CS and
Raab-Traub N: The Epstein-Barr virus BART microRNAs target the
pro-apoptotic protein Bim. Virology. 412:392–400. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Choy EY, Siu KL, Kok KH, Lung RW, Tsang
CM, To KF, Kwong DL, Tsao SW and Jin DY: An Epstein-Barr
virus-encoded microRNA targets PUMA to promote host cell survival.
J Exp Med. 205:2551–2560. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Lei T, Yuen KS, Xu R, Tsao SW, Chen H, Li
M, Kok KH and Jin DY: Targeting of DICE1 tumor suppressor by
Epstein-Barr virus-encoded miR-BART3* microRNA in nasopharyngeal
carcinoma. Int J Cancer. 133:79–87. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Cai L, Ye Y, Jiang Q, Chen Y, Lyu X, Li J,
Wang S, Liu T, Cai H, Yao K, et al: Epstein-Barr virus-encoded
microRNA BART1 induces tumour metastasis by regulating
PTEN-dependent pathways in nasopharyngeal carcinoma. Nat Commun.
6:73532015. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Cai LM, Lyu XM, Luo WR, Cui XF, Ye YF,
Yuan CC, Peng QX, Wu DH, Liu TF, Wang E, et al: EBV-miR-BART7-3p
promotes the EMT and metastasis of nasopharyngeal carcinoma cells
by suppressing the tumor suppressor PTEN. Oncogene. 34:2156–2166.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Hsu CY, Yi YH, Chang KP, Chang YS, Chen SJ
and Chen HC: The Epstein-Barr virus-encoded microRNA MiR-BART9
promotes tumor metastasis by targeting E-cadherin in nasopharyngeal
carcinoma. PLoS Pathog. 10:e10039742014. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Dölken L, Malterer G, Erhard F, Kothe S,
Friedel CC, Suffert G, Marcinowski L, Motsch N, Barth S, Beitzinger
M, et al: Systematic analysis of viral and cellular microRNA
targets in cells latently infected with human gamma-herpesviruses
by RISC immunoprecipitation assay. Cell Host Microbe. 7:324–334.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Iizasa H, Wulff BE, Alla NR, Maragkakis M,
Megraw M, Hatzigeorgiou A, Iwakiri D, Takada K, Wiedmer A, Showe L,
et al: Editing of Epstein-Barr virus-encoded BART6 microRNAs
controls their dicer targeting and consequently affects viral
latency. J Biol Chem. 285:33358–33370. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Nachmani D, Stern-Ginossar N, Sarid R and
Mandelboim O: Diverse herpesvirus microRNAs target the
stress-induced immune ligand MICB to escape recognition by natural
killer cells. Cell Host Microbe. 5:376–385. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Lo AK, To KF, Lo KW, Lung RW, Hui JW, Liao
G and Hayward SD: Modulation of LMP1 protein expression by
EBV-encoded microRNAs. Proc Natl Acad Sci USA. 104:16164–16169.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Lung RW, Tong JH, Sung YM, Leung PS, Ng
DC, Chau SL, Chan AW, Ng EK, Lo KW and To KF: Modulation of LMP2A
expression by a newly identified Epstein-Barr virus-encoded
microRNA miR-BART22. Neoplasia. 11:1174–1184. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Zhang X, Dawson CW, He Z and Huang P:
Immune evasion strategies of the human gamma-herpesviruses:
Implications for viral tumorigenesis. J Med Virol. 84:272–281.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Seto E, Moosmann A, Gromminger S, Walz N,
Grundhoff A and Hammerschmidt W: Micro RNAs of Epstein-Barr virus
promote cell cycle progression and prevent apoptosis of primary
human B cells. PLoS Pathog. 6:e10010632010. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Vereide DT, Seto E, Chiu YF, Hayes M,
Tagawa T, Grundhoff A, Hammerschmidt W and Sugden B: Epstein-Barr
virus maintains lymphomas via its miRNAs. Oncogene. 33:1258–1264.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Ramalingam D, Kieffer-Kwon P and
Ziegelbauer JM: Emerging themes from EBV and KSHV microRNA targets.
Viruses. 4:1687–1710. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Fernandes Q, Merhi M, Raza A, Inchakalody
VP, Abdelouahab N, Zar Gul AR, Uddin S and Dermime S: Role of
Epstein-Barr virus in the pathogenesis of head and neck cancers and
its potential as an immunotherapeutic target. Front in Oncol.
8:2572018. View Article : Google Scholar
|
|
56
|
Albanese M, Tagawa T, Buschle A and
Hammerschmidt W: MicroRNAs of Epstein-Barr virus control innate and
adaptive antiviral immunity. J Virol. 91(pii): e01667–16.
2017.PubMed/NCBI
|
|
57
|
Poling BC, Price AM, Luftig MA and Cullen
BR: The Epstein-Barr virus miR-BHRF1 microRNAs regulate viral gene
expression in cis. Virology. 512:113–123. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Kim Y, Lee S, Kim S, Kim D, Ahn JH and Ahn
K: Human cytomegalovirus clinical strain-specific microRNA
miR-UL148D targets the human chemokine RANTES during infection.
PLoS Pathog. 8:e10025772012. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Haneklaus M, Gerlic M, Kurowska-Stolarska
M, Rainey AA, Pich D, McInnes IB, Hammerschmidt W, O'Neill LA and
Masters SL: Cutting edge: miR-223 and EBV miR-BART15 regulate the
NLRP3 inflammasome and IL-1β production. J Immunol. 189:3795–3799.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Kim S, Lee S, Shin J, Kim Y, Evnouchidou
I, Kim D, Kim YK, Kim YE, Ahn JH, Riddell SR, et al: Human
cytomegalovirus microRNA miR-US4-1 inhibits CD8(+) T cell responses
by targeting the aminopeptidase ERAP1. Nat Immunol. 12:984–991.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Bauman Y, Nachmani D, Vitenshtein A,
Tsukerman P, Drayman N, Stern-Ginossar N, Lankry D, Gruda R and
Mandelboim O: An identical miRNA of the human JC and BK polyoma
viruses targets the stress-induced ligand ULBP3 to escape immune
elimination. Cell Host Microbe. 9:93–102. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Abend JR, Ramalingam D, Kieffer-Kwon P,
Uldrick TS, Yarchoan R and Ziegelbauer JM: Kaposi's
sarcoma-associated herpesvirus microRNAs target IRAK1 and MYD88,
two components of the toll-like receptor/interleukin-1R signaling
cascade, to reduce inflammatory-cytokine expression. J Virol.
86:11663–11674. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Lei X, Bai Z, Ye F, Xie J, Kim CG, Huang Y
and Gao SJ: Regulation of NF-kappaB inhibitor IkappaBalpha and
viral replication by a KSHV microRNA. Nat Cell Biol. 12:193–199.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Liang D, Gao Y, Lin X, He Z, Zhao Q, Deng
Q and Lan K: A human herpesvirus miRNA attenuates interferon
signaling and contributes to maintenance of viral latency by
targeting IKKε. Cell Res. 21:793–806. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Hooykaas MJG, van Gent M, Soppe JA, Kruse
E, Boer IGJ, van Leenen D, Groot Koerkamp MJA, Holstege FCP,
Ressing ME, Wiertz EJHJ and Lebbink RJ: EBV MicroRNA BART16
suppresses type I IFN signaling. J Immunol. 198:4062–4073. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Skinner CM, Ivanov NS, Barr SA, Chen Y and
Skalsky RL: An Epstein-Barr virus microrna blocks interleukin-1
(IL-1) signaling by targeting IL-1 receptor 1. J Virol. 91(pii):
e00530–17. 2017.PubMed/NCBI
|
|
67
|
Dillon CP and Green DR: Molecular cell
biology of apoptosis and necroptosis in cancer. Adv Exp Med Biol.
930:1–23. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Kvansakul M, Caria S and Hinds MG: The
Bcl-2 family in host-virus interactions. Viruses. 9(pii): E2902017.
View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Kang D, Skalsky RL and Cullen BR: EBV BART
microRNAs target multiple pro-apoptotic cellular genes to promote
epithelial cell survival. PLoS Pathog. 11:e10049792015. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Kim H, Choi H and Lee SK: Epstein-Barr
virus miR-BART20-5p regulates cell proliferation and apoptosis by
targeting BAD. Cancer Lett. 356:733–742. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Kim H, Choi H and Lee SK: Epstein-Barr
virus MicroRNA miR-BART20-5p suppresses lytic induction by
inhibiting BAD-mediated caspase-3-dependent apoptosis. J Virol.
90:1359–1368. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Marquitz AR, Mathur A, Shair KH and
Raab-Traub N: Infection of Epstein-Barr virus in a gastric
carcinoma cell line induces anchorage independence and global
changes in gene expression. Proc Natl Acad Sci USA. 109:9593–9598.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Shinozaki-Ushiku A, Kunita A, Isogai M,
Hibiya T, Ushiku T, Takada K and Fukayama M: Profiling of
virus-encoded microRNAs in Epstein-Barr virus-associated gastric
carcinoma and their roles in gastric carcinogenesis. J Virol.
89:5581–5591. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Harold C, Cox D and Riley KJ: Epstein-Barr
viral microRNAs target caspase 3. Virol J. 13:1452016. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Choi H and Lee SK: TAX1BP1 downregulation
by EBV-miR-BART15-3p enhances chemosensitivity of gastric cancer
cells to 5-FU. Arch Virol. 162:369–377. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Ma J, Nie K, Redmond D, Liu Y, Elemento O,
Knowles DM and Tam W: EBV-miR-BHRF1-2 targets PRDM1/Blimp1:
Potential role in EBV lymphomagenesis. Leukemia. 30:594–604. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Bernhardt K, Haar J, Tsai MH, Poirey R,
Feederle R and Delecluse HJ: A viral microRNA cluster regulates the
expression of PTEN, p27 and of a bcl-2 homolog. PLoS Pathog.
12:e10054052016. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Engelman JA, Luo J and Cantley LC: The
evolution of phosphatidylinositol 3-kinases as regulators of growth
and metabolism. Nat Rev Genet. 7:606–619. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Zhang G, Zong J, Lin S, Verhoeven RJ, Tong
S, Chen Y, Ji M, Cheng W, Tsao SW, Lung M, et al: Circulating
Epstein-Barr virus microRNAs miR-BART7 and miR-BART13 as biomarkers
for nasopharyngeal carcinoma diagnosis and treatment. Int J Cancer.
136:E301–E312. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Ferrajoli A, Ivan C, Ciccone M, Shimizu M,
Kita Y, Ohtsuka M, D'Abundo L, Qiang J, Lerner S, Nouraee N, et al:
Epstein-Barr virus microRNAs are expressed in patients with chronic
lymphocytic leukemia and correlate with overall survival.
EBioMedicine. 2:572–582. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Nordström M, Hardell L, Lindström G,
Wingfors H, Hardell K and Linde A: Concentrations of
organochlorines related to titers to Epstein-Barr virus early
antigen IgG as risk factors for hairy cell leukemia. Environ Health
Perspect. 108:441–445. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Sempere LF: Tissue slide-based microRNA
characterization of tumors: How detailed could diagnosis become for
cancer medicine? Expert Rev Mol Diagn. 14:853–869. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Mishra PJ: MicroRNAs as promising
biomarkers in cancer diagnostics. Biomark Res. 2:192014. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Ohshima K, Inoue K, Fujiwara A, Hatakeyama
K, Kanto K, Watanabe Y, Muramatsu K, Fukuda Y, Ogura S, Yamaguchi K
and Mochizuki T: Let-7 microRNA family is selectively secreted into
the extracellular environment via exosomes in a metastatic gastric
cancer cell line. PLoS One. 5:e132472010. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Zhao H, Shen J, Medico L, Wang D,
Ambrosone CB and Liu S: A pilot study of circulating miRNAs as
potential biomarkers of early stage breast cancer. PLoS One.
5:e137352010. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Pigati L, Yaddanapudi SC, Iyengar R, Kim
DJ, Hearn SA, Danforth D, Hastings ML and Duelli DM: Selective
release of microRNA species from normal and malignant mammary
epithelial cells. PLoS One. 5:e135152010. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Wei R, Huang GL, Zhang MY, Li BK, Zhang
HZ, Shi M, Chen XQ, Huang L, Zhou QM, Jia WH, et al: Clinical
significance and prognostic value of microRNA expression signatures
in hepatocellular carcinoma. Clin Cancer Res. 19:4780–4791. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Cheng H, Zhang L, Cogdell DE, Zheng H,
Schetter AJ, Nykter M, Harris CC, Chen K, Hamilton SR and Zhang W:
Circulating plasma MiR-141 is a novel biomarker for metastatic
colon cancer and predicts poor prognosis. PLoS One. 6:e177452011.
View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Subramanian S, Lui WO, Lee CH, Espinosa I,
Nielsen TO, Heinrich MC, Corless CL, Fire AZ and van de Rijn M:
MicroRNA expression signature of human sarcomas. Oncogene.
27:2015–2026. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Osaki M, Takeshita F and Ochiya T:
MicroRNAs as biomarkers and therapeutic drugs in human cancer.
Biomarkers. 13:658–670. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Nair VS, Maeda LS and Ioannidis JP:
Clinical outcome prediction by microRNAs in human cancer: A
systematic review. J Natl Cancer Inst. 104:528–540. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Hu Z, Chen X, Zhao Y, Tian T, Jin G, Shu
Y, Chen Y, Xu L, Zen K, Zhang C and Shen H: Serum microRNA
signatures identified in a genome-wide serum microRNA expression
profiling predict survival of non-small-cell lung cancer. J Clin
Oncol. 28:1721–1726. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Zuo Z, Calin GA, de Paula HM, Medeiros LJ,
Fernandez MH, Shimizu M, Garcia-Manero G and Bueso-Ramos CE:
Circulating microRNAs let-7a and miR-16 predict progression-free
survival and overall survival in patients with myelodysplastic
syndrome. Blood. 118:413–415. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Esquela-Kerscher A and Slack FJ:
Oncomirs-microRNAs with a role in cancer. Nat Rev Cancer.
6:259–269. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Zhang MX, Li J, Shen GP, Zou X, Xu JJ,
Jiang R, You R, Hua YJ, Sun Y, Ma J, et al: Intensity-modulated
radiotherapy prolongs the survival of patients with nasopharyngeal
carcinoma compared with conventional two-dimensional radiotherapy:
A 10-year experience with a large cohort and long follow-up. Eur J
Cancer. 51:2587–2595. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Chung IC, Chen LC, Chung AK, Chao M, Huang
HY, Hsueh C, Tsang NM, Chang KP, Liang Y, Li HP and Chang YS:
Matrix metalloproteinase 12 is induced by heterogeneous nuclear
ribonucleoprotein K and promotes migration and invasion in
nasopharyngeal carcinoma. BMC Cancer. 14:3482014. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Yi W, Li X, Liu Z, Jiang C, Niu D and Xia
Y: A risk score model for the metastasis of level Ib lymph node
based on the clinicopathological features of nasopharyngeal
carcinoma in a large sample. Mol Clin Oncol. 2:789–797. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Shi Q, Shen C, Kong L, Wang X, Ding J, Gao
Y, Xu T and Hu C: Involvement of both cervical lymph nodes and
retropharyngeal lymph nodes has prognostic value for N1 patients
with nasopharyngeal carcinoma. Radiat Oncol. 9:72014. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Wang HZ, Cao CN, Luo JW, Yi JL, Huang XD,
Zhang SP, Wang K, Qu Y, Xiao JP, Li SY, et al: High-risk factors of
parotid lymph node metastasis in nasopharyngeal carcinoma: A
case-control study. Radiat Oncol. 11:1132016. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Tang LL, Tang XR, Li WF, Chen L, Tian L,
Lin AH, Sun Y and Ma J: The feasibility of contralateral lower neck
sparing intensity modulation radiated therapy for nasopharyngeal
carcinoma patients with unilateral cervical lymph node involvement.
Oral Oncol. 69:68–73. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Zhang Y, Li WF, Chen L, Mao YP, Guo R,
Zhang F, Peng H, Liu LZ, Li L, Liu Q and Ma J: Prognostic value of
parotid lymph node metastasis in patients with nasopharyngeal
carcinoma receiving intensity-modulated radiotherapy. Sci Rep.
5:139192015. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Tagawa T, Albanese M, Bouvet M, Moosman A,
Mautner J, Heissmeyer V Zielinski C, Hoser J, Hastreiter M, Hayes
M, et al: Epstein-Barr viral miRNAs inhibit antiviral CD4+ T cell
responses targeting IL-12 and peptide processing. J Exp Med.
213:2065–2080. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Feederle R, Haar J, Bernhardt K,
Linnstaedt SD, Bannert H, Lips H, Cullen BR and Delecluse HJ: The
members of an Epstein-Barr virus microRNA cluster cooperate to
transform B lymphocytes. J Virol. 85:9801–9810. 2011. View Article : Google Scholar : PubMed/NCBI
|