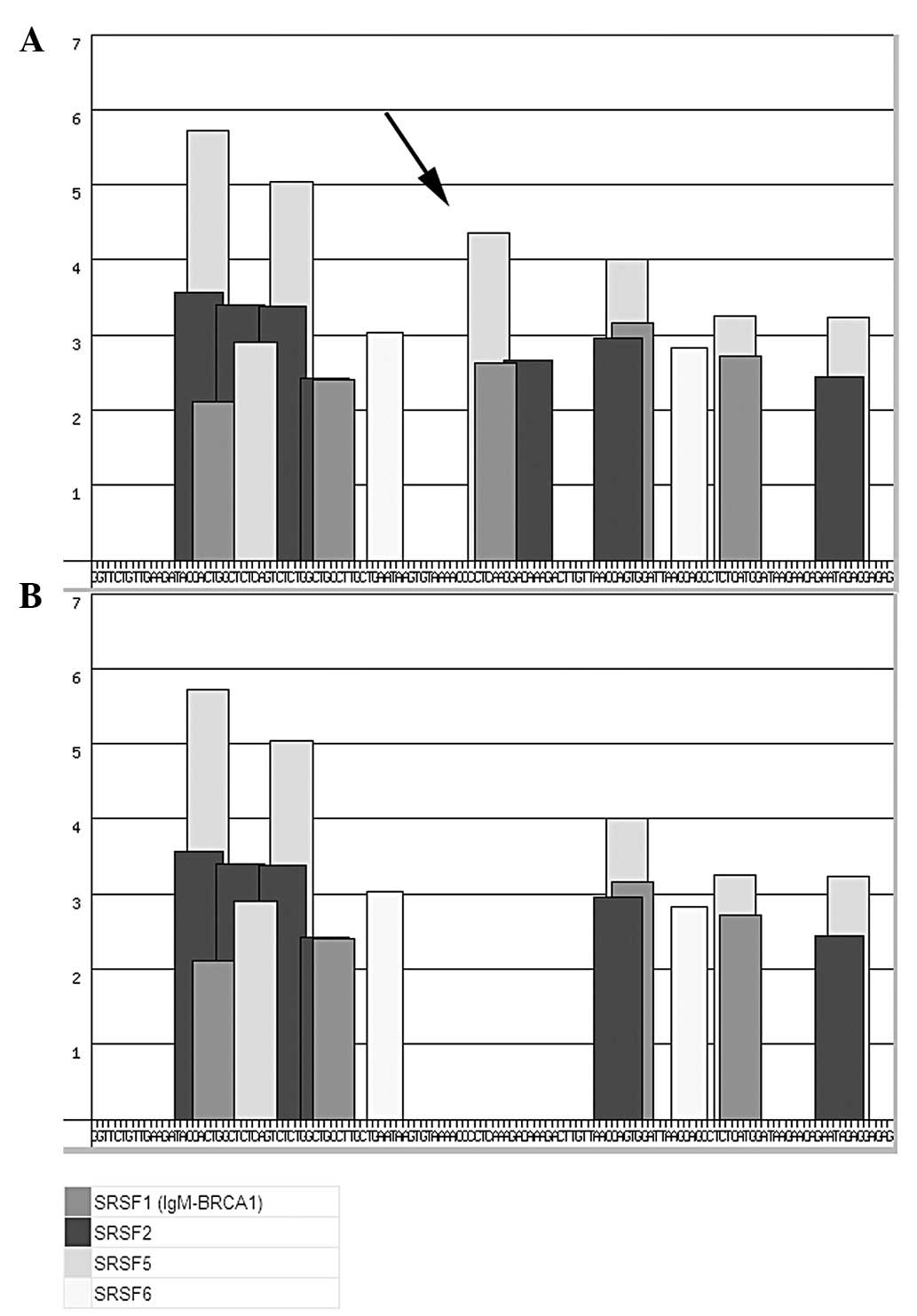
|
1
|
Peltomäki P and Vasen HF: Mutations
predisposing to hereditary nonpolyposis colorectal cancer: database
and results of a collaborative study. The International
Collaborative Group on hereditary nonpolyposis colorectal cancer.
Gastroenterology. 113:1146–1158. 1997.
|
|
2
|
Lynch HT and de la Chapelle A: Genetic
susceptibility to non-polyposis colorectal cancer. J Med Genet.
36:801–818. 1999.PubMed/NCBI
|
|
3
|
Cartegni L, Chew SL and Krainer AR:
Listening to silence and understanding nonsense: exonic mutations
that affect splicing. Nat Rev Genet. 3:285–298. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Pagani F and Baralle FE: Genomic variants
in exons and introns: identifying the splicing spoilers. Nat Rev
Genet. 5:389–396. 2004. View
Article : Google Scholar : PubMed/NCBI
|
|
5
|
Shapiro MB and Senapathy P: RNA splice
junctions of different classes of eukaryotes: sequence statistics
and functional implications in gene expression. Nucleic Acids Res.
15:7155–7174. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Jurica MS and Moore MJ: Pre-mRNA splicing:
awash in a sea of proteins. Mol Cell. 12:5–14. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Liu HX, Zhang M and Krainer AR:
Identification of functional exonic splicing enhancer motifs
recognized by individual SR proteins. Genes Dev. 12:1998–2012.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Blencowe BJ: Exonic splicing enhancers:
mechanism of action, diversity and role in human genetic diseases.
Trends Biochem Sci. 25:106–110. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hastings ML and Krainer AR: Pre-mRNA
splicing in the new millennium. Curr Opin Cell Biol. 13:302–309.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zuo P and Maniatis T: The splicing factor
U2AF35 mediates critical protein-protein interactions in
constitutive and enhancer-dependent splicing. Genes Dev.
10:1356–1368. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kan JL and Green MR: Pre-mRNA splicing of
IgM exons M1 and M2 isdirected by a juxtaposed splicing enhancer
and inhibitor. Genes Dev. 13:462–471. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Cartegni L, Wang J, Zhu Z, Zhang MQ and
Krainer AR: ESE finder: a web resource to identify exonic splicing
enhancers. Nucleic Acid Research. 31:3568–3571. 2003.PubMed/NCBI
|
|
13
|
Smith PJ, Zhang C, Wang J, Chew SL, Zhang
MQ and Krainer AR: An increased specificity score matrix for the
prediction of SF2/ASF-specific exonic splicing enhancers. Hum Mol
Genet. 15:2490–2508. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wang Z, Rolish ME, Yeo G, Tung V, Mawson M
and Burge CB: Systematic identification and analysis of exonic
splicing silencers. Cell. 119:831–845. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Cai Q, Sun MH, Fu G, et al: Mutation
analysis of hMSH2 and hMLH1 genes in Chinese hereditary
nonpolyposis colorectal cancer families. Zhonghua Bing Li Xue Za
Zhi. 32:323–328. 2003.(In Chinese).
|
|
16
|
Wang XL, Yuan Y, Zhang SZ, Cai SR, Huang
YQ, Jiang Q and Zheng S: Clinical and genetic characteristics of
Chinese hereditary nonpolyposis colorectal cancer families. World J
Gastroenterol. 12:4074–4077. 2006.PubMed/NCBI
|
|
17
|
Nomura S, Sugano K, Kashiwabara H, et al:
Enhanced detection of deleterious and other germline mutations of
hMSH2 and hMLH1 in Japanese hereditary nonpolyposis colorectal
cancer kindreds. Biochem Biophys Res Commun. 271:120–129. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kim JC, Kim HC, Roh SA, et al: hMLH1 and
hMSH2 mutations in families with familial clustering of gastric
cancer and hereditary non-polyposis colorectal cancer. Cancer
Detect Prev. 25:503–510. 2001.PubMed/NCBI
|
|
19
|
Nakahara M, Yokozaki H, Yasui W, Dohi K
and Tahara E: Identification of concurrent germline mutations in
hMSH2 and/or hMLH1 in Japanese hereditary nonpolyposis colorectal
cancer kindreds. Cancer Epidemiol Biomarkers Prev. 6:1057–1064.
1997.PubMed/NCBI
|
|
20
|
Park SJ, Lee KA, Park TS, Kim NK, Song J,
Kim BY and Choi JR: A novel missense MSH2 gene mutation in a
patient of a Korean family with hereditary nonpolyposis colorectal
cancer. Cancer Genet Cytogenet. 182:136–139. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Fan Y, Liu X, Zhang H, et al: Variations
in exon 7 of the MSH2 gene and susceptibility to gastrointestinal
cancer in a Chinese population. Cancer Genet Cytogenet.
170:121–128. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yan HL, Hao LQ, Jin HY, et al: Clinical
features and mismatch repair genes analyses of Chinese suspected
hereditary non-polyposis colorectal cancer: a cost-effective
screening strategy proposal. Cancer Sci. 99:770–780. 2008.
View Article : Google Scholar
|
|
23
|
Leung SY, Chan TL, Chung LP, et al:
Microsatellite instability and mutation of DNA mismatch repair
genes in gliomas. Am J Pathol. 153:1181–1188. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wu MS, Lee CW, Shun CT, et al: Distinct
clinicopathologic and genetic profiles in sporadic gastric cancer
with different mutator phenotypes. Genes Chromosomes Cancer.
27:403–411. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kim YM, Choe CG, Cho SK, Jung IH, Chang WY
and Cho M: Three novel germline mutations in MLH1 and MSH2 in
families with Lynch syndrome living on Jeju island, Korea. BMB Rep.
43:693–697. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhao C, Tang WZ, Gao F and Li W: Study on
hMSH2 gene mutation in the carcinogenesis of sporadic colorectal
carcinoma. Journal of Guangxi Medical University. 27:11–14.
2010.
|
|
27
|
Tang R, Hsiung C, Wang JY, et al: Germ
line MLH1 and MSH2 mutations in Taiwanese Lynch syndrome families:
characterization of a founder genomic mutation in the MLH1 gene.
Clin Genet. 75:334–345. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Jin HY, Liu X, Li VK, et al: Detection of
mismatch repair gene germline mutation carrier among Chinese
population with colorectal cancer. BMC Cancer. 8:442008. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Han HJ, Yuan Y, Ku JL, et al: Germline
mutations of hMLH1 and hMSH2 genes in Korean hereditary
nonpolyposis colorectal cancer. J Natl Cancer Inst. 88:1317–1319.
1996. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Sheng JQ, Fu L, Sun ZQ, et al: Mismatch
repair gene mutations in Chinese HNPCC patients. Cytogenet Genome
Res. 122:22–27. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Cui L, Jin HY, Cheng HY, Yan YD, Meng RG
and Yu DH: Genetic detection of Chinese hereditary nonpolyposis
colorectal cancer. World J Gastroenterol. 10:209–213.
2004.PubMed/NCBI
|
|
32
|
Furukawa T, Konishi F, Shitoh K, Kojima M,
Nagai H and Tsukamoto T: Evaluation of screening strategy for
detecting hereditary nonpolyposis colorectal carcinoma. Cancer.
94:911–920. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yuen ST, Chan TL, Ho JW, et al: Germline,
somatic and epigenetic events underlying mismatch repair deficiency
in colorectal and HNPCC-related cancers. Oncogene. 21:7585–7592.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhang CH, He YL, Wang FJ, et al: Detection
of hMSH2 and hMLH1 mutations in Chinese hereditary non-polyposis
colorectal cancer kindreds. World J Gastroenterol. 14:298–302.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zhang Y, Liu X, Fan Y, et al: Germline
mutations and polymorphic variants in MMR, E-cadherin and MYH genes
associated with familial gastric cancer in Jiangsu of China. Int J
Cancer. 119:2592–2596. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Yuan Y, Han HJ, Zheng S and Park JG:
Germline mutations of hMLH1 and hMSH2 genes in patients with
suspected hereditary nonpolyposis colorectal cancer and sporadic
early-onset colorectal cancer. Dis Colon Rectum. 41:434–440. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Yoon SN, Ku JL, Shin YK, et al: Hereditary
nonpolyposis colorectal cancer in endometrial cancer patients. Int
J Cancer. 122:1077–1081. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Sinn DH, Chang DK, Kim YH, et al:
Effectiveness of each Bethesda marker in defining microsatellite
instability when screening for Lynch syndrome.
Hepatogastroenterology. 56:672–676. 2009.PubMed/NCBI
|
|
39
|
Shin YK, Heo SC, Shin JH, et al: Germline
mutations in MLH1, MSH2 and MSH6 in Korean hereditary non-polyposis
colorectal cancer families. Hum Mutat. 24:3512004. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Zhang XM, Li JT, Zhu M, Wu XL, Gao P, Zhou
P and Wang YP: Study on the relationship between genetic
polymorphism Val384Asp in hMLH1 gene and the risk of four different
carcinomas. Zhonghua Liu Xing Bing Xue Za Zhi. 25:978–981. 2004.(In
Chinese).
|
|
41
|
Han HJ, Maruyama M, Baba S, Park JG and
Nakamura Y: Genomic structure of human mismatch repair gene, hMLH1,
and its mutation analysis in patients with hereditary non-polyposis
colorectal cancer (HNPCC). Hum Mol Genet. 4:237–242. 1995.
View Article : Google Scholar
|
|
42
|
Wang CF, Zhou XY, Sun MH, et al: Two novel
germline mutations of MLH1 in hereditary nonpolyposis colorectal
cancer family. Zhonghua Bing Li Xue Za Zhi. 35:68–72. 2006.(In
Chinese).
|
|
43
|
Sheng JQ, Chan TL, Chan YW, et al:
Microsatellite instability and novel mismatch repair gene mutations
in northern Chinese population with hereditary non-polyposis
colorectal cancer. Chin J Dig Dis. 7:197–205. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Wei W, Liu F, Liu L, et al: Distinct
mutations in MLH1 and MSH2 genes in hereditary non-polyposis
colorectal cancer (HNPCC) families from China. BMB Rep. 44:317–322.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Ng PC and Henikoff S: Predicting
deleterious amino acid substitutions. Genome Res. 11:863–874. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Kumar P, Henikoff S and Ng PC: Predicting
the effects of coding non-synonymous variants on protein function
using the SIFT algorithm. Nat Protoc. 4:1073–1082. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Liu HX, Cartegni L, Zhang MQ and Krainer
AR: A mechanism for exon skipping caused by nonsense or missense
mutations in BRCA1 and other genes. Nat Genet. 27:55–58. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Fackenthal JD, Cartegni L, Krainer AR and
Olopade OI: BRCA2 T2722R is a deleterious allele that causes exon
skipping. Am J Hum Genet. 71:625–631. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Gorlov IP, Gorlova OY, Frazier ML and Amos
CI: Missense mutations in hMLH1 and hMSH2 are associated with
exonic splicing enhancers. Am J Hum Genet. 73:1157–1161. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Gorlov IP, Gorlova OY, Frazier ML and Amos
CI: Missense mutations in cancer suppressor gene TP53 are
colocalized with exonic splicing enhancers (ESEs). Mutat Res.
554:175–183. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Nielsen KB, Sørensen S, Cartegni L, et al:
Seemingly neutral polymorphic variants may confer immunity to
splicing-inactivating mutations: a synonymous SNP in exon 5 of MCAD
protects from deleterious mutations in a flanking exonic splicing
enhancer. Am J Hum Genet. 80:416–432. 2007. View Article : Google Scholar
|
|
52
|
Cartegni L and Krainer AR: Disruption of
an SF2/ASF dependent exonic splicing enhancer in SMN2 causes spinal
muscular atrophy in the absence of SMN1. Nat Genet. 30:377–384.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Auclair J, Busine MP, Navarro C, et al:
Systematic mRNA analysis for the effect of MLH1 and MSH2 missense
and silent mutations on aberrant splicing. Hum Mutat. 27:145–154.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Pagenstecher C, Wehner M, Friedl W, et al:
Aberrant splicing in MLH1 and MSH2 due to exonic and intronic
variants. Hum Genet. 119:9–22. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Oliveira J, Soares-Silva I, Fokkema I, et
al: Novel synonymous substitution in POMGNT1 promotes exon skipping
in a patient with congenital muscular dystrophy. J Hum Genet.
53:565–572. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Raponi M, Kralovicova J, Copson E, et al:
Prediction of single-nucleotide substitutions that result in exon
skipping: identification of a splicing silencer in BRCA1 exon 6.
Hum Mutat. 32:436–444. 2011. View Article : Google Scholar : PubMed/NCBI
|