Introduction
miRNAs are small [20–24 nucleotides (nt)] non-coding
RNA gene products that have become known as important regulators of
various cellular processes by post-transcriptionally modulating
gene expression (1,2). Currently, more than several hundred
unique mature human miRNAs are known (3). miRNAs have been reported to be
involved in tumorigenesis acting as oncogenes (4–6) or
suppressors (7,8). Several other reports have described
altered expression of miRNAs in cancer tissues compared to normal
tissues, suggesting that these miRNAs could potentially represent
novel clinical and prognostic markers (9–12). In
particular, miR-155, which is considerd as an oncomiR, has been
found to be up-regulated in B cell lymphoma and carcinoma of head
and neck, breast, lung, pancreas, kidney and colon (9,13–17).
To date, several studies have demonstrated the
association of elevated miR-155 with late stage and poor overall
survival in several types of malignancy (9,18). It
is well known that the prognosis of breast cancer patients is
closely related with the clinicopathological features including
tumor size, histological grade and the presence of lymph node or
distant metastases (19,20). These clinicopathological features
alone or in combination, enable the identification of individuals
who are at increased risk of dying of breast cancer and also who
may benefit from aggressive treatment (21). However, there is rare evidence
indicating the correlation of miR-155 expression with
clinicopathological features in human breast carcinoma, which may
help us understand the role of miR-155 in this disease.
Therefore, in this study, we used the real-time
RT-PCR method to detect the expression of miR-155, which we
correlated with clinicopathological features in 42 infiltrating
ductal carcinomas and 3 infiltrating lobular carcinomas.
Additionally, in order to analyze the effect of miR-155 on cell
growth and to investigate the potential mechanism of miR-155 in
breast cancer, we utilized the antisense technique to inhibit
miR-155 expression in vitro.
Materials and methods
Cell line and breast tumor specimens
The breast cancer cell line (HS578T) was obtained
from ATCC and grown according to ATCC recommended culture
conditions. Fresh samples from 45 cases of human breast cancer and
paired normal adjacent tissues (>5 cm from cancer tissue) were
obtained from the Department of Oncology, The First Affiliated
Hospital of Wenzhou Medical College (Wenzhou, China) between
February 2009 and June 2009. The samples used were not subjected to
preoperative radiotherapy and/or chemotherapy and all patients were
treated by modified radical mastectomy. Before RNA extraction,
sections were stained with H&E for histological diagnosis and
tumor cell evaluation. Only those cases with a population of at
least 70% tumor cells in the section were used in this study.
Tissues were preserved by snap-freeze and stored at −80°C. The
diagnosis and histological grade of each case were independently
confirmed by two pathologists based on the WHO classification. The
clinical stage was classified according to the American Joint
Committee on Cancer (AJCC) tumor-lymph node-metastasis (TNM)
classification system (22). The
relationship between the clinicopathological characteristics of the
patients and miR-155 expression are summarized in Table II.
 | Table IIRelationship between the miR-155
expression level and clinicopathological parameters of breast
cancer. |
Table II
Relationship between the miR-155
expression level and clinicopathological parameters of breast
cancer.
| Variable | N | Relative expression
of miR-155a | P-valueb |
|---|
| Menstrual
status | | | 0.640 |
| Premenopausal | 20 | 0.365
(0.333–0.421) | |
|
Post-menopausal | 25 | 0.355
(0.308–0.403) | |
| Tumor size
(cm)c | | | 0.530 |
| ≤2 | 19 | 0.356
(0.316–0.401) | |
| >2 | 26 | 0.361
(0.334–0.420) | |
| TNM stage | | | 0.002 |
| I | 8 | 0.316
(0.286–0.350) | |
| II | 24 | 0.358
(0.332–0.416) | |
| III | 13 | 0.417
(0.367–0.446) | |
| ER status | | | 0.041 |
| Positive | 30 | 0.367
(0.349–0.424) | |
| Negative | 15 | 0.318
(0.299–0.401) | |
| PR status | | | 0.029 |
| Positive | 26 | 0.398
(0.354–0.423) | |
| Negative | 19 | 0.335
(0.313–0.399) | |
| Her-2 status | | | 0.647 |
| Positive | 14 | 0.398
(0.313–0.420) | |
| Negative | 31 | 0.359
(0.337–0.419) | |
| Lymph node
status | | | 0.034 |
| Metastasis | 20 | 0.383
(0.355–0.437) | |
| No metastasis | 25 | 0.355
(0.314–0.399) | |
| Proliferation index
(Ki-67) (%) | | | 0.019 |
| ≤10 | 21 | 0.353
(0.364–0.398) | |
| >10 | 24 | 0.387
(0.355–0.437) | |
| Pathologic
type | | | 0.559 |
| Ductal | 42 | 0.360
(0.331–0.420) | |
| Lobular | 3 | 0.330
(0.277–0.442) | |
Synthesis of miR-155 ASO sequences and
transfection
The mature miRNA sequences are available from the
miRNA registry. The sequences of miRNA ASO were designed, according
to the principle of sequences complementary to the mature mRNA. The
ASO and the scrambled negative control (SCR) sequences used in this
study are listed in Table I. Both
of them were chemically synthesized and 2′-OMe modified by Shanghai
GenePharma Co., Ltd. (Shanghai, China) and stored at −20°C.
Twenty-four hours before transfection, HS578T cells in the
exponential phase of growth were seeded in 96- or 6-well plates
(Costar) and allowed to grow overnight. The cells were then
transfected with oligonucleotides using Lipofectamine™ 2000 reagent
(Invitrogen) in Opti-MEM I for 6 h. Transfection complexes were
prepared according to the manufacturer’s instructions. At the end
of transfection, the cells were incubated in medium containing 10%
fetal calf serum (FCS). Transfection efficiency was detected by
flow cytometry.
 | Table ISequences of oligonucleotide and
primers for the analysis of miR-155 expression. |
Table I
Sequences of oligonucleotide and
primers for the analysis of miR-155 expression.
| Gene name | Sequence
(5′→3′) |
|---|
| miR-155 ASO |
ACCCCUAUCAAGAUUAGCAUUAA |
| 5′FAM SCR |
CAGUACUUUUGUGUAGUACAA |
| Stem-loop RT
primer |
CTCAACTGGTGTCGTGGGGCAATTCAGTTGAGCCCCTATC |
| miR-155-F |
TGCCTCCAACTGACTCCTAC |
| miR-155-R |
GCGAGCACAGAATAATACGAC |
| U6 snRNA-F |
CTCGCTTCGGCAGCACA |
| U6 snRNA-R |
AACGCTTCACGAATTTGCGT |
Total RNA extraction and real-time PCR
for quantitative analysis of miR-155
HS578T cells were incubated in 6-well plates and
transfected with 75 nM oligonucleotides using the Lipofectamine
2000 reagent for 24 h. The same process was followed as described
above. Total RNA from the tissues or treated HS578T cells was
isolated using TRIzol reagent (Invitrogen) according to the
manufacturer’s instructions. RNA purity and concentration were
controlled by UV spectrophotometry (A260:A280>1.8).
The primers for the analysis of miR-155 expression
were designed by the Shanghai Sangon Biological Engineering
Technology and Services Co., Ltd. (Shanghai, China) and are
summarized in Table I. Mixtures of
1 μg of total RNAs together with 50 nM reverse primer, 2 units of
RNAase inhibitor (Takara Bio), 5 units of M-MLV reverse
transcriptase (Takara Bio) and 0.5 μM dNTP were used for each RT
reaction. The reaction parameters were incubation at 25°C for 10
min, 42°C for 60 min, 52°C for 15 min, 70°C for 15 min and then a
hold at −20°C. At the same time, to generate the cDNA template for
the endogenous control PCR reactions, first strand cDNA was
synthesized using 1 μg of RNA from the same samples for stem-loop
reverse transcription and oligo(dT) as the primer. The reaction
parameters were incubation at 42°C for 30 min, 70°C for 15 min and
then a hold at −20°C.
The qPCR was performed on the Applied Biosystems
7500 detection system. For quantitation of miR-155, the 25 μl PCR
included 1 μl of the RT product of miR-155, 1X SYBR-Green I
Mastermix (Toyobo Co., Ltd.), 0.5 μM specific forward primer of
miR-155 and 0.5 μM reverse primer. For the endogenous control, U6
snRNAs, 1 μl of cDNAs synthesized by using oligo(dT) were used as a
template. The reaction parameters were incubation at 95°C for 10
min, then 40 cycles of 95°C for 15 sec and 60°C for 1 min. To
calculate the relative concentration, miR-155 and U6 CT values for
all samples were obtained. A normalized expression for each sample
was obtained by dividing the Ct value of miR-155 by the same
sample’s U6 Ct and designated as ΔCt. This value was then
transformed by the formula 2−ΔCt. Furthermore, the
(ΔΔCt) method was used in comparing miR-155 expression in each
group of treated HS578T cells or matched non-tumor tissue to cancer
tissues.
Cell viability and apoptosis assays
The effect of miR-155 ASO on HS578T cell viability
was determined by the
2-(2-methoxy-4-nitrophenyl)-3-(4-nitrophenyl)-5-(2,4-disulfo-phenyl)-2H-tetrazolium,
monosodium salt (WST-8) assay kit (CCK-8, Dojindo, Kumamoto,
Japan). Twenty-four hours before transfection, 1×104
HS578T cells/well were seeded in 96-well plates and allowed to grow
overnight. The cells were then transfected with three different
concentrations of miR-155 ASO (25, 50 and 75 nM) and the highest
concentration of SCR siRNA (75 nM) using Lipofectamine 2000
according to the manufacturer’s protocol. After 24 h, WST-8 was
added into each well for 1 h before the measurement according to
the manufacturer’s instructions. The absorbance at 450 nm was
measured by a microplate reader. The following formula was used for
calculating the inhibition rate. Inhibition rate = (1-absorbance of
treated cells/control cells) × 100% (23). The apoptosis assay was performed
with the Annexin V-FITC/PI Apoptosis Detection kit (Roche). HS578T
cells were transfected with miR-155 ASO or SCR siRNA as previously
described for 6 h, and incubated in medium containing 10% FCS for
another 24 h in 6-well plates. Cells were collected and
double-stained with FITC-conjugated Annexin V and propidium iodide
(PI). For each sample, data from approximately 1×104
cells were recorded in the list mode on logarithmic scales.
Apoptosis and necrosis were analyzed by quadrant statistics on
PI-negative, Annexin V-positive cells and both positive cells,
respectively.
Statistical analysis
For all data, statistical analysis was performed in
SPSS 17.0 for Windows (SPSS, Inc.). The miR-155 expression levels
were characterized by their median and ranges from the 25th to the
75th percentile. The Wilcoxon test was used for comparing two
paired groups (tumor and paired non-tumor), the Mann-Whitney U test
for two independent groups and the Kruskall-Wallis test for three
independent groups (relationship between miR-155 expression level
and TNM stage of breast cancer). The one-way ANOVA test was
performed to investigate the differences in the obtained results of
the WST-8 array and apoptosis analysis. All tests were two-tailed
and the significance level was set at P<0.05.
Results
Expression of miR-155 in tumor tissue and
matched non-tumor tissue
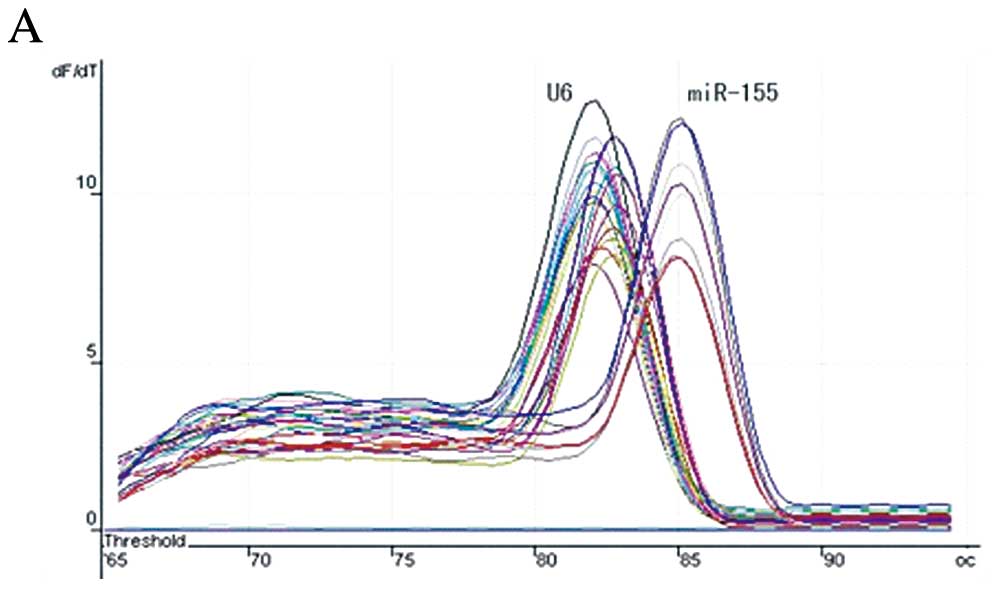
The melting-curves of miR-155 and U6 snRNA were
sharply defined curves with a narrow peak (Fig. 1A), indicating that pure, homogeneous
PCR products were produced. As shown in the amplification curves
(Fig. 1B), the Ct value of miR-155
in tumor tissue was lower than that of miR-155 in non-tumor tissue,
which means the expression level of miR-155 in tumor samples was
higher than that in the controls. The median of the relative
expression of miR-155 (2−ΔΔCt) was 0.360 (25th-75th
percentile, 0.328–0.420) in tumor samples, with that in non-tumor
control samples set at 0.135 (25th–75th percentile, 0.003–0.199)
(Fig. 1C). The difference of
expression of miR-155 between the tumor and the control samples was
statistically significant (P<0.001, Wilcoxon test).
Association between miR-155 expression
level and clinicopathological parameters
The up-regulated expression of miR-155 was
associated with advanced clinical TNM stage (P=0.002,
Kruskall-Wallis test), lymph node positivity (P=0.034, Mann-Whitney
U test) and high proliferation index (Ki-67 >10%) (P=0.019,
Mann-Whitney U test). Furthermore, the miR-155 expression levels
were closely related to ER and PR status (P=0.041 and 0.029,
respectively, by the Mann-Whitney U test). However, no significant
relationship was found between the expression of miR-155 and the
menstrual status (P=0.640), size of primary tumor (P=0.530), Her-2
status (P=0.647) and pathologic type (P=0.559) using the
Mann-Whitney U test (Table
II).
miR-155 ASO down-regulation of miRNA
expression
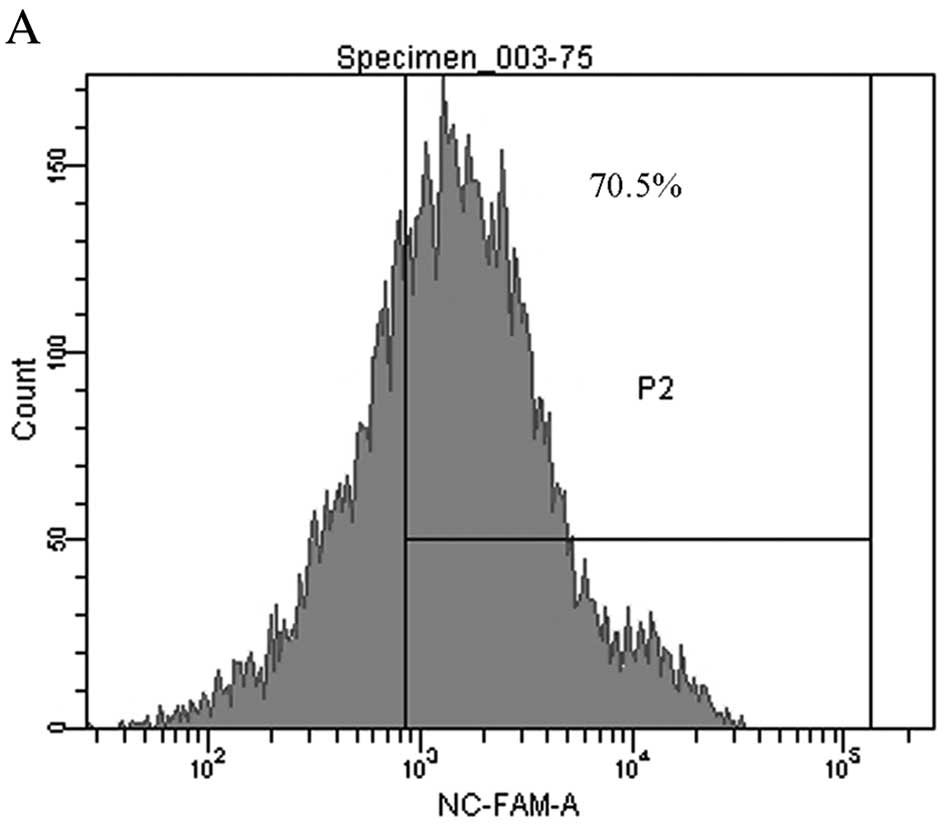
Eight hours after transfection with 75 nM 5′FAM SCR,
the transfection efficiency was detected by flow cytometry
(Fig. 2A).To validate whether
miR-155 ASO decreased miR-155 levels in treated HS578T cells,
miR-155 and U6 snRNA expression was determined by real-time RT-PCR.
The fold change for the miR-155 expression level was calculated
using the 2−ΔΔCt method. As shown in Fig. 2B, the 2−ΔΔCt value of
HS578T cells treated with anti-miR-155 was 0.052, which showed that
the level of miR-155 in HS578T cells was down-regulated by miR-155
ASO.
miR-155 ASO inhibition of HS578T cells
viability
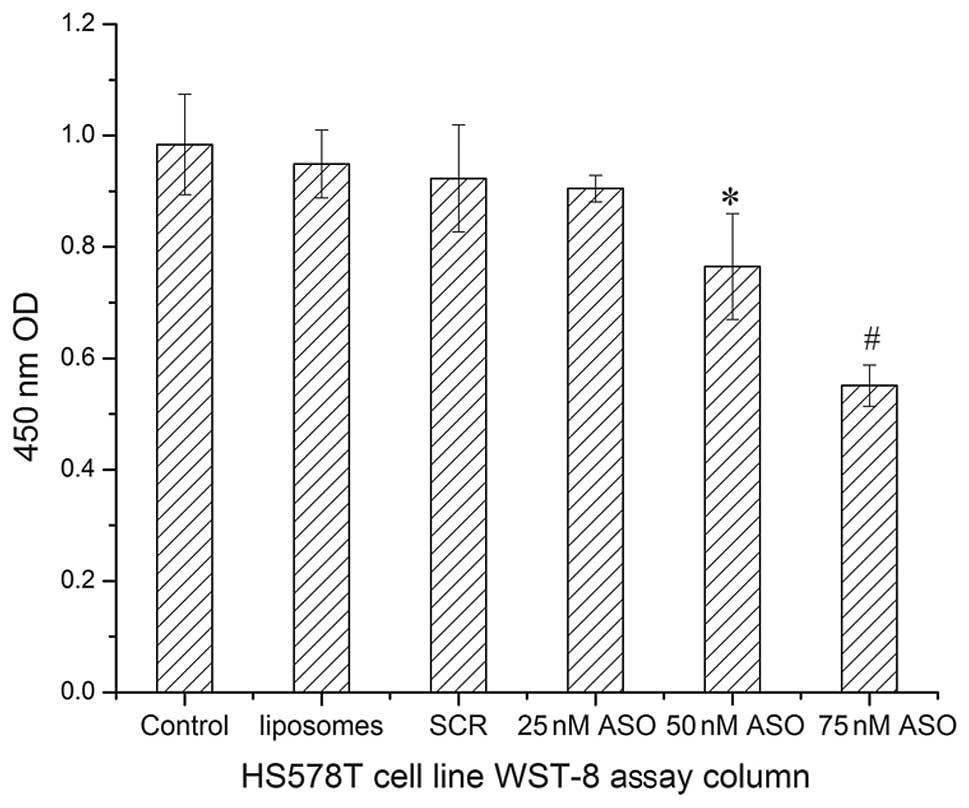
In this study, we determined the influence of
miR-155 ASO on cell viability by WST-8 assay. Optical densities at
450 nm were obtained for 4 groups: 0.984±0.090 for control group,
0.949±0.061 for liposomes group, 0.923±0.096 for SCR group and
0.905±0.024, 0.765±0.095 and 0.551±0.037, respectively for 25, 50
and 75 nM concentrations of miR-155 ASO (Fig. 3). These results demonstrated that
the optical density and therefore the cell viability was similar in
the control, SCR, 25 nM miR-155 ASO and liposome groups (P=0.237,
one-way ANOVA test). However, there were significant differences
between the optical density in the 50 nM miR-155 ASO and 75 nM
miR-155 ASO group and the control group (P=0.000 for both by
one-way ANOVA). The optical density and cell viability gradually
decreased with the increase of miR-155 ASO concentration
(0.905±0.024, 0.765±0.095 and 0.551±0.037, respectively). At 75 nM
concentration of miR-155 ASO, the optical density and cell
viability were nearly half of these parameters in the control
group. These data indicate that a higher concentration of miR-155
ASO had a higher toxicity effect on HS578T cells and could decrease
the cell viability and proliferation.
miR-155 ASO promotion of HS578T cell
apoptosis
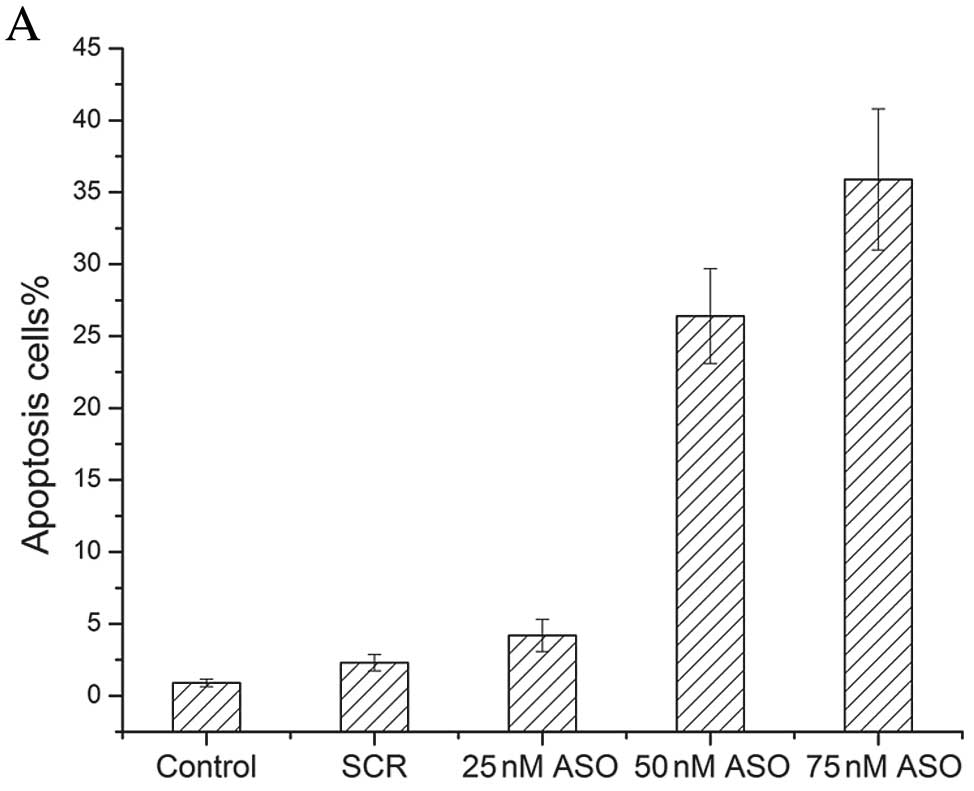
To explore the effects of miR-155 ASO on cell
apoptosis, miR-155 ASO treatment was investigated in HS578T cells.
Apoptotic HS578T cells were detected by double staining with
Annexin V and PI. The results demonstrated that miR-155 ASO could
induce cell apoptosis. Along with the increase of concentration of
miR-155 ASO, the apoptosis rate of HS578T cells gradually
increased. The double-stained images are shown in Fig. 4.
Discussion
As previous research has indicated, miR-155 acts as
a multifunctional miRNA in many pathophysiological process, such as
immunology (24,28), inflammation (25,26),
hematopoiesis (27),
angiocardiopathy (28) and
carcinogenesis. Interestingly, miR-155 may act as a bridge between
inflammation and malignancy, which may provide new insight in
carcinogenesis (29–31). miR-155 is one of the most prominent
miRNAs involved in tumor development and progression. Diverse
studies have shown that miR-155 is overexpressed in various tumor
types. Not surprisingly, it was found to be significantly
up-regulated in breast cancer in both our and previous other
studies (15,29,32).
However, there is rare evidence focused on the relationship between
miR-155 expression and clinicopathological features in breast
cancer.
In the first stage of our study, we detected the
expression of miR-155 in all 45 carcinomas and paired non-tumor
tissues by real-time RT-PCR. The amplification and melting curve,
as shown in Fig. 1, indicate the
method is specific and sensitive enough for detection of miR-155.
Furthermore, to reduce the error caused by gene expression
differences between different individuals, we used matched
non-tumor tissue as control and used 2−ΔΔCt to represent
the level of miR-155 expression in tumors relative to matched
non-tumor samples. As expected, our results confirmed that miR-155
expression was significantly up-regulated in breast cancer compared
with the matched non-tumor tissue.
Our study next focused on the potential correlation
between the expression level of miR-155 and various breast cancer
clinicopathological characteristics. The data showed that high
levels of miR-155 appear to be significantly correlated with
advanced clinical stage, lymph node metastases, and higher
proliferation index (Ki-67 >10%). All of them are well known as
poor prognostic factors of breast cancer patients (33–35).
Interestingly, we next found that patients with ER-positive or
PR-positive tumors have higher miR-155 expression levels than those
that are ER-negative or PR-negative. This is in part consistent
with the findings of Zhu et al (36) and Lu and Tsourkas (37), whose experimental material was
either serum or cells but not tissues. However, it should be noted
that both ER and PR are protective factors of patients with breast
cancer (38–40). Therefore, the representation of the
oncomiR-like miR-155 is contradictory to the protective function of
ER and PR. And we may hypothesize that it is miR-155 that results
in the aggressive behavior in carcinomas that ER- or PR-positive.
These results indicate that, as an independent risk factor, miR-155
could serve as a prognostic marker for survival of breast cancer
patients.
The precise molecular mechanisms behind the altered
expression of miR-155 in breast cancer remain poorly understood. To
our knowledge, this is the first report to describe the
significance of miR-155 to the clinical stage, lymph node
metastasis, hormone receptor status of breast cancer patients. It
was recently reported that miR-155 mediates lymphoblastoid cell
lines (LCLs); suppression of this miRNA, which is highly expressed
in LCLs, was associated with decreased cell proliferation and
increased apoptosis (41). In solid
tumors, such as breast cancer and lung cancer, similar findings of
the function for miR-155 were obtained (29,42).
Furthermore, Kong et al (15) discovered that miR-155 is a critical
therapeutic target and is closely related with chemosensitivity in
breast cancer. Two recent studies reported two additional direct
miR-155 targets, FOXO3a (15) and
suppressor of cytokine signaling 1 (29), respectively, both of which function
as protective factors in breast cancer patients, demonstrating that
this miRNA acts as an oncomiR in breast cancer.
Therefore, in this study, we utilized miR-155 ASO
for repression of HS578T cell growth and proliferation.
Transfection of HS578T cells with either miR-155 ASO or SCR were
performed successfully with at least 70% efficiency. The results of
real-time PCR suggested that the synthesized miR-155 ASO could
effectively down-regulate this miRNA expression. The WST-8 assay
was performed and the results indicated that although 25 and 50 nM
of miR-155 ASO have a toxic effect on HS578T cells, 75 nM of
miR-155 ASO could strongly repress tumor cell proliferation.
Moreover, apoptosis analysis demonstrated that the apoptosis rates
of the group of 50 and 75 nM of miR-155 ASO were significantly
higher than the other three groups.
In conclusion, we demonstrated that overexpression
of miR-155, one of the most significantly altered miRNAs in breast
cancer, is related to clinical stage, lymph node metastasis, higher
Ki-67 and hormone receptor status of breast cancer patients.
Although the precise molecular mechanism of the ectopic expression
of miR-155 in breast cancer requires further clarification, our
data suggest that miR-155 may be a promising candidate as a
molecular biomarker and a potential therapeutic target for breast
cancer intervention.
Acknowledgements
This study was supported by grants from the Wenzhou
Science and Technology Bureau (Y20080081, Y20100008).
References
|
1
|
Ambros V: MicroRNA pathways in flies and
worms: growth, death, fat, stress, and timing. Cell. 113:673–676.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bartel DP: MicroRNAs: target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Griffiths-Jones S, Grocock RJ, van Dongen
S, Bateman A and Enright AJ: miRBase: microRNA sequences, targets
and gene nomenclature. Nucleic Acids Res. 34:D140–D144. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
He L, Thomson JM, Hemann MT, et al: A
microRNA polycistron as a potential human oncogene. Nature.
435:828–833. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Tam W and Dahlberg JE: miR-155/BIC as an
oncogenic microRNA. Genes Chromosomes Cancer. 45:211–212. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Papagiannakopoulos T, Shapiro A and Kosik
KS: MicroRNA-21 targets a network of key tumor-suppressive pathways
in glioblastoma cells. Cancer Res. 68:8164–8172. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hammond SM: MicroRNAs as tumor
suppressors. Nat Genet. 39:582–583. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Tazawa H, Tsuchiya N, Izumiya M and
Nakagama H: Tumor-suppressive miR-34a induces senescence-like
growth arrest through modulation of the E2F pathway in human colon
cancer cells. Proc Natl Acad Sci USA. 104:15472–15477. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yanaihara N, Caplen N, Bowman E, et al:
Unique microRNA molecular profiles in lung cancer diagnosis and
prognosis. Cancer Cell. 9:189–198. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Mattie MD, Benz CC, Bowers J, et al:
Optimized high-throughput microRNA expression profiling provides
novel biomarker assessment of clinical prostate and breast cancer
biopsies. Mol Cancer. 5:242006. View Article : Google Scholar
|
|
11
|
Chen Y and Stallings RL: Differential
patterns of microRNA expression in neuroblastoma are correlated
with prognosis, differentiation, and apoptosis. Cancer Res.
67:976–983. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ru Y, Dancik GM and Theodorescu D:
Biomarkers for prognosis and treatment selection in advanced
bladder cancer patients. Curr Opin Urol. 21:420–427. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Eis PS, Tam W, Sun L, et al: Accumulation
of miR-155 and BIC RNA in human B cell lymphomas. Proc Natl Acad
Sci USA. 102:3627–3632. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wald AI, Hoskins EE, Wells SI, Ferris RL
and Khan SA: Alteration of microRNA profiles in squamous cell
carcinoma of the head and neck cell lines by human papillomavirus.
Head Neck. 33:504–512. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kong W, He L, Coppola M, Guo J, Esposito
NN, Coppola D and Cheng JQ: MicroRNA-155 regulates cell survival,
growth, and chemosensitivity by targeting FOXO3a in breast cancer.
J Biol Chem. 285:17869–17879. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Volinia S, Calin GA, Liu CG, et al: A
microRNA expression signature of human solid tumors defines cancer
gene targets. Proc Natl Acad Sci USA. 103:2257–2261. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Juan D, Alexe G, Antes T, et al:
Identification of a microRNA panel for clear-cell kidney cancer.
Urology. 75:835–841. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Du ZM, Hu LF, Wang HY, Yan LX, Zeng YX,
Shao JY and Ernberg I: Upregulation of MiR-155 in nasopharyngeal
carcinoma is partly driven by LMP1 and LMP2A and downregulates a
negative prognostic marker JMJD1A. PLoS One. 6:e191372011.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Bundred NJ: Prognostic and predictive
factors in breast cancer. Cancer Treat Rev. 27:137–142. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Devi KR, Kuruvila S and Musa MM:
Pathological prognostic factors in breast carcinoma. Saudi Med J.
21:372–375. 2000.PubMed/NCBI
|
|
21
|
Wiseman SM, Makretsov N, Nielsen TO, et
al: Coexpression of the type 1 growth factor receptor family
members HER-1, HER-2, and HER-3 has a synergistic negative
prognostic effect on breast carcinoma survival. Cancer.
103:1770–1777. 2005. View Article : Google Scholar
|
|
22
|
Singletary SE, Allred C, Ashley P, et al:
Staging system for breast cancer: revisions for the 6th edition of
the AJCC Cancer Staging Manual. Surg Clin North Am. 83:803–819.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhang RG, Zhang RP, Wang XW and Xie H:
Effects of cisplatin on telomerase activity and telomere length in
BEL-7404 humanhepatoma cells. Cell Res. 12:55–62. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
O’Connell RM, Taganov KD and Boldin MP:
MicroRNA-155 is induced during the macrophage inflammatory
response. Proc Natl Acad Sci USA. 104:1604–1609. 2007.PubMed/NCBI
|
|
25
|
O’Connell RM, Kahn D, Gibson WS, et al:
MicroRNA-155 promotes autoimmune inflammation by enhancing
inflammatory T cell development. Immunity. 33:607–619. 2010.
|
|
26
|
Bhattacharyya S, Balakathiresan NS,
Dalgard C, et al: Elevated miR-155 promotes inflammation in cystic
fibrosis by driving hyperexpression of interleukin-8. J Biol Chem.
286:11604–11615. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
O’Connell RM, Rao DS, Chaudhuri AA, et al:
Sustained expression of microRNA-155 in hematopoietic stem cells
causes a myeloproliferative disorder. J Exp Med. 205:585–594.
2008.PubMed/NCBI
|
|
28
|
Urbich C, Kuehbacher A and Dimmeler S:
Role of microRNAs in vascular diseases, inflammation, and
angiogenesis. Cardiovasc Res. 79:581–588. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Jiang S, Zhang HW, Lu MH, et al:
MicroRNA-155 functions as an OncomiR in breast cancer by targeting
the suppressor of cytokine signaling 1 gene. Cancer Res.
70:3119–3127. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Pedersen IM, Otero D, Kao E, Miletic AV,
et al: Onco-miR-155 targets SHIP1 to promote TNFalpha-dependent
growth of B cell lymphomas. EMBO Mol Med. 1:288–295. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Tili E, Croce CM and Michaille JJ:
miR-155: on the crosstalk between inflammation and cancer. Int Rev
Immunol. 28:264–284. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Iorio MV, Ferracin M, Liu CG, et al:
MicroRNA gene expression deregulation in human breast cancer.
Cancer Res. 65:7065–7070. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wrba F, Reiner A, Markis-Ritzinger E,
Holzner JH, Reiner G and Spona J: Prognostic significance of
immunohistochemical parameters in breast carcinomas. Pathol Res
Pract. 183:277–283. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Carter CL, Allen C and Henson DE: Relation
of tumor size, lymph node status, and survival in 24,740 breast
cancer cases. Cancer. 63:181–187. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
De Azambuja E, Cardoso F, de Castro G Jr,
et al: Ki-67 as prognostic marker in early breast cancer: a
meta-analysis of published studies involving 12,155 patients. Br J
Cancer. 96:1504–1513. 2007.PubMed/NCBI
|
|
36
|
Zhu W, Qin W, Atasoy U and Sauter ER:
Circulating microRNAs in breast cancer and healthy subjects. BMC
Res Notes. 2:892009. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Lu J and Tsourkas A: Imaging individual
microRNAs in single mammalian cells in situ. Nucleic Acids Res.
37:e1002009. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Lee YT and Markland FS: Steroid receptors
study in breast carcinoma. Med Pediatr Oncol. 5:153–166. 1978.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Paone JF, Abeloff MD, Ettinger DS, Arnold
EA and Baker RR: The correlation of estrogen and progesterone
receptor levels with response to chemotherapy for advanced
carcinoma of the breast. Surg Gynecol Obstet. 152:70–74.
1981.PubMed/NCBI
|
|
40
|
Clark GM and McGuire WL: Prognostic
factors in primary breast cancer. Breast Cancer Res Treat.
3(Suppl): S69–S72. 1983. View Article : Google Scholar
|
|
41
|
Linnstaedt SD, Gottwein E, Skalsky RL,
Luftig MA and Cullen BR: Virally induced cellular microRNA miR-155
plays a key role in B-cell immortalization by Epstein-Barr virus. J
Virol. 84:11670–11678. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Qin A, Zhou Y, Sheng M, Fei G, Ren T and
Xu L: Effects of microRNA-155 on the growth of human lung cancer
cell line 95D in vitro. Zhongguo Fei Ai Za Zhi. 14:575–580.
2011.PubMed/NCBI
|