Introduction
The incidence and mortality of breast cancer has
been rising in women over the past 15 years, and breast cancer has
ranked first in women in China (1).
Most of the treatment failures and deaths of breast cancer,
particularly the more invasive hormone-dependent cases, were caused
by tumor metastasis (2). Signal
transducer and activator of transcription-3 (Stat3) is known to
have frequently been aberrantly activated in a large percentage of
breast cancers, and has been involved in numerous tumor biological
processes, including cell proliferation, survival, tumor
angiogenesis, invasion and migration (3,4).
Evidences supports that selectively suppressing constitutive Stat3
signaling is an effective approach in inhibiting cancer-associated
processes and cancer progression (5,6). It
has been reported that primary breast tumor growth and brain
metastasis could be suppressed by Stat3 inhibition (7). The established strategies targeting
Stat3 include tyrosine kinase inhibitors (e.g. tyrphostin AG490),
antisense oligonucleotides, decoy oligonucleotides and dominant
negative Stat3 protein (8–13). In recent years, gene silencing by
small interference RNA (siRNA) is a well-known and promising
approach used to repress target gene expression (14,15).
However, as it is prone to degradation and has poor cellular
permeability, naked siRNA cannot enter the intracellular
environment where biological function of siRNA molecules occurs.
Recently, various siRNA-delivery systems, liposomes, polymers,
peptides (16) and membrane
penetrating peptides (MPPs) have been highlighted. MPPs rich in
basic amino acids, such as arginine and lysine, key motifs for the
efficient delivery of extracellular molecules into cells are
attractive peptide-mediated delivery systems (17).
PR39, a porcine cathelicidin is rich in proline
(49%) and arginine (26%) and has been shown to be involved in
antimicrobial activities (18),
suppressing DNA and protein synthesis (19,20).
In addition to versatile biological functions, there is evidence
that PR39 could penetrate cell membranes rapidly and interact with
the SH3 domains of p47phox (21), p130Cas (22), and the α7 subunit of the 20S
proteasome (23) because of its
proline and arginine rich composition. Therefore, we hypothesized
that PR39 could be exploited as a novel MMP, a carrier which would
deliver siRNA into cells cytoplasm to knock down target gene
expression.
In this study, we utilized PR39 to deliver siRNA
selectively silencing Stat3 in a mouse breast cancer cell line,
4T1, to repress Stat3 expression as well as downstream components
of the Stat3 pathway. To our knowledge, this study provides the
first evidence that PR39, as a vector, plays an important role in
delivery of siRNA into 4T1 cells.
Materials and methods
Synthesis of PR39 and siRNA
Full length PR39 (RRRPR
PPYLPRPRPPPFFPPRLPPRIPPGFPPRFPPRFP-NH2 and FITC-PR39
(FITC-RRRPRPPYLPRPRPPPFFPPRLPPR IPPGFPPRFPPRFP-NH2) were
prepared by Shanghai Sangon Biological Engineering Technology and
Services Corporation (Shanghai, China). The synthetic peptide PR39
was at least 95% pure by high performance liquid chromatography
(HPLC). PR39 was dissolved in phosphate-buffered saline (PBS, pH
7.4) to appropriate concentrations (4 mM). The small interference
RNA oligo used for Stat3 gene silencing (sense:
5′-GGACGACUUUGAUUUCAACtt-3′, antisense: 5′-GUU
GAAAUCAAAGUCGUCCtg-3′) (24) and
negative control (sense: 5′-UUCUCCGAACGUGUCACGUTT-3′, antisense:
5′-ACGUGACACGUUCGGAGAATT-3′) were purchased from Shanghai
GenePharma Co., Ltd. (Shanghai, China). 5′Cy3-labeled siRNA was
ordered from Guangzhou Ribobio Co., Ltd. (Guangzhou, China). The
siRNA was dissolved in sterilized distilled H2O to the
concentration of 20 μM.
Cell lines and antibodies
The 4T1 cells supplied by our laboratory were
cultured in Dulbecco’s modified Eagle’s medium (DMEM, high glucose,
Gibco, USA) with 10% fetal bovine serum (HyClone, USA) at 37°C in a
5% CO2 atmosphere (Heraeus, Germany). Anti-Stat3 was
purchased from Becton-Dickinson, USA; anti-MMP-9 was obtained from
Bioworld (USA); anti-β-actin was from Santa Cruz Biotechnology,
Inc. (USA); HRP-conjugated secondary antibody was purchased from
Zhongshan Golden Bridge Biotechnology Co., Ltd. (China).
Preparation of PR39/siRNA complex and gel
shift assay
The Stat3 scrambled siRNA was diluted with
appropriate PBS to a concentration of 100 nM, and PR39 was added
into PBS. To determine the optimal relative concentration at which
the PR39/siRNA complex formation would be advanced, 100 nM of siRNA
was incubated with different amounts of PR39 at 25°C for 15 min in
PBS, with the siRNA/PR39 concentration ratio ranging from 0:1 to
1:90. The Scr siRNA/PR39 complexes were detected by electrophoresis
on a 2% agarose gel (Invitrogen Life Technologies, USA) with
GoldenView in TBE buffer at 180 V for 15 min. Then, stained bands
were visualized under UV light and photographed using the Bio-Rad
Chemi Doc XRS imaging system (Bio-Rad, USA).
Observation of labeled PR39/siRNA
complexes by fluorescence microscope
4T1 cells were seeded on 10-mm cover slips with
5×104 cells/cover slip 18 h before transfection with the
5′Cy3-siRNA. The 5′Cy3-siRNA (2.5 μl) was diluted into 50 μl
serum-free DMEM. PR39 (1.125 μl) was diluted in 50 μl serum-free
DMEM, mixed gently and incubated for 5 min at room temperature.
5′Cy3-siRNA was combined with the diluted PR39. They were mixed
gently and incubated for 30 min at room temperature to form a
complex. This complex was added to the cells with 400 μl serum-free
DMEM. Then 4T1 cells were rinsed thrice with PBS after 6 h of
transfection and fixed in methanol for 20 min at −20°C. The cells
were washed twice with PBS and DAPI fluorescent stain (Beyotime,
China) which was applied to the cells for 5 min at room
temperature. The cells were rinsed three times with PBS and
observed under a fluorescence microscope (Nikon, Japan).
Transient transfection of 4T1 cells with
Stat3 siRNA
Stat3 siRNA and scrambled control were transfected
with PR39 (concentration ratio 1:90) or Lipofectamine 2000 reagent
(Lipo 2000; Invitrogen, USA). All steps were performed following
the manufacturer’s instructions. The 4T1 cells suspended in
complete DMEM medium were seeded at 6×105 cells/well in
6-well plates (Costar, USA) and allowed to attach overnight before
transfection. The siRNA (10 μl) was diluted into 250 μl serum-free
DMEM. PR39 (4.5 μl) or Lipo 2000 (5 μl) was diluted in 250 μl
serum-free DMEM, mixed gently and incubated for 5 min at room
temperature. Then, Stat3 siRNA was combined with the diluted PR39
or Lipo 2000. They were mixed gently and incubated to form a
complex for 30 min at room temperature. Then the complex was added
to the cells with 1500 μl serum-free DMEM. After 6 h transfections,
cells were rinsed with PBS and the medium was replaced with fresh
complete growth medium. The cells were incubated at 37°C in a 5%
CO2 atmosphere, and further analysis was performed 24,
48 or 72 h post-transfection.
Cell viability assay
4T1 cells were seeded at a density of
6×103/well into 96-well plates (Costar, USA) and treated
with various concentrations of PR39 or various treatment
combinations for 24 h. The next day, cells were incubated in
methylthiazole tetrazolium (MTT, Sigma, USA) solution for 4 h. The
spectrophotometric absorbance was measured in a microplate reader
(Bio-Rad) at 490 nm. Absorbance rates obtained by untreated cells
were considered as 100% cell survival. The relative cell viability
was calculated according to the equation (Abssample −
Absblank)/(Abscontrol − Absblank)
× 100. Each assay was repeated at least three times, using three
wells per drug concentration in each experimental condition.
Flow cytometry
About 5×105 4T1 cells/well were seeded in
6-well plate before various treatments. The Stat3-specific siRNA
complex was added to the 4T1 cells as above. The 4T1 cells from the
control or treated group were trypsinized and suspended in 70%
ethanol at 4°C at least overnight. Then samples were kept on ice
and analyzed on a FACSCalibur flow cytometer
(Becton-Dickinson).
RNA extraction and reverse
transcription-polymerase chain reaction (RT-PCR)
Total cellular RNA was extracted by the Trizol
Reagent (Takara, Japan) according to the manufacturer’s
instructions. Then, cDNA was obtained from the cellular RNA. For
the reverse transcriptase reaction, 1 μg of total RNA was mixed
with oligo(dt)15 primer and Maloney murine leukemia virus (M-MLV)
reverse transcriptase (Takara). Then the mixture was incubated at
42°C for 60 min and 70°C for 15 min. The sequences of PCR primers
(Invitrogen, China) were: Stat3 forward, 5′-AGAGAAGCAGCAGATGTTGG
AGCA-3′, and reverse, 5′-ATCCTGCATGTCTCCTTGGCT CTT-3′ (PCR product,
148 bp) (25); MMP-9 forward,
5′-ACAG CCAACTATGACCAG-3′ and reverse; 5′-TGCCACCAGGA ACAGG-3′ (PCR
product, 249 bp). β-actin forward, 5′-CCA CTGCCGCATCCTCTTCCTC-3′,
and reverse, 5′-TCCTGCTT GCTGATCCACATCT-3′ (PCR product, 400 bp).
The transcribed cDNA was mixed with each primer and Taq DNA
polymerase (Takara, Japan), and then amplified. PCR was performed
as follows: after incubation at 94°C for 5 min, Stat3, MMP-9 and
β-actin underwent 35 cycles of reaction (94°C for 30 sec, 61°C of
Stat3; 58°C of MMP-9 and β-actin for 30 sec, 72°C for 40 sec).
After cycling, the samples were incubated at 72°C for 10 min. The
PCR products were analyzed on 1.5% agarose gel containing
GoldenView. Stained bands were visualized under UV light and were
photographed.
Western blot analysis
4T1 cells were harvested from 6-well plates at 80%
confluency. Cells were washed with cold PBS and lysed with cell
lysis buffer containing protease inhibitor (Beyotime). Whole cell
lysates were centrifuged at 13000 r for 30 min at 4°C. Protein
concentrations were determined by the BCA reaction (Beytime
Biotechnology, China). Proteins (50 μg of protein/lane) from
control or treated cells were separated by SDS-PAGE and transferred
to polyvinylidene difluoride membrane (PVDF) membranes with a
Bio-Rad semi-dry transfer system (Bio-Rad). The membranes were
blocked with 5% (v/v) non-fat milk overnight and then incubated
with primary antibody for 12 h at 4°C. After washes, the membrane
were incubated with appropriate antibody conjugated with
horseradish peroxidase as secondary antibody for 1 h at 37°C,
followed by three washes with TBST (Tris-buffer saline containing
0.1% Tween 20). Observations of signals were obtained by enhanced
chemiluminescence (ECL reagent, Millipore, USA) according to the
manufacturer’s instructions. Intensities of the bands obtained from
the RT-PCR and western blot analysis were analyzed using the
Quantity One software (Bio-Rad).
Invasion assay
The 4T1 cell invasive ability with or without
treatment was examined using the membrane Transwell culture system.
Transwell membrane (8 μm pore size, 6.5 mm diameter; Sigma) coated
with Matrigel (Sigma) overnight at 4°C were used for the invasion
assay. The 4T1 cells from the control or treated groups after 48 h
were trypsinized, centrifuged, and resuspended at
2.5×105/ml in DMEM without FBS. Then, cells
(2.5×104/well) were seeded onto the upper chamber of
pre-coated Transwell membranes. The lower chamber of the Transwells
contained DMEM with 10% FBS. The cells on the upper chamber and
Matrigel coating the membranes were swabbed with Q-tip after 18 h
incubation. Then, the membranes were fixed with 4% paraformaldehyde
and stained with 0.1% crystal violet. The cells attaching to the
lower surface of the polycarbonate filter were counted under light
microscopy (magnification, ×200). The experiments were performed in
thrice in triplicate. The 4T1 cells obtained from the Transwell
assay were analyzed using the Image J software (NIH, USA).
Migration assay
The 4T1 cells from the control or treated group were
trypsinized and suspended in DMEM medium without FBS. Transwell
membranes were used. Then, cells (2×104/well) were
seeded onto the upper chamber of Transwells. Lower wells of the
Transwell chamber contained DMEM with 10% FBS as chemotactic
medium. Then, the chamber was incubated in a humidified incubator
for 3 h. The medium was removed from the upper chamber and the
filtered cells were fixed with 4% paraformaldehyde and stained with
0.1% crystal violet. The migrated 4T1 cells were evaluated under
the microscope, and random fields were scanned (five fields per
filter) for the cells obtained at the lower membrane side only. The
number of 4T1 cells obtained from the Transwell assay were analyzed
using the Image J software.
Statistical analysis
The repeated-measures ANOVA test was used between
mutiple comparisons analysis with the SPSS17.0 software. P-values
<0.05 were taken to denote significant differences.
Results
Complex formation of PR39 with siRNA
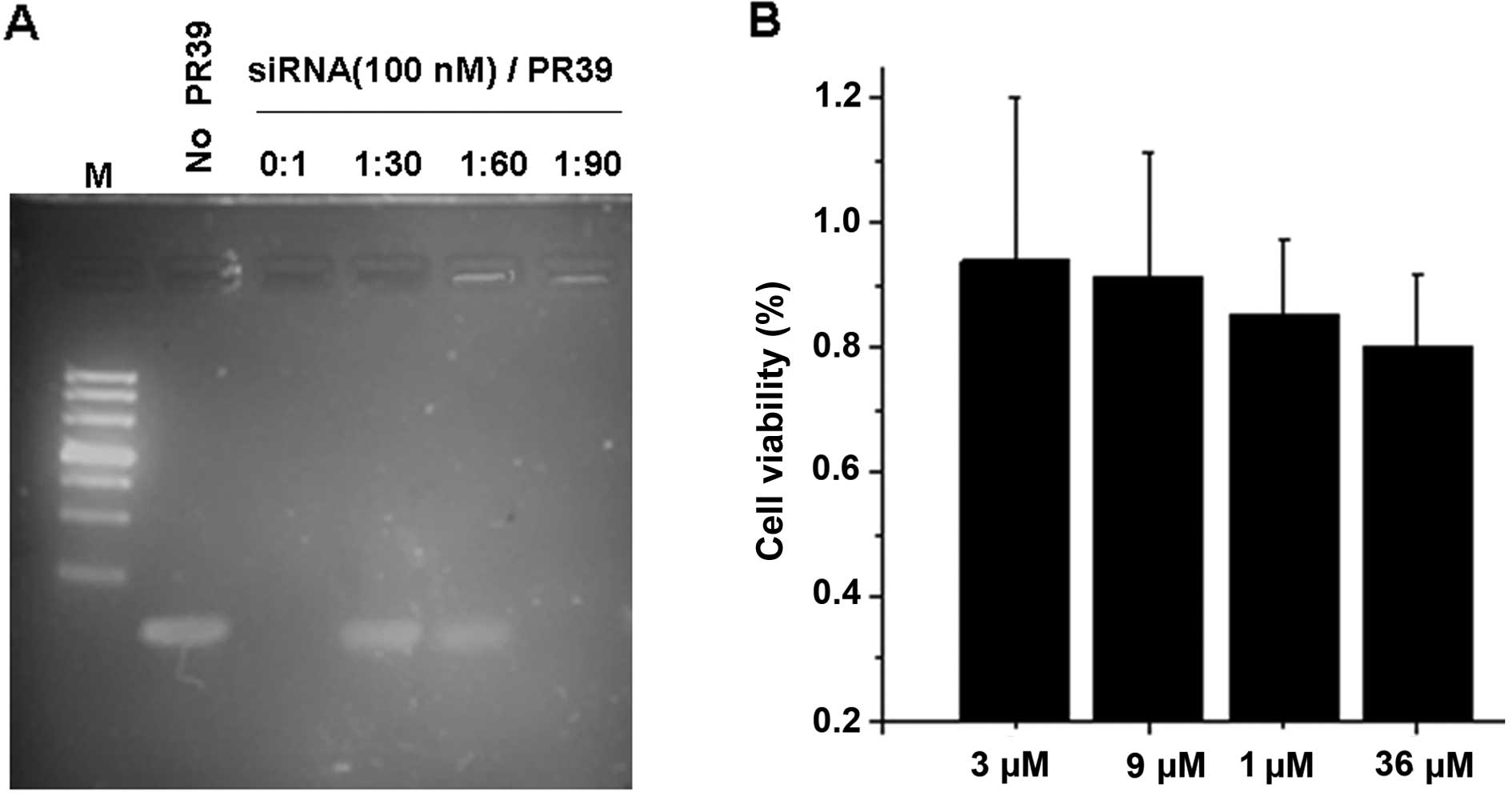
In order to ascertain whether PR39 and siRNA could
form a complex, 100 nM scrambled siRNA was mixed with PR39 at
different concentration ratios. As shown by a gel-shift assay PR39
was able to bind siRNA in a dose-dependent manner (Fig. 1A), and the 1:90 molar ratio of siRNA
to PR39 was an optimal condition. We measured the cytotoxicity of
PR39 usig the MTT assay (Fig. 1B),
and observed that PR39 at 3 μM to 36 μM had no effect on the cell
viability of 4T1 cells. These results indicate that PR39 could
interact with siRNA and form complexes, and it was barely nontoxic
to cells.
Cellular colocalization of the
FITC-PR39/Cy3-siRNA complex
To determine whether PR39 could deliver siRNA into
the cytoplasm, 4T1 cells were transfected with Cy3-labeled siRNA
complexed to FITC-labeled PR39. After 6 h treatment, the
intracellular colocalization of the double-labeled PR39/siRNA
complex in 4T1 cells was assessed by fluorescence microscopy
(Fig. 1C).
Optimal silencing effect of PR39/Stat3
siRNA
To detect the gene-silencing effect of the Stat3
siRNA delivered by PR39, 4T1 cells stably expressing high levels of
Stat3 were transfected with Stat3-siRNA/PR39. Stat3 mRNA expression
was determined after 24–72 h of incubation with an increasing molar
ratio from 0:1 to 1:180 Stat3-siRNA/PR39. A maximum gene silencing
effect assessed by mRNA and protein levels was observed after 48 h
of treatment, and the optimal ratio of siRNA to PR39 was 1:90
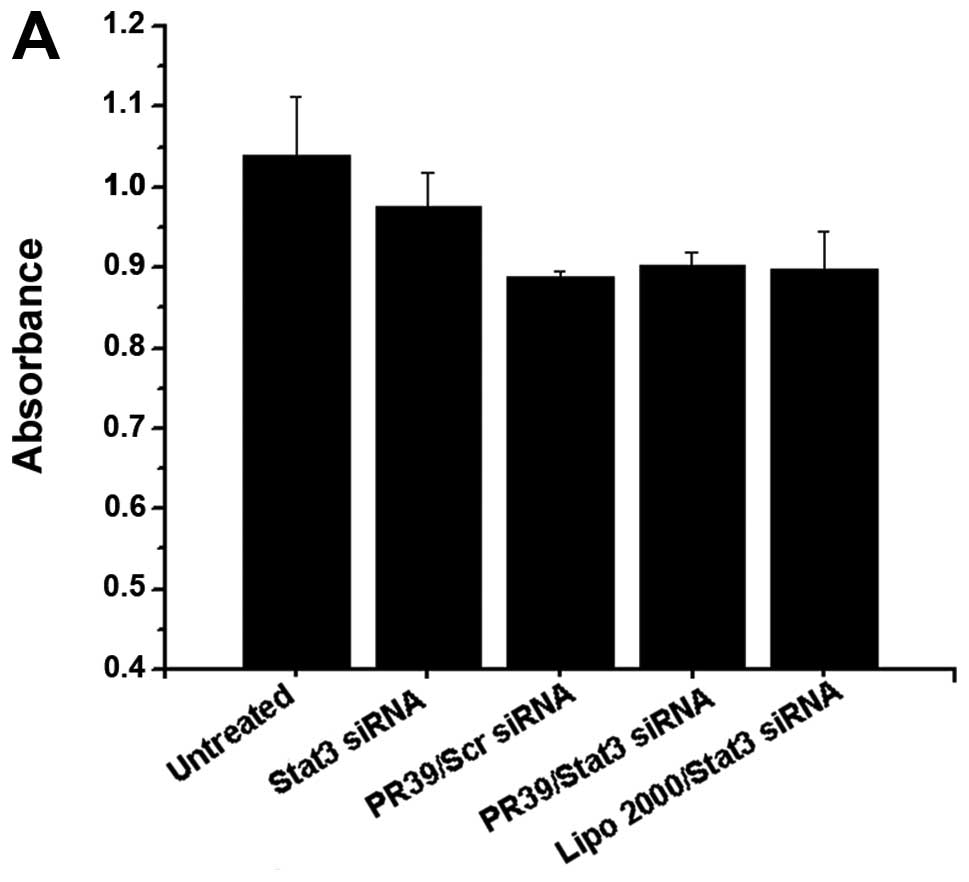
(Fig. 2A). Lipo 2000/Stat3 siRNA
was used as a positive control, and it seemed more potent than
PR39/siRNA in silencing Stat3 expression (Fig. 2B and D).
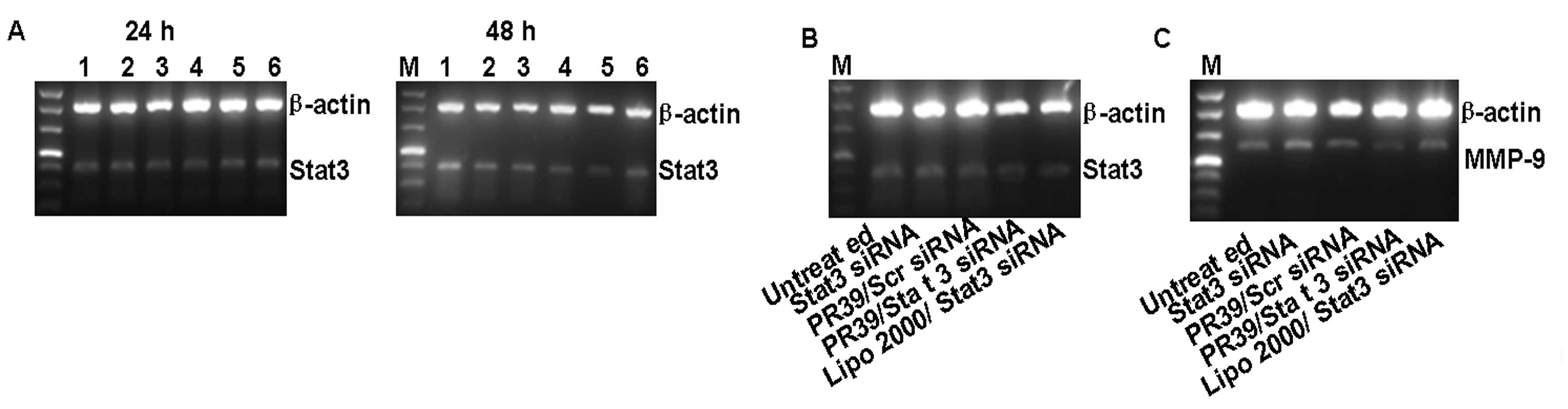 | Figure 2Gene silencing was analyzed by RT-PCR
and Western blotting. (A) 4T1 cells stably expressing Stat3 were
treated with Stat3 siRNA/PR39 of molar ratios ranging from 0:1 to
1:180, and Stat3 expression was analyzed at 24 or 48 h. Lane 1,
Untreated group; lane 2, 100 nM Stat3 siRNA; lane 3, Stat3
siRNA/PR39 1:30; lane 4, Stat3 siRNA/PR39 1:60; lane 5, Stat3
siRNA/PR39 1:90; lane 6, Stat3 siRNA/PR39 1:180. The maximum gene
silencing effect was found after 48 h of treatment, and the optimal
ratio of siRNA to PR39 was 1:90. 4T1 cells were examined for (B)
Stat3 mRNA and (D) protein levels after treatment with PR39/Stat3
siRNA complex for 48 h. Untreated group, Stat3 siRNA and PR39/Scr
siRNA were used as negative control; Lipo 2000/Stat3 siRNA was used
as a positive control. MMP-9 mRNA (C) and protein levels (D) were
determined after treatment with the PR39/Stat3 siRNA complex for 48
h. Lipo 2000/Stat3 siRNA, was used as a positive control, and
seemed more potent than PR39/siRNA in silencing Stat3 expression
but had no effect on its downstream target molecular, MMP-9. (E-G)
4T1 cells were assayed for Stat3 and MMP-9 mRNA and protein levels
after single PR39 treatment for 48 h assessing whether single PR39
could result in MMP-9 downregulation. |
Proliferation and cell cycle of 4T1 cells
is not affected by Stat3 knockdown
Cell proliferation was measured by the MTT and FCM
assays. The results suggest that the proliferation and cell cycle
of 4T1 cells were not affected by Stat3 knockdown (Fig. 3A and B). Cyclin D1 protein levels
were not influenced by the Stat3 reduction (data not shown).
Inhibition of 4T1 cell invasion and
migration with PR39/Stat3 siRNA
The effect of Stat3 siRNA on the invasiveness and
motility of 4T1 cells was determined using the Transwell assay. The
cells treated with PR39/Scr siRNA, PR39/Stat3 siRNA were
significantly fewer compared to the naked Stat3 siRNA and the
untreated group (Fig. 3C-F). More
interesting, although Lipo 2000/Stat3 siRNA displayed more potent
activity than PR39/siRNA in silencing Stat3 expression, the
invasive and migration capacity of the cells after treatment with
Lipo 2000/Stat3 siRNA was not less than that of the PR39/siRNA
group.
In addition, matrix matalloproteinase-9 (MMP-9) has
been shown to be one of the Stat3 downstream target genes, which
plays an important role in tumor cells invasion and migration.
Therefore, MMP-9 expression was examined after 48 h transfection
with PR39/Stat3 siRNA and Lipo 2000/Stat3 siRNA. These observations
suggested that MMP-9 expression was significantly inhibited by the
PR39/siRNA group compared to the Lipo 2000/Stat3 siRNA group
(Fig. 2C and D). Given that
previous reports demonstrated that PR39 gene transduction altered
the invasive activity and actin structure in human hepatocellular
carcinoma cells (26), we
investigated the effect of PR39 alone on cell invasion and
migration. As shown in Fig. 3G and
J, PR39 (9 or 18 μM) could significantly inhibit 4T1 cell
invasion and migration, and it was estimated that PR39 and Stat3
siRNA could have a synergistic effect in the invasion and migration
of 4T1 cells.
Discussion
siRNA shows potential as a therapeutic tool against
genes causing many diseases, but the effectiveness of these siRNAs
are limited by their poor cellular permeability and insufficient
stability profile. Currently, Lipofectamine transfection is one of
the most popular methods of siRNA delivery into cell cytoplasm.
However, the Lipofectamine transfection is confined to specific
cell types and is toxic to cells and animals (27). In recent years, cell penetrating
peptides have been effective carriers delivering siRNA in
vitro. Specifically, an arginine- rich intracellular delivery
peptide could noncovalently deliver macromolecules, and translocate
into animal cells and tissues (28). For example, the nona-arginine (R9)
was able to noncovalently deliver siRNA into mammalian cells
(29), and efficient RNA
interference was also obtained by an arginine peptide (R15) in
vitro and in vivo (30).
Zhang et al reported that human antimicrobial peptide LL-37
could deliver nucleic acids into the host cells as a
cell-penetrating peptide (31). It
is of interst to investigate whether with abundance of cationic
amino acids, PR39 could be used as a novel siRNA carrier. Thus, we
initially investigated the complex formation of PR39 with siRNA and
its cellular colocalization. Then, we optimized the ratio of the
PR39/siRNA complex, cell/complex incubation period and the
concentration of siRNA. The results suggest that PR39 could form a
complex with siRNA and translocate siRNA into 4T1 cells. The
optimal ratio of siRNA to PR39 was 1:90 with a maximum gene
silencing effect observed after 48 h treatment. Except for mice
breast cancer cells, the cellular localization by PR39/siRNA
appeared in HBL100, Hep3B and K562 cells (data not shown). These
results showed that cationic PR39 could noncovalently contact with
negative siRNA and translocate the complex into the cell cytoplasm.
At present, the mechanisms underlying the cellular translocation of
PR39 are poorly understood. It has been deduced that electrostatic
attraction plays an important role in the strong binding and
interaction with cancer cell membranes, caused by the interaction
between the negatively charged component of the cancer cell surface
and the positively charged cationic antimicrobial peptides
(32).
To investigate the knockdown gene efficiency of
PR39-mediated siRNA, Stat3 siRNA/PR39 was introduced to 4T1 cells,
and Stat3 mRNA and protein levels were determined with RT-PCR and
western blot analysis. As indicated in our studies, the Stat3
siRNA/PR39 complex silenced approximately 50% of the gene
expression of Stat3, and the expression knockdown lasted for 72 h.
However, approximately 70% gene reduction was observed with Lipo
2000 transfection used as a positive control (Fig. 2B and D). Lipo 2000/Stat3 siRNA
displayed more potent activity than the PR39/siRNA in silencing
Stat3 expression. Veldhoen et al also reported that a novel
carrier peptide termed MPGα mediated delivery of siRNA might be
less efficient compared with Lipo 2000 (33).
In order to further examine the influence of Stat3
gene silencing on malignant biological features of tumor cells, 4T1
cells proliferation, cell cycle, invasion and migration were
investigated. The results suggested that Stat3 knockdown could not
result in 4T1 cell proliferation inhibition and cell cycle arrest
(Fig. 3A and B), while invasion and
migration of 4T1 cells was strongly inhibited (Fig. 3C-F). Notably, although Lipo
2000/Stat3 siRNA displayed more potent activity than PR39/stat3
siRNA in silencing Stat3 expression, the PR39/Stat3 siRNA complex
showed a stronger suppression of 4T1 cell invasion and migration
along with MMP-9 expression. Besides, PR39/Scr siRNA complex also
demonstrated stronger suppression in cell invasion and migration,
compared with naked siRNA. We hypothesized that single PR39 may
play a role in cell invasion and migration. We present evidence
that single PR39 inhibited invasive and migration activity of 4T1
cells and on the reduction of MMP-9 expression (Fig. 2E-G, Fig.
3G-J). Previous studies also reported that PR39 could alter the
invasive activity and actin structure in human hepatocellular
cancer cells (26). Therefore, it
was shown that PR39 and Stat3 siRNA could play synergistic roles in
the invasion and migration of 4T1 cells (Fig. 3C-F).
In conclusion, we used PR39 as a novel siRNA
delivery system, which could interact with siRNA, form complexes,
and mediate delivery of siRNA into the cytoplasm to silence the
target gene. The results also suggest that in addition to its
anticarcinogenic activity, single PR39 may play a role in cell
invasion and migration. PR39 and Stat3 siRNA displayed synergistic
biological effects on inhibiting cell invasion and migration of 4T1
cells, which were more dominent compared to the Lipo 2000 delivery
system. Although further evidence is required to determine the
exact mechanisms, our study highlights the potential of PR39 to
mediate siRNA to the 4T1 cells.
Acknowledgements
We thank Dr Yang’an Wen at the First Affiliated
Hospital, Chongqing Medical University for providing technical
support. This work was supported by a grant from National Natural
Science Foundation of China (no. 30971131); and by a grant from the
Foundation of National Key Discipline in Laboratory Medicine (no.
2010104). This study was supported in part by the Key Laboratory of
Diagnostic Medicine Designated by the Ministry of Education,
Chongqing Medical University.
References
|
1
|
Yang SY: Sniper the hyperplasia of mammary
glands and escort for the women health. Med People. 34–35.
2007.
|
|
2
|
Jun JY, Griffith JW, Bruggeman R, et al:
Effects of polyamine depletion by alpha-difluoromethylornithine on
in vitro and in vivo biological properties of 4T1
murine mammary cancer cells. Breast Cancer Res Treat. 107:33–40.
2008.PubMed/NCBI
|
|
3
|
Garcia R, Yu CL, Hudnall A, et al:
Constitutive activation of Stat3 in fibroblasts transformed by
diverse oncoproteins and in breast carcinoma cells. Cell Growth
Differ. 8:1267–1276. 1997.PubMed/NCBI
|
|
4
|
Yu H and Jove R: The STATs of cancer – new
molecular targets come of age. Nat Rev Cancer. 4:97–105. 2004.
|
|
5
|
Frank DA: STAT3 as a central mediator of
neoplastic cellular transformation. Cancer Lett. 251:199–210. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Darnell JE Jr: Transcription factors as
targets for cancer therapy. Nat Rev Cancer. 2:740–749. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Chiu WT, Lee HT, Huang FJ, et al:
Caveolin-1 upregulation mediates suppression of primary breast
tumor growth and brain metastases by stat3 inhibition. Cancer Res.
71:4932–4943. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Blaskovich MA, Sun J, Cantor A, et al:
Discovery of JSI-124 (cucurbitacin I), a selective Janus
kinase/signal transducer and activator of transcription 3 signaling
pathway inhibitor with potent antitumor activity against human and
murine cancer cells in mice. Cancer Res. 63:1270–1279. 2003.
|
|
9
|
Leong PL, Andrews GA, Johnson DE, et al:
Targeted inhibition of Stat3 with a decoy oligonucleotide abrogates
head and neck cancer cell growth. Proc Natl Acad Sci USA.
100:4138–4143. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Meydan N, Grunberger T, Dadi H, et al:
Inhibition of acute lymphoblastic leukaemia by a Jak-2 inhibitor.
Nature. 379:645–648. 1996. View
Article : Google Scholar : PubMed/NCBI
|
|
11
|
Mora LB, Buettner R, Seigne J, et al:
Constitutive activation of Stat3 in human prostate tumors and cell
lines: direct inhibition of Stat3 signaling induces apoptosis of
prostate cancer cells. Cancer Res. 62:6659–6666. 2002.PubMed/NCBI
|
|
12
|
Nakajima K, Yamanaka Y, Nakae K, et al: A
central role for Stat3 in IL-6-induced regulation of growth and
differentiation in M1 leukemia cells. EMBO J. 15:3651–3658.
1996.PubMed/NCBI
|
|
13
|
Ni Z, Lou W, Leman ES, et al: Inhibition
of constitutively activated Stat3 signaling pathway suppresses
growth of prostate cancer cells. Cancer Res. 60:1225–1228.
2000.PubMed/NCBI
|
|
14
|
de Fougerolles A, Vornlocher HP,
Maraganore J and Liberman J: Interfering with disease: a progress
report on siRNA-based therapeutics. Nat Rev Drug Discov. 6:443–453.
2007.PubMed/NCBI
|
|
15
|
Akhtar S and Benter I: Toxicogenomics of
non-viral drug delivery systems for RNAi: potential impact on
siRNA-mediated gene silencing activity and specificity. Adv Drug
Deliv Rev. 59:164–182. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Bumcrot D, Manoharan M, Koteliansky V and
Sah DW: RNAi therapeutics: a potential new class of pharmaceutical
drugs. Nat Chem Biol. 2:711–719. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Nakamura Y, Kogure K, Futaki S and
Harashima H: Octaarginine-modified multifunctional envelope-type
nano device for siRNA. J Control Release. 119:360–367. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Agerberth B, Lee JY, Bergman T, et al:
Amino acid sequence of PR-39. Isolation from pig intestine of a new
member of the family of proline-arginine-rich antibacterial
peptides. Eur J Biochem. 202:849–854. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Boman HG, Agerberth B and Boman A:
Mechanisms of action on Escherichia coli of cecropin P1 and
PR-39, two antibacterial peptides from pig intestine. Infect Immun.
61:2978–2984. 1993.PubMed/NCBI
|
|
20
|
Storici P and Zanetti M: A cDNA derived
from pig bone marrow cells predicts a sequence identical to the
intestinal antibacterial peptide PR-39. Biochem Biophys Res Commun.
196:1058–1065. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Shi J, Ross CR, Leto TL and Blecha F:
PR-39, a proline-rich antibacterial peptide that inhibits phagocyte
NADPH oxidase activity by binding to Src homology 3 domains of
p47phox. Proc Nat Acad Sci USA. 93:6014–6018. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chan YR and Gallo RL: PR-39, a
syndecan-inducing antimicrobial peptide, binds and affects
p130(Cas). J Biol Chem. 273:28978–28985. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Gao Y, Lecker S, Post MJ, et al:
Inhibition of ubiquitin-proteasome pathway–mediated I kappa B alpha
degradation by a naturally occurring antibacterial peptide. J Clin
Invest. 106:439–448. 2000.
|
|
24
|
Alshamsan A, Hamdy S, Samuel J, et al: The
induction of tumor apoptosis in B16 melanoma following STAT3 siRNA
delivery with a lipid-substituted polyethylenimine. Biomaterials.
31:1420–1428. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ye Y, Yin DT, Chen L, et al:
Identification of Piwil2-like (PL2L) proteins that promote
tumorigenesis. PLoS One. 5:e134062010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ohtake T, Fujimoto Y, Ikuta K, et al:
Proline-rich antimicrobial peptide, PR39 gene transduction altered
invasive activity and actin structure in human hepatocellular
carcinoma cells. Br J Cancer. 81:393–403. 1999. View Article : Google Scholar
|
|
27
|
Ohki EC, Tilkins ML, Ciccarone VC and
Price PJ: Improving the transfection efficiency of post-mitotic
neurons. J Neurosci Methods. 112:95–99. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wang YH, Chen CP, Chan MH, et al:
Arginine-rich intracellular delivery peptides noncovalently
transport protein into living cells. Biochem Biophys Res Commun.
346:758–767. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wang YH, Hou YW and Lee HJ: An
intracellular delivery method for siRNA by an arginine-rich
peptide. J Biochem Biophys Methods. 70:579–586. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Kim SW, Kim NY, Choi YB, et al: RNA
interference in vitro and in vivo using an arginine
peptide/siRNA complex system. J Control Release. 143:335–343.
2010.PubMed/NCBI
|
|
31
|
Zhang X, Oglecka K, Sandgren S, et al:
Dual functions of the human antimicrobial peptide LL-37-target
membrane perturbation and host cell cargo delivery. Biochim Biophys
Acta. 1798:2201–2208. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Hoskin DW and Ramamoorthy A: Studies on
anticancer activities of antimicrobial peptides. Biochim Biophys
Acta. 1778:357–375. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Veldhoen S, Laufer SD, Trampe A and Restle
T: Cellular delivery of small interfering RNA by a non-covalently
attached cell-penetrating peptide: quantitative analysis of uptake
and biological effect. Nucleic Acids Res. 34:6561–6573. 2006.
View Article : Google Scholar : PubMed/NCBI
|