Introduction
Oligodendrogliomas (ODGs) are classified by the
World Health Organization (WHO) as low-grade gliomas (OII or OIII)
(1). Grade II oligodendrogliomas
(OII) are rare and slow-growing tumors, and affected patients show
long-term survival when treated with surgical resection at
diagnosis, even though the majority eventually relapse (2). Presently accepted guidelines for
treatment suggest that adjuvant radio- and/or chemotherapy be
delayed until there is clinical evidence of disease progression in
OII patients (3). However, it has
been shown that their risk of relapse may be defined at an earlier
stage (4). Traditionally, a low
risk of recurrence among low-grade glioma patients has been related
to clinical variables, such as age (≤40 years) at diagnosis,
seizures at presentation, absence of neurological deficits, a
Karnofsky performance status of ≥70, absence of enhancement at
CT/MRI, a pre-operative tumor size of ≤5–6 cm and the tumor not
crossing the midline (5). Moreover,
a well-accepted molecular factor of prognostic significance is the
allelic loss [loss of heterozygosity (LOH)] of chromosome 1p,
together with 19q (6). Combined
deletions of both arms were observed in up to 80% of ODG patients
with prolonged survival (7–9), suggesting that these unstable
chromosomal regions carry critical genes whose silencing may define
their clinical history. Both the physical loss of a genetic trait
and epigenetic silencing, via CpG island promoter methylation, may
downregulate or suppress the expression of significant genes in
cancer cells compared to normal ones. The epigenetic profiling of
brain tumors has indeed significance as a prognostic index
(10): e.g., the epigenetic
silencing of O6-methylguanine methyltransferase
(MGMT, 10q26) correlates with responsiveness to alkylating
agents and radiotherapy in glioblastoma (GBM, WHO grade IV)
(11–13). The 19q region, frequently lost in
ODG tumors, harbors putative tumor suppressor genes, such as the
epithelial membrane protein 3 (EMP3, 19q13.3) (14–19).
The function of EMP3 remains unclear; however, its homology to
peripheral myelin protein 22 (PMP22) suggests that the protein is
implicated in cell cycle regulation and cell-to-cell interactions
(17). The EMP3 promoter
contains a CpG island, which is methylated in both high-grade
astrocytoma and neuroblastoma brain tumors associated with poorer
prognosis (14). Demethylating
agents have been shown to restore EMP3 expression in
neuroblastoma cell lines, with consequent lower in vitro
colony formation and reduction of tumor growth in nude mice
(14). A methylated EMP3
promoter was found by Li et al in a group of ODG samples
with different histological classifications; however, no
association between the EMP3 gene promoter methylation, the
corresponding protein expression and 19q deletion was found
(20). However, Kunitz et al
found a significant association between EMP3 promoter
methylation and the allelic loss of 19q, consistent with the
reduced EMP3 mRNA expression (21). All these findings suggest that
EMP3 methylation may occur in ODG together with LOH of
chromosome 19q (LOH 19q), although the clinical significance of
their co-occurrence remains to be clarified.
We previously described a cohort of OII patients in
whom LOH 19q was present in the subgroup at a higher risk of
relapse (22). Given this
preliminary evidence, in this study, we evaluated the EMP3
gene promoter in the same cohort of patients, to investigate
whether CpG methylation of the residual allele promoter is
consistent with the physical loss of the homologous allele promoter
and whether a correlation can be established between EMP3
cyto- and epigenetic loss and a higher risk of relapse. In
addition, we examined the MGMT and cyclooxygenase 2
(COX2), an isozyme of prostaglandin-endoperoxidase synthase
2 (1q25.2q25.3) gene promoter methylation in these patients.
MGMT is a gene of interest in high-grade gliomas, namely
GBM, as the methylation of its promoter predicts chemosensitivity
to alkylating agents; it is also under evaluation in low-grade
gliomas (11–13,23).
The COX2 gene promoter methylation provides epigenetic
information regarding genetic regions that are more stable than the
19q arms, both in low- and high-grade gliomas. The methylation of
the 3 gene promoters, EMP3, MGMT and COX2, was also
evaluated in a small group of primary WHO grade IV gliomas, in
order to validate a different epigenetic profile between low- and
high-grade gliomas (21). A further
characterization between these 2 subgroups was provided by
isocitrate dehydrogenase 1 (IDH1) genotyping, as the
IDH1 mutation is rare in primary GBM but frequent in OII
(24).
Materials and methods
Patients and tumor specimens
This study was carried out in a small homogeneous
cohort of WHO grade II oligodendroglioma (OII) patients, an
infrequent low-grade glioma subtype. Study participants gave their
informed consent to this study. The protocols were approved by the
institutional ethics committee in accordance with the ethical
standard of Declaration of Helsinki (1964). Samples from 23
patients were collected over a period of 10 years as previously
described (22). The age and gender
of patients are reported in Table
I. Seventeen GBM (WHO grade IV) samples were collected from 15
patients (2 of which were relapses). There were 9 female patients
(60%); median age at the time of diagnosis was 62 years (range,
43–80) (Table II). Tumor samples
were embedded in paraffin. All histological diagnoses were reviewed
by 2 experienced neuropathologists (S.C. and L.R.).
 | Table ISummary of OII patient personal data
(age and gender), presence of loss of heterozygosity (LOH) of 1p
and 19q, EMP3, MGMT and COX2 promoter
methylation status and IDH1 genotyping. |
Table I
Summary of OII patient personal data
(age and gender), presence of loss of heterozygosity (LOH) of 1p
and 19q, EMP3, MGMT and COX2 promoter
methylation status and IDH1 genotyping.
| No. | Age | Gender | LOH 1p | LOH 19q | EMP3 | MGMT | COX2 | IDH1 |
|---|
| 1 | 33 | Male | + | + | M | M | U | R132H |
| 2 | 50 | Male | + | + | M | M | U | wt |
| 3 | 43 | Female | + | + | M | M | U | R132H |
| 4 | 35 | Male | + | + | M | M | U | NI |
| 5 | 51 | Female | + | + | M | NS | U | NI |
| 6 | 38 | Female | + | − | M | NS | U | wt |
| 7 | 42 | Female | + | + | M | M | NS | R132H |
| 8 | 37 | Male | − | + | M | M | U | wt |
| 9 | 59 | Female | + | + | M | M | U | wt |
| 10 | 37 | Male | − | − | M | M | U | R132H |
| 11 | 33 | Female | + | − | M | M | U | R132H |
| 12 | 66 | Male | + | + | M | NS | U | R132H |
| 13 | 30 | Male | − | − | M | M | U | R132H |
| 14 | 70 | Male | + | + | M | NS | U | NI |
| 15 | 42 | Female | + | + | M | M | U | R132H |
| 16 | 37 | Male | + | NI | M | M | U | wt |
| 17 | 38 | Male | + | + | M | M | U | R132H |
| 18 | 37 | Female | − | + | U | M | U | wt |
| 19 | 56 | Female | − | − | M | M | U | R132H |
| 20 | 69 | Male | + | + | M | M | U | R132H |
| 21 | 45 | Female | + | + | M | M | U | NI |
| 22 | 28 | Female | + | − | M | NS | U | R132H |
| 23 | 39 | Female | + | + | M | M | U | R132G |
 | Table IISummary of GBM patient personal data
(age and gender), presence of EMP3, MGMT and
COX2 promoter methylation status and IDH1
genotyping. |
Table II
Summary of GBM patient personal data
(age and gender), presence of EMP3, MGMT and
COX2 promoter methylation status and IDH1
genotyping.
| No. | Age | Gender | EMP3 | MGMT | COX2 | IDH1 |
|---|
| 1 | 56 | Female | U | U | U | wt |
| 2 | 56 | Female | U | U | U | wt |
| 3 | 73 | Male | U | M | U | wt |
| 4 | 49 | Male | U | U | U | wt |
| 5 | 43 | Female | U | U | U | wt |
| 6 | 52 | Female | U | M | U | NI |
| 7 | 68 | Female | U | U | U | wt |
| 8 | 57 | Male | U | M | U | wt |
| 9 | 59 | Female | M | M | U | wt |
| 10 | 80 | Female | U | M | U | wt |
| 11 | 74 | Female | U | U | U | wt |
| 12 | 69 | Female | U | U | U | wt |
| 13 | 62 | Male | M | U | U | wt |
| 14 | 62 | Male | U | U | U | wt |
| 15 | 74 | Male | M | U | U | NI |
| 16 | 71 | Female | U | U | U | NI |
| 17 | 55 | Male | U | U | U | wt |
Genomic DNA extraction
DNA extraction from the paraffin-embedded tissue was
performed using the standard procedure with phenol-chloroform from
5 slices (5-μm thick). The paraffin was dissolved with xylene, and
any remaining xylene was washed out with ethanol. After overnight
protein digestion with proteinase K, the DNA was separated from the
organic phase with phenol-chloroform, precipitated with ethanol and
resuspended in Tris-EDTA (TE) buffer.
Analysis of gene promoter
methylation
The DNA was processed with sodium bisulfite,
converting unmethylated cytosine to uracil, as previously described
(11). Briefly, 1 μg of DNA was
denatured with sodium hydroxide and modified with sodium bisulfite.
The DNA samples were then purified with a commercial kit (Wizard
DNA Clean-Up System; A7280; Promega, Madison, WI, USA), and again
treated with sodium hydroxide to complete the conversion to uracil.
Finally, DNA was precipitated with ethanol and resuspended in 30 μl
of TE buffer. The methylation status of the genes of interest was
determined by methylation-specific PCR (MSP), using 2 sets of
primers (Table III), specific for
either methylated or unmethylated DNA (25). The total volume for the MSP reaction
was 15 μl, comprising 4 mM MgCl2, 0.5 mM of each dNTP,
0.2 μM of each primer, 0.5 unit of Hot Start DNA polymerase and 3
μl of bisulfite-treated DNA. The methylation of the MGMT and
EMP3 gene promoters was spot-checked in selected GBM samples
by bisulfite sequencing, cloning PCR products using the
pGEM®-T Easy Vector System (A1360, Promega). Selective
positive clones were sequenced and products were aligned using the
BioEdit sequence alignment editor.
 | Table IIISequences of primers used for the
promoter methylation analysis. |
Table III
Sequences of primers used for the
promoter methylation analysis.
| Gene | Primer | Sequence 5′-3′ | Annealing
temperature (°C) |
|---|
| MGMT | UM-F |
TTTGTGTTTTGATGTTTGTAGGTTTTTGT | 62 |
| UM -R |
AACTCCACACTCTTCCAAAAACAAAACA | |
| M-F |
TTTCGACGTTCGTAGGTTTTCGC | |
| M-R |
GCACTCTTCCGAAAACGAAACG | |
| EMP3 | UM -F |
GAAGAGATGTAGAAGGAGAGTGAGT | 62 |
| UM -R |
CTTATCCCTCACTCAAACCTCCATA | |
| M-F |
GACGTAGAAGGAGAGCGAGC | |
| M-R |
CCTCGCTCGAACCTCCGTA | |
| COX2 | UM -F |
GAGAGGGGATTTTTTGTGTTTTT | 60 |
| UM -R |
CCCAAACACTTCCAAAAACC | |
| M-F |
GAGGGGATTTTTTGCGTTTTC | |
| M-R |
CCGAACGCTTCCGAAAAC | |
| MGMT | BS-F |
TTTTGATTAGGGGAGIGGT | 57 |
| BS-R |
TCTATACCTTAATTTACC | |
| EMP3 | BS-F |
GGGAGTAAGAGAGAAGGAGGTTTAG | 60 |
| BS-R |
TTAAAAAATCCCAACCCTAAATAAC | |
Analysis of the IDH1 mutation
IDH1 alterations of the mutational hotspot
codon R132 were assessed by sequencing of PCR-amplified fragments.
Primers used were 5′-ACCAAAT GGCACCATACGA-3′ (forward) and
5′-TTCATACCTTGC TTAATGGGTGT-3′ (reverse).
Data analysis
Disease progression-free interval (DPI) was defined
as the time between surgery and either disease progression or the
last follow-up examination. DPI curves were plotted using the
Kaplan-Meier method with GraphPad Prism (Version 4.0, GraphPad
Software, San Diego, CA, USA).
Results
Analysis of gene promoter
methylation
Analysis of gene promoter methylation was performed
via MSP, amplifying the 5′-CpG island close to the transcription
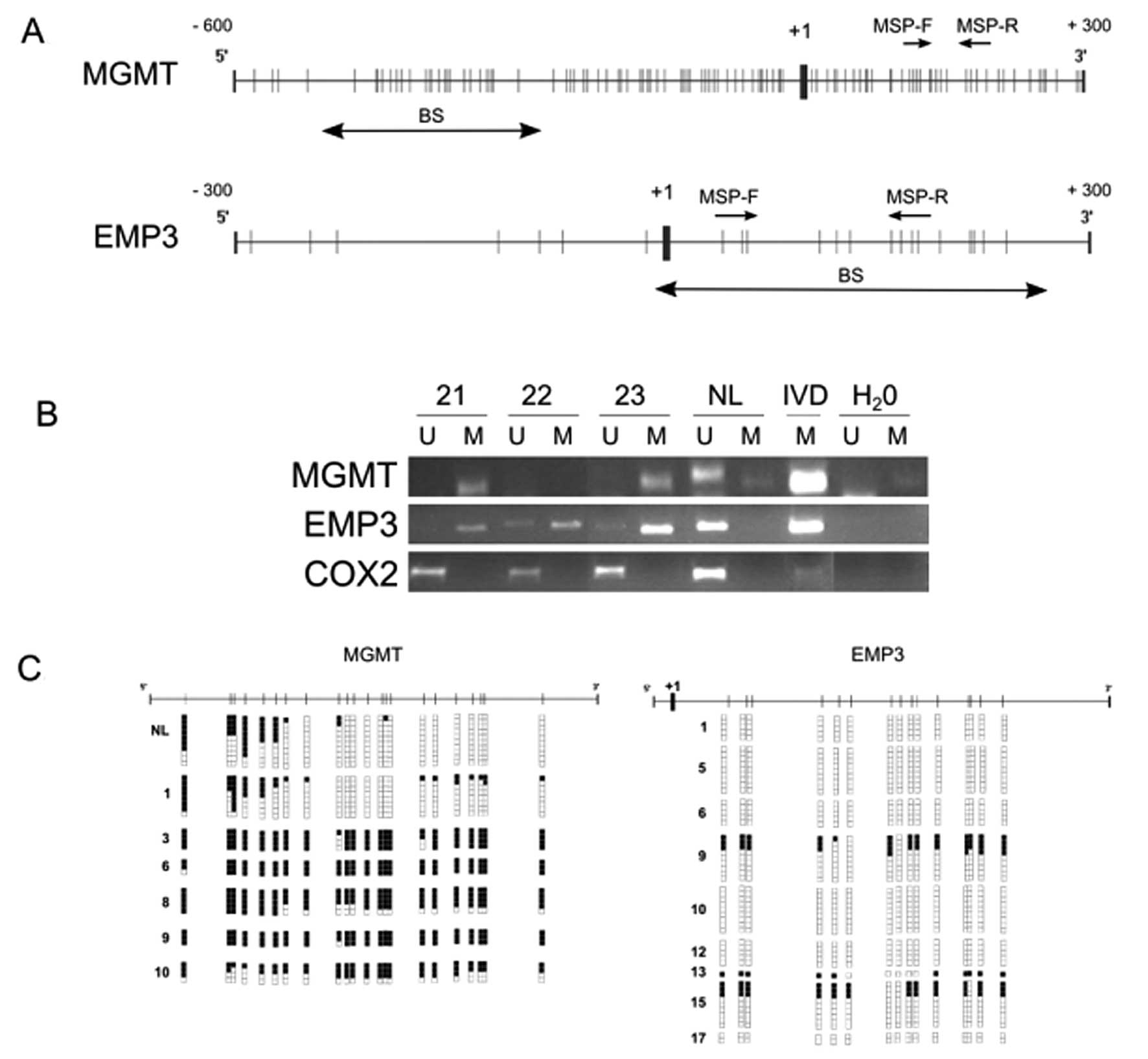
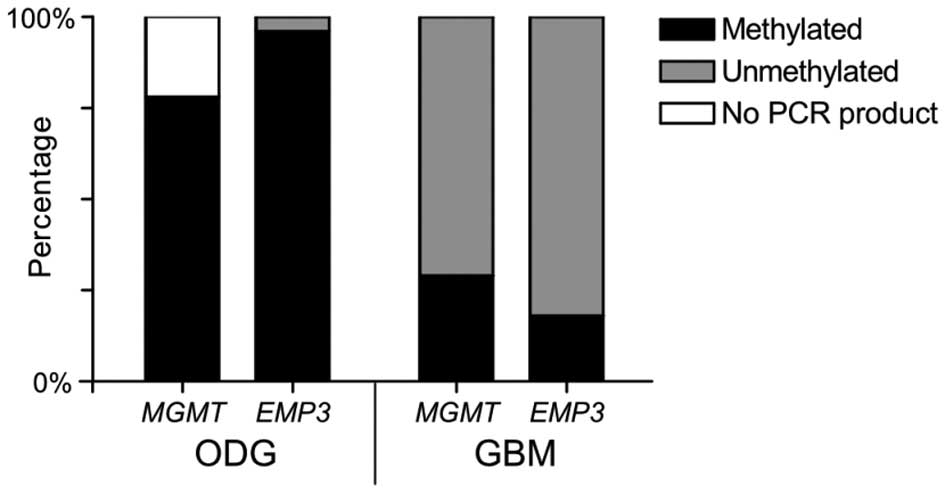
start site of each gene of interest (Fig. 1A and 1B). Among our 23 OII patients,
the EMP3 gene promoter was methylated in 22 samples and
unmethylated in 1 sample; MGMT was methylated in 18 samples
and no PCR product was scored in 5 cases; COX2 was
unmethylated in 22 samples and no PCR product was detected in 1
sample (Fig. 2 and Table I).
As regards our 17 GBM patients, our results showed
that the EMP3 gene promoter was methylated in 3 and
unmethylated in 14 samples; MGMT was methylated in 5 and
unmethylated in 12 samples; COX2 was unmethylated in all the
samples (Fig. 2 and Table II).
As a quality control strategy, selected GBM samples
in which the MGMT and EMP3 gene promoter methylation
was assayed by MSP were re-analyzed by bisulfite sequencing,
confirming the MSP results (Fig.
1C).
Analysis of IDH1 mutation
Analysis of the IDH1 mutational hotspot codon
R132 was evaluated in the OII and GBM patients. Among our 23 OII
patients, 13 samples showed mutated IDH1. In 12 cases, a
Arg→His amino acid substitution (codon CGT→CAT change) was observed
at residue 132; only 1 sample showed a Arg→Gly mutation (CGT→GGT).
The wild-type genotype was observed in 6 samples. Four cases were
not informative (Table I).
In our 17 GBM patients, wild-type IDH1 was
found in 14 samples, none showed a mutated gene and 3 cases were
not informative (Table II).
EMP3 gene promoter methylation vs. LOH
19q
1p/19q allelic loss in OII samples was evaluated by
microsatellite amplification as described in our previous study
(22). LOH 1p was scored in 18
samples out of the total 23 (Table
I). LOH 19q was present in 16 samples out of 22 (1 sample being
not informative) (Table I).
Concomitant LOH 1p/19q was observed in 14 out of 22 samples. Of the
16 LOH 19q samples, 15 had the EMP3 gene promoter
methylation, while only 1 exhibited the unmethylated EMP3
gene promoter (Table I). Moreover,
the 6 samples with conserved 19q chromosome arms had a methylated
EMP3 promoter (Table I).
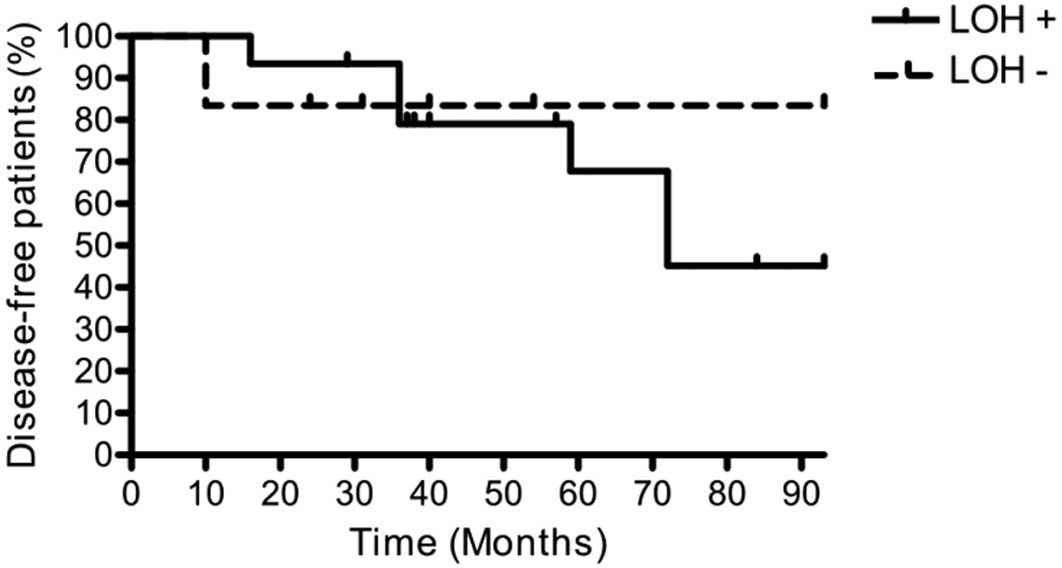
LOH 19q and DPI
DPI was evaluated in the 21 OII patients with
methylated EMP3 promoter. Among them, 15 showed LOH 19q,
while 6 showed conserved chromosome arms. Relapses were observed in
7 LOH 19q patients and in 1 patient who retained both chromosome
arms. Analysis of the DPI curves indicated a different risk of
relapse depending on LOH 19q status. When this chromosome
alteration was present, 55% of patients relapsed, compared to 17%
when both chromosome arms were conserved (Fig. 3).
Discussion
We previously described a group of 23 OII patients
evaluated for the presence of LOH 1p19q (22). OII is a neoplastic subclass of
cancer with an overall more favorable outcome than other brain
tumors. However, this outcome is heterogeneous and unpredictable at
the individual level (26); thus,
the policy to delay adjuvant therapy until clinical evidence of
disease progression (3) may fail to
treat some patients harboring aggressive tumors. Individuating
features indicating the patient's risk of relapse at diagnosis
would ensure a better clinical treatment. In our previous study
(22), we identified a subgroup of
patients with a shorter disease-free interval and LOH 19q, a
cytogenetic alteration frequently observed in OII (27,28)
but not in GBM (29). We thus
decided to evaluate in the same patients the methylation status of
the EMP3 gene promoter located in the 19q chromosome region
(19q13.3), which has gained increasing interest as it is considered
a candidate tumor suppressor gene for solid tumors (15), including brain tumors. The
hypermethylated status of the EMP3 gene promoter in
low-grade gliomas is known as a molecular feature distinguishing
them from highgrade gliomas (20,21).
Thus, a reduced expression level of EMP3 resulting from the
methylation of its gene promoter has been reported in OII (21), whereas the hyperexpression of
EMP3 has been shown in GBM (30,31).
Further support of the critical role of EMP3 methylation in
distinct glioma subtypes is provided by the observation of its
different frequency in primary GBM (pGBM), vs. secondary GBM (sGBM)
and grade II and III astrocytomas (AII and AIII) (21). The higher occurrence in the second
group supports the idea that sGBM arises from the progression of
anaplastic astrocytomas, while pGBM develops de novo via the
dysregulation of different genetic pathways (21). This alternative methylation pattern
of EMP3 in distinct glioma histotypes (GBM vs. OII) is
consistent with the data obtained examining another important tumor
suppressor gene, MGMT; its promoter methylation is currently
a recognized prognostic marker of: i) disease progression in WHO
grade III glioma (23); and ii)
response to temozolomide in GBM (11–13).
In our study, the methylation of the EMP3 gene promoter was
detected in 96% of OII patients, but only in 18% of GBM samples
(Fig. 2). All the informative OII
samples showed the MGMT gene promoter methylation, compared
to the 29% observed in GBM (Fig.
2). Further differentiation between these 2 subgroups was
provided by IDH1 genotyping: mutated IDH1 was present in
approximately 2/3 of the informative OII samples but absent in all
the GBM samples; these results are in agreement with those from
previous studies (24). The
unmethylated status of the COX2 gene promoter in both low-
and high-grade gliomas confirms the stability of the genetic region
1q, conserved in both these tumor types, and supports a correlation
between chromosome instability and CpG island methylation.
Given the findings in the literature reporting a
reduced expression level of EMP3 resulting from the
methylation of its gene promoter (21,32),
we thus assumed that in OII patients with LOH 19q, methylation of
the gene copy located on the residual chromosome arm would imply
its silencing, with an unfavorable outcome. Further support of this
hypothesis is provided by the report of a more aggressive tumor
phenotype associated with EMP3 promoter methylation in
neuroblastoma patients (14), and
by the finding that the re-expression of EMP3, induced by
demethylating agents, is associated with lower tumorigenicity in
neuroblastoma cell lines (demonstrated by lower in vitro
colony formation and reduced tumor size of xenografts) (14). Our results indicate that the
EMP3 gene promoter methylation is a hallmark in OII patients
and that when this event occurs in the presence of LOH 19q, a
complete (cyto- and epigenetic) functional loss of both EMP3
alleles occurs. Its putative role as a tumor suppressor gene fits
well with our findings regarding DPI: a higher risk of relapse was
observed in patients with LOH 19q/EMP3 methylation (55%) vs.
the patients who, although harboring methylated EMP3, had
both 19q chromosome arms conserved (17%) (Fig. 3). The small sample size, collected
over a 10-year period at our clinical institution, due to the low
frequency of this neoplastic disease, limits the statistical power
of this result; nevertheless, our observation discloses a potential
molecular explanation for the higher risk of relapse of OII
patients with deleted 19q.
In conclusion, the results from our study on OII
patients highlight the fact that LOH 19q determines the complete
loss of EMP3 function, as the conserved allele is frequently
hypermethylated, and this may represent the molecular basis of the
higher risk of relapse in this subgroup of OII patients. Testing
this hypothesis in a larger number of patients is of clinical
relevance to highlight the presence of EMP3 gene promoter
methylation together with LOH 19q as an indication for treatment
ab initio with adjuvant therapy in order to improve the
overall survival of these patients.
Aknowledgements
We are grateful to Ms. Kristina Mayberry for the
careful critical editing of the manuscript.
References
|
1
|
Louis DN, Ohgaki H, Wiestler OD, Cavenee
WK, Burger PC, Jouvet A, et al: The 2007 WHO classification of
tumours of the central nervous system. Acta Neuropathol.
114:97–109. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
El-Hateer H, Souhami L, Roberge D, Maestro
RD, Leblanc R, Eldebawy, et al: Low-grade oligodendroglioma: an
indolent but incurable disease? J Neurosurg. 111:265–271. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Mason WP: Oligodendroglioma. Curr Treat
Options Neurol. 7:305–314. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Shaw EG, Berkey B, Coons SW, Bullard D,
Brachman D, Buckner JC, et al: Recurrence following
neurosurgeon-determined gross-total resection of adult
supratentorial low-grade glioma: results of a prospective clinical
trial. J Neurosurg. 109:835–841. 2008. View Article : Google Scholar
|
|
5
|
Pignatti F, van den Bent M, Curran D,
Debruyne C, Sylvester R, Therasse P, Afra D, Cornu P, Bolla M,
Vecht C and Karim AB; European Organization for Research and
Treatment of Cancer Brain Tumor Cooperative Group; European
Organization for Research and Treatment of Cancer Radiotherapy
Cooperative Group. Prognostic factors for survival in adult
patients with cerebral low-grade glioma. J Clin Oncol.
20:2076–2084. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Jenkins RB, Blair H, Ballman KV, Giannini
C, Arusell RM, Law M, et al: At(1;19)(q10;p10) mediates the
combined deletions of 1p and 19q and predicts a better prognosis of
patients with oligodendroglioma. Cancer Res. 66:9852–9861. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Felsberg J, Erkwoh A, Sabel MC, Kirsch L,
Fimmers R, Blaschke B, et al: Oligodendroglial tumours: refinement
of candidate regions on chromosome arm 1p and correlation of 1p/19q
status with survival. Brain Pathol. 14:121–130. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Okamoto Y, Di Patre PL, Burkhard C,
Horstmann S, Jourde B, Fahey M, et al: Population-based study on
incidence, survival rates, and genetic alterations of low-grade
diffuse astrocytomas and oligodendrogliomas. Acta Neuropathol.
108:49–56. 2004. View Article : Google Scholar
|
|
9
|
van den Bent MJ, Looijenga LH, Langenberg
K, Dinjens W, Graveland W, Uytdewilligen L, et al: Chromosomal
anomalies in oligodendroglial tumours are correlated with clinical
features. Cancer. 97:1276–1284. 2003.PubMed/NCBI
|
|
10
|
Natsume A, Kondo Y, Ito M, Motomura K,
Wakabayashi T and Yoshida J: Epigenetic aberrations and therapeutic
implications in gliomas. Cancer Sci. 101:1331–1336. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Esteller M, Hamilton SR, Burger PC, Baylin
SB and Herman JG: Inactivation of the DNA repair gene
O6-methylguanine-DNA methyltransferase by promoter
hypermethylation is a common event in primary human neoplasia.
Cancer Res. 59:793–797. 1999.PubMed/NCBI
|
|
12
|
Esteller M, Garcia-Foncillas J, Andion E,
Goodman SN, Hidalgo OF, Vanaclocha V, et al: Inactivation of the
DNA-repair gene MGMT and the clinical response of gliomas to
alkylating agents. N Engl J Med. 343:1350–1354. 2000. View Article : Google Scholar
|
|
13
|
Hegi ME, Diserens AC, Gorlia T, Hamou MF,
de Tribolet N, Weller M, et al: MGMT gene silencing and benefit
from temozolomide in glioblastoma. N Engl J Med. 352:997–1003.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Alaminos M, Dávalos V, Ropero S, Setién F,
Paz MF, Herranz M, et al: EMP3, a myelin-related gene located in
the critical 19q13.3 region, is epigenetically silenced and
exhibits features of a candidate tumour suppressor in glioma and
neuroblastoma. Cancer Res. 65:2565–2571. 2005. View Article : Google Scholar
|
|
15
|
Fumoto S, Tanimoto K, Hiyama E, Noguchi T,
Nishiyama M and Hiyama K: EMP3 as a candidate tumor suppressor gene
for solid tumors. Expert Opin Ther Targets. 13:811–822. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Qu M, Jiao H, Zhao J, Ren ZP, Smits A,
Kere J and Nistér M: Molecular genetic and epigenetic analysis of
NCX2/SLC8A2 at 19q13.3 in human gliomas. Neuropathol Appl
Neurobiol. 36:198–210. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Taylor V and Suter U: Epithelial membrane
protein-2 and epithelial membrane protein-3: two novel members of
the peripheral myelin protein 22 gene family. Gene. 175:115–120.
1996. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Tews B, Felsberg J, Hartmann C, Kunitz A,
Hahn M, Toedt G, et al: Identification of novel
oligodendroglioma-associated candidate tumour suppressor genes in
1p36 and 19q13 using microarray-based expression profiling. Int J
Cancer. 119:792–800. 2006. View Article : Google Scholar
|
|
19
|
Yim JH, Kim YJ, Ko JH, Cho YE, Kim SM, Kim
JY, et al: The putative tumour suppressor gene GLTSCR2 induces
PTEN-modulated cell death. Cell Death Differ. 14:1872–1879. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Li KK, Pang JC, Chung NY, Ng YL, Chan NH,
Zhou L, et al: EMP3 overexpression is associated with
oligodendroglial tumours retaining chromosome arms 1p and 19q. Int
J Cancer. 120:947–950. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kunitz A, Wolter M, van den Boom J,
Felsberg J, Tews B, Hahn M, et al: DNA hypermethylation and
aberrant expression of the EMP3 gene at 19q13.3 in human gliomas.
Brain Pathol. 17:363–370. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Molinari C, Iorio P, Medri L, Ballardini
M, Guiducci G, Cremonini AM, et al: Chromosome 1p and 19q
evaluation in low-grade oligodendrogliomas: A descriptive study.
Int J Mol Med. 25:145–151. 2010.PubMed/NCBI
|
|
23
|
Brandes AA, Tosoni A, Cavallo G, Reni M,
Franceschi E, Bonaldi L, et al: Correlations between
O6-methylguanine DNA methyltransferase promoter
methylation status, 1p and 19q deletions, and response to
temozolomide in anaplastic and recurrent oligodendroglioma: a
prospective GICNO study. J Clin Oncol. 24:4746–4753.
2006.PubMed/NCBI
|
|
24
|
Ichimura K, Pearson DM, Kocialkowski S,
Bäcklund LM, Chan R, Jones DT and Collins VP: IDH1 mutations are
present in the majority of common adult gliomas but are rare in
primary glioblastomas. Neuro Oncol. 11:341–347. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Herman JG, Graff JR, Myöhänen S, Nelkin BD
and Baylin SB: Methylation-specific PCR: a novel PCR assay for
methylation status of CpG islands. Proc Natl Acad Sci USA.
93:9821–9826. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Giannini C, Burger PC, Berkey BA,
Cairncross JG, Jenkins RB, Mehta M, et al: Anaplastic
oligodendroglial tumours: refining the correlation among
histopathology, 1p 19q deletion and clinical outcome in Intergroup
Radiation Therapy Oncology Group Trial 9402. Brain Pathol.
18:360–369. 2008. View Article : Google Scholar
|
|
27
|
Bello MJ, Leone PE, Vaquero J, de Campos
JM, Kusak ME, Sarasa JL, et al: Allelic loss at 1p and 19q
frequently occurs in association and may represent early oncogenic
events in oligodendroglial tumours. Int J Cancer. 64:207–210. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Smith JS, Alderete B, Minn Y, Borell TJ,
Perry A, Mohapatra G, et al: Localization of common deletion
regions on 1p and 19q in human gliomas and their association with
histological subtype. Oncogene. 18:4144–4152. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kaneshiro D, Kobayashi T, Chao ST, Suh J
and Prayson RA: Chromosome 1p and 19q deletions in glioblastoma
multiforme. Appl Immunohistochem Mol Morphol. 17:512–516. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Nigro JM, Misra A, Zhang L, Smirnov I,
Colman H, Griffin C, et al: Integrated array-comparative genomic
hybridization and expression array profiles identify clinically
relevant molecular subtypes of glioblastoma. Cancer Res.
65:1678–1686. 2005. View Article : Google Scholar
|
|
31
|
Ruano Y, Mollejo M, Ribalta T, Fiaño C,
Camacho FI, Gómez E, et al: Identification of novel candidate
target genes in amplicons of Glioblastoma multiforme tumors
detected by expression and CGH microarray profiling. Mol Cancer.
5:392006. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Fumoto S, Hiyama K, Tanimoto K, Noguchi T,
Hihara J, Hiyama E, et al: EMP3 as a tumor suppressor gene for
esophageal squamous cell carcinoma. Cancer Lett. 274:25–32. 2009.
View Article : Google Scholar : PubMed/NCBI
|