Introduction
Lung cancer is the leading cause of cancer-related
death worldwide including China (1). In most patients, non-small cell lung
cancer (NSCLC), which accounts for 70–80% of lung cancer cases, is
generally diagnosed at an advanced stage accompanied by extensive
invasion or lymphatic metastasis. Successful clinical strategies
are limited, and the prognosis of NSCLC patients is still very
poor. Only 15% of lung cancer patients live more than 5 years after
diagnosis. Lung carcinogenesis is a multistep process, which is
currently believed to be associated with the activation of
oncogenes and the inactivation of tumor-suppressor genes (2). To date, the molecular mechanisms
underlying the progression of NSCLC are still poorly understood.
Therefore, investigations into the molecular mechanisms involved in
NSCLC development have major importance and may be helpful to
develop novel therapeutic targets and strategies for the treatment
of human NSCLC.
microRNAs (miRNAs) are endogenously expressed
non-coding RNA molecules that cause mRNA translation inhibition or
degradation via direct base-pairing interactions, which mainly
repress the expression of several putative targets
post-transcriptionally (3). With
the development of high throughput technologies such as microarray
platforms, more and more miRNAs have been found to be abnormally
expressed in a variety of cancers (4,5).
Moreover, emerging evidence demonstrates that miRNAs are abnormally
expressed in various types of cancers, and act as oncogenes or
tumor-suppressor genes to control many fundamental cellular
processes, including tumorigenesis, cellular differentiation,
proliferation, apoptosis, tumor invasion, metastasis and
therapeutic response (6,7). Furthermore, altered expression of
certain miRNAs can also serve as effective biomarkers for
predicting prognosis in multiple tumor types (8–10).
Lung cancer is associated with unique dysregulated miRNAs (4,11);
among them, miRNA-148a has been found to be one of the
downregulated miRNAs (11).
Currently, there is a consensus that expression
change of miRNA-148a is very important for cancer development.
Studies have shown that miRNA-148a is downregulated in cancers,
including chronic lymphocytic leukemia (12), hepatoblastoma (13), breast (14), gastrointestinal carcinoma (15–19)
and pancreatic ductal adenocarcinoma (20). It has been shown to suppress tumor
growth, angiogenesis, invasion and dissemination and promote
apoptosis in different types of human tumors (14,18,20,21).
Furthermore, low levels of miRNA-148a were observed to be
associated with advanced T stage, increased tumor size and lymph
node metastasis in gastrointestinal cancers (17,22).
miRNA-148a was also negatively associated with tumor
differentiation in adenocarcinoma of the esophagus (9). Yet, in NSCLC, there are no data
available evaluating the association between miRNA-148a expression
and clinicopathological features and clinical outcomes. Moreover
the possible molecular mechanisms in NSCLC metastasis are still not
well elucidated.
In this study, our data suggest that DNA methylation
is involved in the downregulation of miRNA-148a in NSCLC. Further
studies have demonstrated that decreased miRNA-148a expression is
closely associated with lymph node metastasis, advanced clinical
stage and poor survival, and is an independent prognostic factor
for overall survival. Moreover, miRNA-148a induces demethylation of
E-cadherin promoters and upregulates E-cadherin expression by
repressing expression of DNA methyltransferase 1 (DNMT1), which in
turn leads to suppression of the migratory and invasive abilities
of lung cancer cells.
Materials and methods
Ethical approval of the study
protocol
The present study was approved by the Ethics
Committee of Subei Hospital (Jiangsu, China), and written informed
consent was obtained from all patients.
Study population
Ten pairs of formalin-fixed, paraffin-embedded
(FFPE) NSCLC tissues (4 patients with squamous cell carcinoma and 6
adenocarcinoma) and their corresponding non-tumorous lung tissues
were collected from May to October, 2012 at Subei Hospital. Another
48 FFPE NSCLC tissues were obtained from February, 2008 to
December, 2009. On enrollment, tissue samples used in this study
were collected prior to treatment with chemoradiotherapy. Each
sample was confirmed by histopathologic evaluation through
hematoxylin and eosin (H&E) staining. Clinical data were
extracted at the time of resection, and patients who entered into
the registry were prospectively followed up for the ascertainment
of vital status through April 30, 2012.
Cell culture
Human lung cancer A549 and H1299 cells and normal
bronchial epithelial BEAS-2B cells were purchased from the Cell
Resource Center, Shanghai Institute of Biochemistry. BEAS-2B cells
were derived by transforming human bronchial epithelial cells with
an adenovirus 12-simian virus 40 construct (23). The cell lines were maintained at
37°C in a humidified air atmosphere containing 5% carbon dioxide in
RPMI-1640 (Wisent, Quebec, Canada) supplemented with 10% fetal
bovine serum (Wisent).
RNA isolation
For cultured cells, total RNA was extracted using
TRIzol reagent (Invitrogen Life Technologies, Carlsbad, CA, USA).
Total tissue RNA was extracted from FFPE tissue sections using the
miRNeasy FFPE kit (Qiagen, Valencia, CA, USA) according to the
manufacturer’s instructions. Briefly, all paraffin was removed from
freshly cut FFPE tissue sections by treating with deparaffinization
solution. Samples were under protease digestion to release RNA from
the sections and short incubation to reverse formalin cross-linking
of the released nucleic acids. DNA was removed from the RNA sample
by a final DNase digestion step. Total RNA including miRNA was
eventually dissolved in 20 μl of RNase-free water. The
concentration of RNA was measured using a NanoDrop-1000 (Thermo
Fisher Scientific, Waltham, MA, USA), and RNA integrity was
determined by 1.5% agarose gel electrophoresis.
Expression analysis of miRNA-148a by
quantitative real-time polymerase chain reaction (qRT-PCR)
cDNA synthesis for miRNA-148a validation was
performed according to the manufacturer’s recommendation. Briefly,
RNA was reverse-transcribed (RT) using a ReverAid First Strand cDNA
kit (Thermo Fisher Scientific) in combination with a stem-loop
primer for miRNA-148a. Under our experimental conditions, we used
U6 snRNA as an internal control for normalizing the expression
levels of miRNA-148a. The RT primers were designed as follows:
miRNA-148a, 5′-GTCGTATCCAG TGCAGGGTCCGAGGTATTCGCACTGGATACGACACA
AAG-3′ and U6, 5′-TGGTGTCGTGGAGTCG-3′. In addition, 1.0 μg total
RNA was combined with 4.0 μl reaction buffer (5X), 1.0 μl RiboLock
RNase inhibitor, 2.0 μl dNTP mix (10 mM each), 1.0 μl ReverAid
M-MuLV reverse transcriptase in a total reaction volume of 20 μl.
Reactions were carried out using the ABI Prism 7900HT Fast
Real-Time PCR system (Applied Biosystems, Foster City, CA, USA)
using the following conditions: 25°C for 5 min, 42°C for 60 min,
70°C for 5 min, and then hold at 4°C. After the RT reaction, miRNA
expression was detected using LightCycler® 480
SYBR-Green I Master on the LightCycler® 480 Real-Time
PCR system (both from Roche Applied Science, Indianapolis, IN,
USA). cDNA products were diluted at 20X, and the 10-μl PCR mixture
contained 1 μl diluted RT product, 5 μl SYBR-Green Master Mix, 2 μl
RNase-free water, 1 μl forward, and 1 μl reverse primers. The PCR
primers for miRNA-148a and U6 were designed as follows: miRNA-148a
forward, 5′-TCAGTGCACTACAGAACTT TGT-3′ and reverse,
5′-GCTGTCAACGATACGCTACGT-3′; U6 forward, 5′-CTCGCTTCGGCAGCACA-3′
and reverse, 5′-AACGCTTCACGAATTTGCGT-3′. The reaction was incubated
at 95°C for 10 min followed by 40 cycles of 95°C for 15 sec and
65°C for 60 sec. The relative miRNA-148a expression was calculated
using the 2−ΔCt method, where ΔCt is the difference in
threshold cycles (Ct) for the target and reference = Ct(miRNA-148a)
− Ct(U6). The RT and PCR primers were synthesized by Shenggong
Biotech Co., Ltd. (Shanghai, China).
DNA isolation and bisulfite reaction
Total tissue DNA was extracted from FFPE tissue
scrolls using a Qiagen EpiTect Plus FFPE Bisulfite kit (Qiagen)
according to the manufacturer’s instructions. Briefly, FFPE tissue
scrolls were deparaffinized, followed by proteinase digestion and
decrosslinking. The DNA bisulfite reaction was then set up and
performed using the ABI Prism 7900HT Fast Real-Time PCR system.
Upon completion of the bisulfite conversion, modified DNA was
purified by several steps of clean-up and eventually eluted. The
modified DNA concentration was measured using a NanoDrop-1000 and
stored at −20°C for further analysis.
Methylation analysis of the DNA region
encoding miRNA-148a in tissue samples by quantitative
methylation-specific polymerase chain reaction (qMS-PCR)
As previously described, the methylation rate of
miRNA-148a was determined by qMS-PCR (24). We subjected 10 ng of the modified
DNA to PCR amplification with ABI Prism 7900HT Fast Real-Time PCR
system and primers: miRNA-148a forward (5-TGGGTA
TTTGTTTTTGTTGATTG-3) and reverse (5-ACTACACTTA AACCCCCTCTAACC-3).
Reactions were carried out using the following conditions: 90°C for
5 min, 40 cycles of 95°C for 15 sec, 60°C for 30 sec and 72°C for
15 sec; final extension of 10 min at 72°C. The PCR products were
diluted into water at 500X, and 1 μl diluted products was then
subjected to qMS-PCR with LightCycler® 480 SYBR-Green I
Master. The RT primers were designed as follows:
unmethylated-specific miRNA-148a forward, 5-TATGATTTGTTTTATTATTGG
TT-3 and reverse, 5-AACACTAACAACATCAACAACC-3; methylated-specific
miRNA-148a forward, 5-TGATTCGTTT TATTATCGGTC-3 and reverse,
5-AACACTAACGACATC GACG-3. The reaction was conducted at 95°C for 10
min followed by 40 cycles of 95°C for 15 sec and 60°C for 60 sec in
the LightCycler® 480 Real-Time PCR system. The
methylation level was calculated with the formula 2−ΔCt,
where ΔCt is the difference in the threshold cycle values for
methylated and unmethylated = Ct(methylated) − Ct(unmethylated).
The methylation level was expressed as a percentage.
Treatment with 5-aza-2′-deoxycytidine in
A549 and H1299 cells
For treatment with DNA methylation inhibitor
5-aza-2′-deoxycytidine (5-aza-dC; Sigma, St. Louis, MO, USA), A549
and H1299 cells were seeded into 24-well plates on day 0 and
exposed to 5-aza-dC at a final concentration of 5 μmol/l from day 1
to 3 and then harvested for qRT-PCR assay of miRNA-148a expression
on day 4.
Cell transfection
miRNA-148a mimics and negative control
oligonucleotides were synthesized by GenePharma Co., Ltd.
(Shanghai, China). Cell transfection was performed using
Lipofectamine 2000 reagents (Invitrogen Life Technologies)
according to the manufacturer’s protocol. Twenty-four hours after
transfection, cells were collected for qRT-PCR analysis and
functional assay.
Wound-healing assay
miRNA-148a mimic-transfected cells were grown to
confluence in a 24-well plate and wounded by dragging a 200-μl
pipette tip through the monolayer. Cells were then washed to remove
cellular debris and allowed to migrate for 24 h. Images were
captured at 0 and 24 h after wounding under an Eclipse Ti-U (Nikon,
Kanagawa, Japan) inverted microscope. The relative surface traveled
by the leading edge was assessed using Image-Pro Plus version 6.0
software. Three independent experiments were performed.
Cell migration and invasion assays
For the migration assays, 2.5×104 cells
were added into the upper chamber of the insert (8-μm pore size;
Corning Incorporated, Corning, NY, USA). For the invasion assays,
cells were added into the upper chamber of the insert pre-coated
with Matrigel (BD Biosciences, Bedford, MA, USA). In both assays,
cells were plated in medium without serum, and medium containing
10% fetal bovine serum in the lower chamber served as a
chemoattractant. After 40 h of incubation, the cells that did not
migrate or invade through the pores were carefully wiped off with
cotton wool. The inserts were then stained with 20% methanol and
0.2% crystal violet, imaged with an Eclipse Ti-U inverted
microscope, and counted by Image-Pro Plus version 6.0.
Western blotting
Seventy-two hours after transfection, cells were
collected and homogenized in lysis buffer (50 mM Tris-HCl, pH 8.0,
150 mM NaCl, 0.1% SDS, 1% Nonidet P-40, 0.5% sodium deoxycholate,
0.02% sodium azide, 100 mg/l PMSF, 1 mg/l aprotinin) and
centrifuged at 11,000 × g for 15 min. The supernatant was collected
and the protein content was determined by BCA protein reagent
(Pierce Chemical Co., Rockford, IL, USA). The proteins (50 μg/lane)
were separated on 10% SDS-PAGE gels and transferred to PVDF
membranes. Membranes were blocked with 5% nonfat milk and incubated
with primary antibodies against DNMT1 (Abcam, Cambridge, MA, USA),
E-cadherin and β-actin (Santa Cruz Biotechnology, Inc., Santa Cruz,
CA, USA) overnight at 4°C. After washing, the membranes were
subjected to continued incubation with goat anti-rabbit or
anti-mouse secondary antibody (Santa Cruz Biotechnology, Inc.) and
visualized with enhanced chemiluminescence.
Methylation-specific PCR (MSP) for
promoters of E-cadherin
Genomic DNA was obtained from the cell lines and
modified with sodium bisulfite as described above. The modified
DNAs were specifically amplified by methylated and unmethylated
primers of E-cadherin as previously described (25). The PCR products were separated by
high-resolution 4% agarose E-gels (Invitrogen Life Technologies)
and visualized under UV illumination.
Statistical analysis
The results were analyzed using the SPSS 13.0
statistical software. Data are shown as means ± standard deviation
(SD), and the Student’s t-test was used for statistical analysis.
Correlation between miRNA-148a expression and the methylation level
of the DNA region encoding miRNA-148a in tissues was evaluated by
Pearson’s correlation. Descriptive statistics for the study
population were generated by miRNA-148a expression for demographic
and clinicopathologic characteristics. Categorical variables were
compared using analysis of variance test. Comparing factors for
miRNA-148a expression included gender, age, pathology, smoking
status, tumor size, histologic grade, T stage, lymph node
metastasis and clinical stage. Univariate Kaplan-Meier method was
performed to estimate disease-free survival and overall survival
according to miRNA-148a expression. Survival differences according
to expression were analyzed using the log-rank test. For the
analysis of disease-free and overall survival, patients who died
prior to recurrence were considered censored at death.
Multivariable Cox proportional hazards model analysis of factors
potentially related to survival was used to identify the
significant influence of miRNA-148a expression on survival,
adjusted for gender, age, histologic grade, T stage and smoking
status. P<0.05 was considered to indicate a statistically
significant result.
Results
miRNA-148a is downregulated in lung
cancer cell lines and tumor tissues
The expression levels of miRNA-148a in A549 and
H1299 lung cancer cells and in BEAS-2B normal lung bronchial
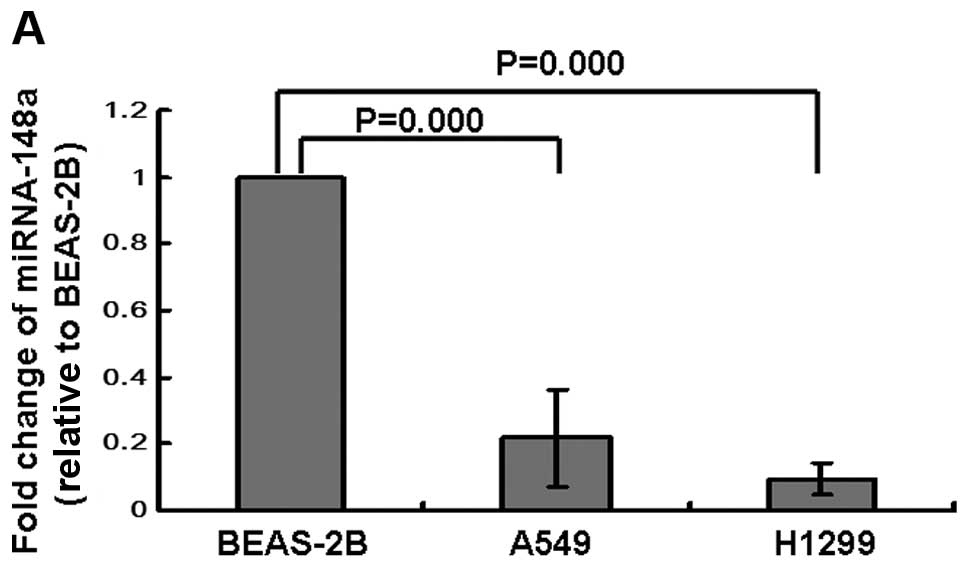
epithelial cells were determined by qRT-PCR. Our results showed
that the levels of miRNA-148a were dramatically downregulated in
lung cancer A549 and H1299 cells when compared with BEAS-2B cells
(Fig. 1A). Next, we analyzed the
expression levels of miRNA-148a in 10 pairs of FFPE lung cancer
tissue specimens and matched adjacent normal lung tissues.
Consistent with the results in the cell lines, the expression
levels of miRNA-148a in lung cancer tissues were significantly
decreased when compared to that in the matched normal lung tissues
(Fig. 1B).
Hypermethylation of the miRNA-148a gene
results in the silencing of miRNA-148a in lung cancer
There are many factors that reduce the expression of
miRNAs. CpG plot software (http://bioweb.pasteur.fr) reported two CpG islands
located close to the miRNA-148a gene, and the hypermethylation of
the DNA region encoding miRNA-148a was reported to be responsible
for its repression (24). We
retrospectively tested miRNA-148a expression in 48 FFPE lung cancer
tissues using qRT-PCR. We also analyzed the methylation status of
the DNA region encoding miRNA-148a in the same population by means
of qMS-PCR assay and found that 38 tumor tissue samples were
available for further analysis. Among the analyzed tissues, we
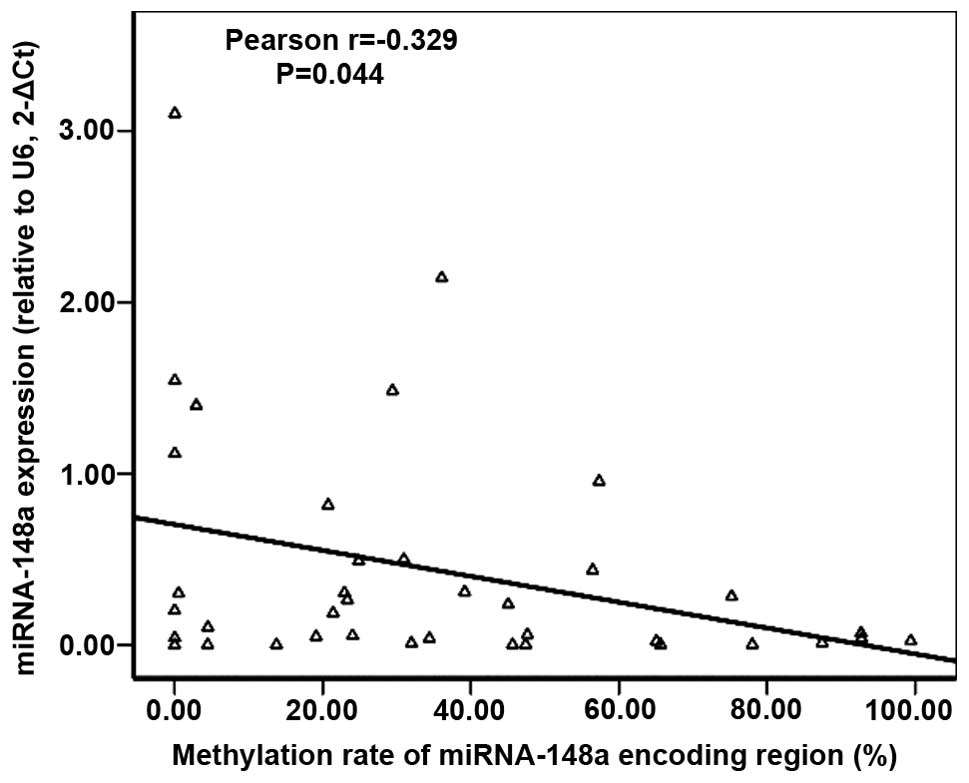
found an inverse correlation between the expression level of
miRNA-148a and the methylation rates of the miRNA-148a encoding
region in lung cancer tissues as evaluated by Pearson’s regression
(P=0.044, Fig. 2). The correlation
coefficient was −0.329. To further identify whether miRNA-148a was
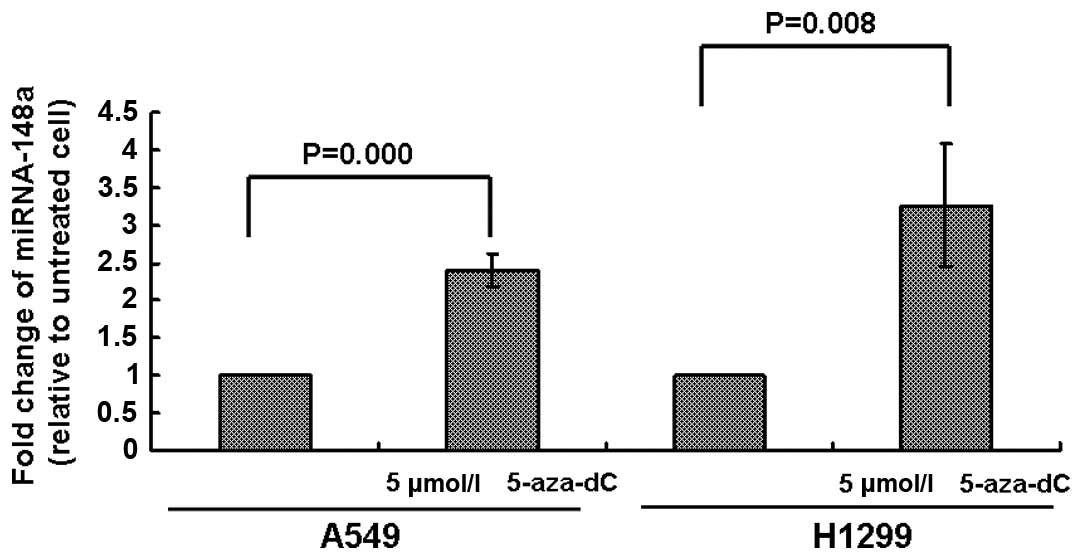
aberrantly inhibited by DNA hypermethylation, we treated A549 and
H1299 cells with 5 μmol/l 5-aza-dC for 72 h and then analyzed the
expression of miRNA-148a. As shown in Fig. 3, the expression of miRNA-148a was
increased in both A549 and H1299 cells, suggesting that DNA
methylation was involved in the silencing of miRNA-148a in lung
cancer.
Decreased expression of miRNA-148a is
associated with lymph node metastasis and advanced clinical
stage
To determine whether miRNA-148a expression is
associated with clinicopathologic features of lung cancer, we
further analyzed the relationship between miRNA-148a expression and
clinicopathologic characteristics of lung cancer. We found that
patients with lymph node metastasis and advanced clinical stage had
a significantly lower expression of miRNA-148a (P=0.043 and 0.018
respectively, Table I). There were
no significant differences between expression of miRNA-148a and
other clinicopathologic characteristics of the lung cancer cases
including gender, age, pathology, smoking status, tumor size,
histologic grade and T stage (Table
I).
 | Table IAssociations between the expression
of miRNA-148a and clinicopathologic features in patients with
NSCLC. |
Table I
Associations between the expression
of miRNA-148a and clinicopathologic features in patients with
NSCLC.
|
Characteristics | No. of
patients | Relative miRNA-148a
expression (2−ΔCt) | F-value | P-value |
|---|
| Gender |
| Male | 37 | 0.5922±0.7646 | 2.346 | 0.132 |
| Female | 11 | 0.2227±0.4066 | | |
| Age (years) |
| ≤60 | 21 | 0.5849±0.8000 | 0.435 | 0.513 |
| >60 | 27 | 0.4473±0.6454 | | |
| Pathology |
| Squamous | 25 | 0.6437±0.8492 | 1.946 | 0.170 |
|
Adenocarcinoma | 23 | 0.3595±0.5036 | | |
| Smoking status |
| Non-smoker | 20 | 0.2941±0.4823 | 3.222 | 0.079 |
| Smoker | 28 | 0.6599±0.8136 | | |
| Tumor size
(cm) |
| ≤4 | 34 | 0.6289±0.7964 | 3.568 | 0.065 |
| >4 | 14 | 0.2128±0.3045 | | |
| Histologic
grade |
| I | 12 | 0.5404±0.4651 | 0.033 | 0.856 |
| II–III | 36 | 0.4965±0.7830 | | |
| pT stage |
| T1 | 25 | 0.5904±0.7226 | 0.701 | 0.407 |
| T2 + T3 | 23 | 0.4174±0.7060 | | |
| Lymph node
metastasis |
| Negative | 26 | 0.6977±0.8781 | 4.329 | 0.043 |
| Positive | 22 | 0.2827±0.3468 | | |
| pTNM stage |
| I | 20 | 0.8437±0.9377 | 4.387 | 0.018 |
| II | 17 | 0.2876±0.3205 | | |
| III | 11 | 0.2361±0.4015 | | |
Low levels of miRNA-148a expression are
associated with shortened survival
To further evaluate the correlation between
miRNA-148a expression and the prognosis of NSCLC patients,
Kaplan-Meier survival analysis and multivariable Cox proportional
hazards analysis were performed. In this study, one patient died
from pneumonia shortly after surgery. Thus, according to the median
relative miRNA-148a expression (0.2638) in the remaining 47 tumor
tissues, patients were classified into 2 groups: the low miRNA-148a
expression group (n=24) with miRNA-148a expression less than or
equal to the median; and the high miRNA-148a group (n=23), with
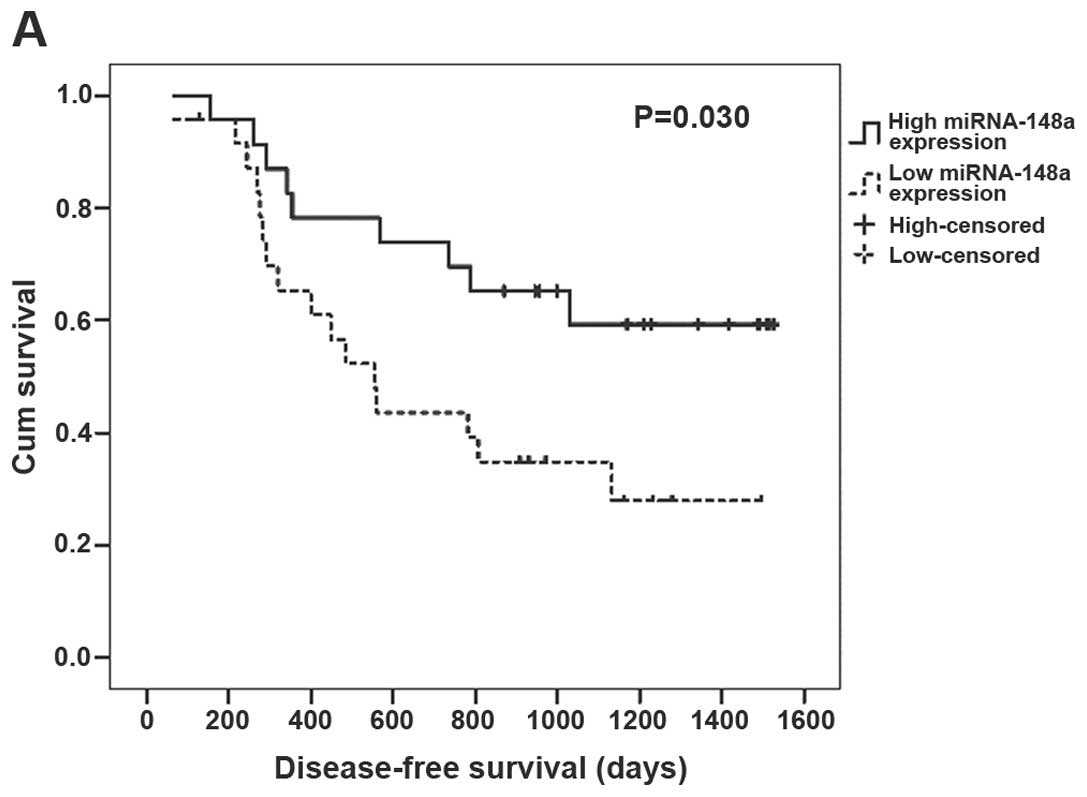
miRNA-148a expression greater than the median. In the univariate
Kaplan-Meier analysis, patients with low expression of miRNA-148a
had a significantly shorter disease-free survival (P=0.030,
Fig. 4A) and overall survival
(P=0.006, Fig. 4B) when compared
with the patients with high expression of miRNA-148a. In the
multivariable Cox proportional hazards analysis, adjusting for
gender, age, histologic grade, smoking status and T stage, our
results showed that patients with low expression of miRNA-148a had
a higher risk of tumor-related death (hazard ratio, 5.018; 95% CI,
1.257–20.028, P=0.022, Table II)
compared with the patients with high expression of miRNA-148a;
expression of miRNA-148a was not significantly associated with
disease-free survival (P=0.178).
 | Table IICox proportional hazards model
analysis of adjusted hazard ratios for disease-free and overall
survival according to miRNA-148a expression in NSCLC patients. |
Table II
Cox proportional hazards model
analysis of adjusted hazard ratios for disease-free and overall
survival according to miRNA-148a expression in NSCLC patients.
| miRNA-148a
expression | HR | 95% CI
(lower-upper) | P-value |
|---|
| Disease-free
survival | | | 0.178 |
| High | 1.000 | | |
| Low | 1.927 | 0.742–5.006 | |
| Overall
survival | | | 0.022 |
| High | 1.000 | | |
| Low | 5.018 | 1.257–20.028 | |
miRNA-148a suppresses lung cancer A549
and H1299 cell invasion in vitro
Since we observed that the downregulation of
miRNA-148a in lung cancer was closely associated with lymph node
metastasis and poor survival, we postulated that overexpression of
miRNA-148a in lung cancer cells may exert inhibitory effects on
cell invasion and metastasis. We measured the difference in the
migratory capability of lung cancer A549 and H1299 cells using
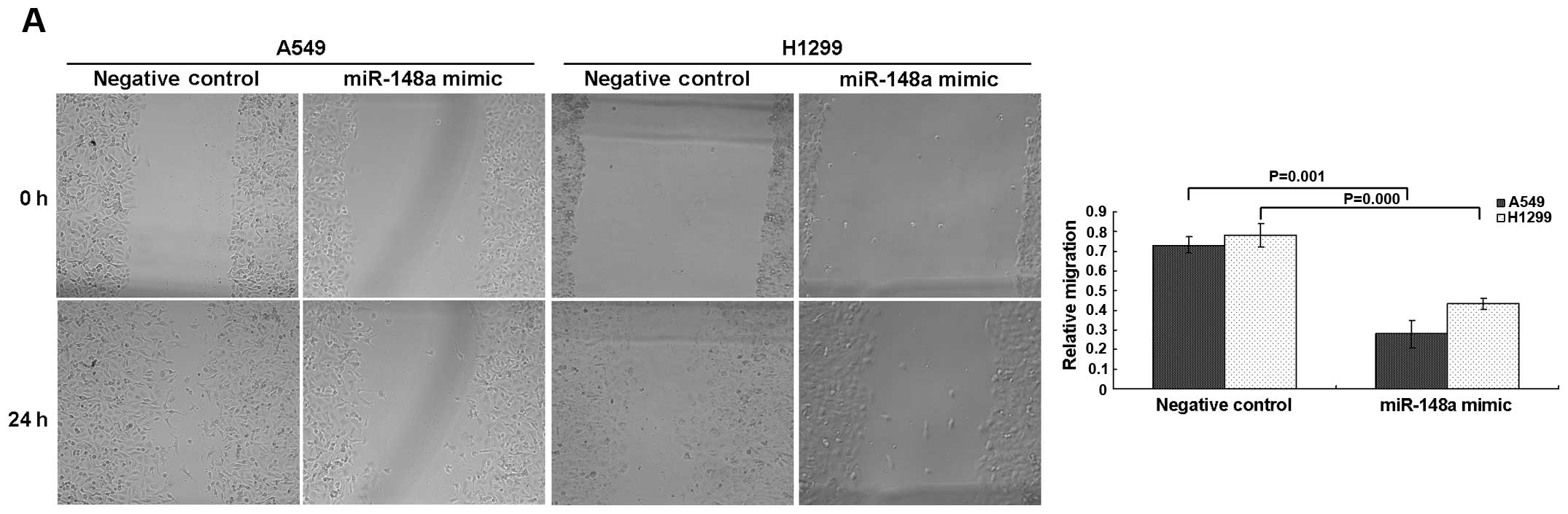
transient transfection. The wound-healing assay showed that
miRNA-148a mimic-transfected A549 and H1299 cells had a
significantly lower capability of migration than the negative
control cells (Fig. 5A). Moreover,
overexpression of miRNA-148a significantly suppressed the migratory
and invasive abilities of cells as determined by Transwell
migration and Matrigel invasion assays (Fig. 5B). These results strongly suggest
that miRNA-148a downregulation was associated with high migration
capability of lung cancer cells.
miRNA-148a downregulates DNMT1 expression
in lung cancer cells
It was predicted that both the 3′UTR and the gene
coding DNA sequence (CDS) of DNMT1 had putative miRNA-148a binding
elements, and miRNA-148a was found to inhibit DNMT1 protein
expression in CD4+ T cells and gastric cancer cells
(19,26). To explore the molecular mechanism of
miRNA-148a in lung cancer metastasis, we examined whether the
expression of DNMT1 was regulated by miRNA-148a in lung cancer
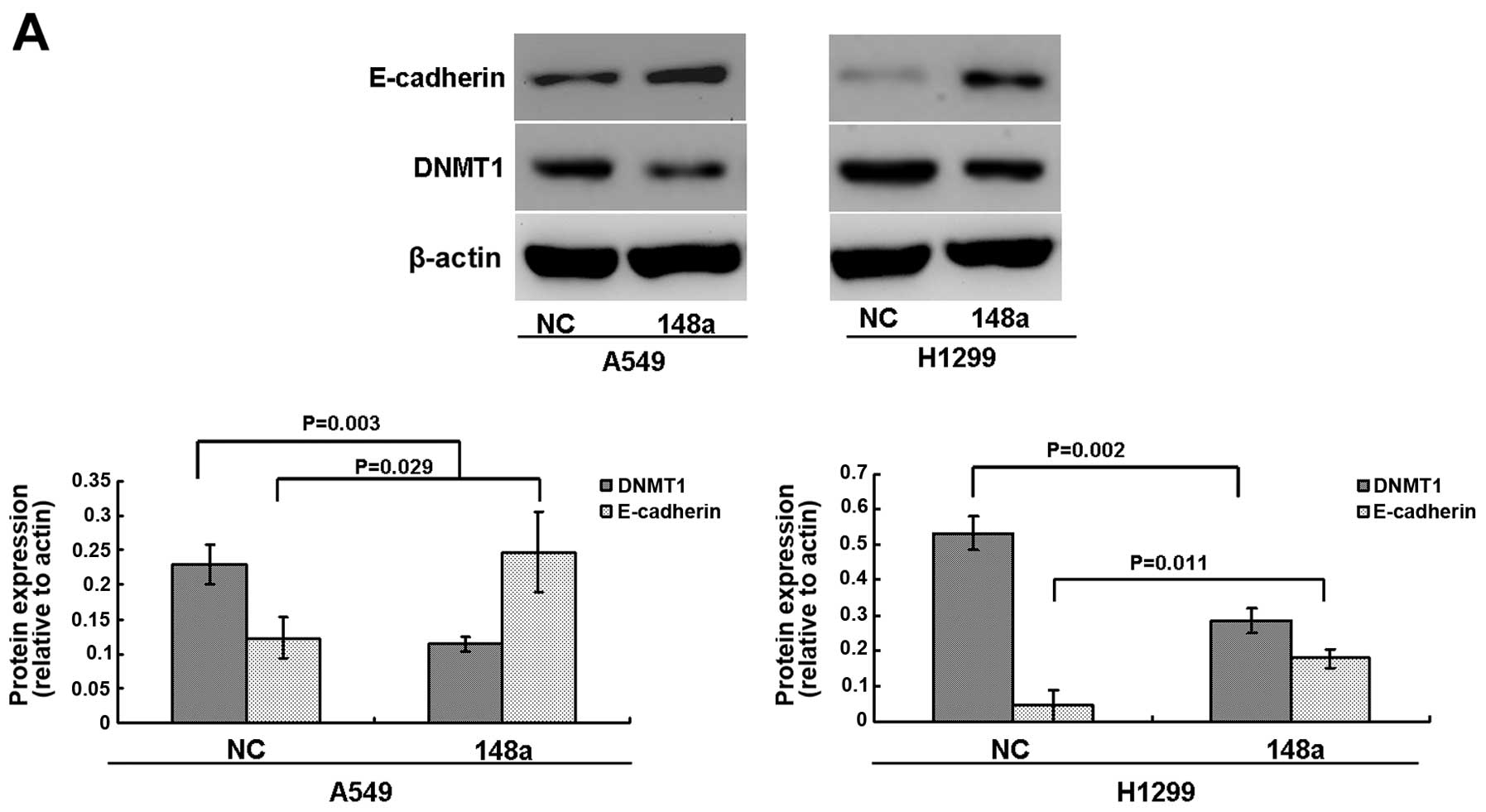
cells. As shown in Fig. 6A, our
results showed that the protein levels of DNMT1 were significantly
inhibited in both A549 and H1299 cells after transfection with the
miRNA-148a mimics.
miRNA-148a induces the overexpression of
methylation-sensitive gene E-cadherin in lung cancer cells
In lung cancer, a number of methylation-sensitive
genes including E-cadherin that are linked to lung cancer
metastasis are downregulated, and E-cadherin was found to be
demethylated and overexpressed following treatment with the DNMT
inhibitor in lung cancer cells (25). As described above, miRNA-148a
inhibited DNMT1 protein expression in lung cancer cells. We next
examined whether miRNA-148a induces the overexpression of
E-cadherin. Results showed that the protein levels of E-cadherin
were significantly upregulated in A549 and H1299 cells after
transfection with miRNA-148a mimics (Fig. 6A). We further used MSP to determine
the methylation status of the E-cadherin promoter post-transfection
of miRNA-148a mimics in lung cancer cells. Consistent with a
previous study (25), the results
showed that the A549 and H1299 cells had a nearly complete
methylation of the E-cadherin promoter (Fig. 6B). We also showed that restoration
of E-cadherin was significantly associated with its promoter
demethylation in the A549 and H1299 cell lines (Fig. 6B). Taken together, our results
suggest that the inhibition of migration by miRNA-148a may be
associated with DNMT1-mediated epigenetic regulation of
E-cadherin.
Discussion
Studies have revealed that miRNAs constitute a
robust and complicated regulatory network by post-transcriptional
regulation of almost one third of human genes. Aberrant expression
of miRNAs has been reported in several tumor types and may play a
critical role in carcinogenesis and tumor progression. Furthermore,
dysregulation of miRNA expression has also been correlated with
outcome or prognosis in many cancer types (10). Currently, miRNAs are able to be
successfully detected in many sample types, including fresh frozen
tissue samples, FFPE and blood samples. Published comparisons from
research using fresh frozen tissue samples versus FFPE for qRT-PCR
and microarray analysis have demonstrated a good correlation for
miRNA expression profiles derived from both sample types. Since
FFPE samples are easily stored in hospitals and remain stable for
many years, FFPE samples are believed to provide a feasible
alternative to fresh frozen tissue samples for miRNA expression
analysis (9,27–29).
In the present study, we used FFPE samples to detect the expression
of miRNA-148a in NSCLC. Our qRT-PCR results showed that the
expression of miRNA-148a was successfully detected in NSCLC FFPE
samples stored in our hospital, which enabled further evaluation of
the clinical value of miRNA-148a expression in NSCLC.
Recently, Watanabe et al(11) assessed the expression of miRNA-148a
in primary tumor samples and found that expression of miRNA-148a
was lower when compared to that in normal lung tissue. Since
miRNA-148a is widely accepted to be an miRNA with tumor-suppressor
activity in different types of cancer (20–22),
there are only limited data available concerning the role of
miRNA-148a in NSCLC. In our study, the miRNA-148a expression data
were gathered from 10 pairs of FFPE tissue samples and 3 cell lines
(A549, H1299 and BEAS-2B cells). The results demonstrated a
significant difference in expression between tumor and normal
samples.
There are many factors that reduce the expression of
miRNAs. DNA methylation of CpG islands is one of the important
regulatory mechanisms for gene expression, which has also been
found to be responsible for inactivating miRNA expression including
miRNA-148a (19). In a previous
study, miRNA-148a was found to be silenced by specific
hypermethylation in many types of cancers when compared with normal
tissues. The involvement of miRNA-148a hypermethylation was also
frequently observed in human primary malignancies such as colon,
breast, head and neck carcinomas and melanomas (21). In addition the aberrant
hypermethylation of the miRNA-148a coding region may lead to
downregulation of miRNA-148a expression in pancreatic ductal
adenocarcinomas (24). We examined
retrospectively miRNA-148a expression in another 48 FFPE lung
cancer tissues, and our present study showed that low expression of
miRNA-148a was associated with a high methylation level of the
miRNA-148a coding region in lung cancer tissues. Intriguingly,
miRNA-148a was upregulated by the treatment of the DNMT inhibitor
5-aza-dC in A549 and H1299 cells, which suggest that methylation of
miRNA-148a may be one possible mechanism which leads to silencing
of miRNA-148a in human lung cancer.
Currently, the association of altered miRNA
expression and cancerogenesis, tumor progression as well as patient
survival is well established (7,30,31).
In patients with esophageal squamous cell cancer, Hummel et
al(9) found a significant
negative association between miRNA-148a expression and the risk of
tumor recurrence and tumor-related death. In our study, we found a
correlation between decreased expression of miRNA-148a and clinical
signs of a more aggressive and advanced tumor stage in NSCLC
patients. Our present study also showed that low miRNA-148a
expression was significantly associated with a shortened
disease-free and overall patient survival. Furthermore,
multivariable Cox proportional hazards analysis also indicated that
the expression of miRNA-148a was an independent prognostic factor
for overall survival.
To further determine the role of miRNA-148a in tumor
progression and metastatic behavior in lung cancer A549 and H1299
cell lines, wound-healing and Transwell migration assays were
carried out. Our functional studies showed that overexpression of
miRNA-148a suppressed cell migration and invasion in vitro.
Similar to our results, Lujambio et al(21) found that the reintroduction of
miRNA-148a in SIHN-011B (head and neck cancer) cancer cells
inhibited tumor motility and metastasis in vitro and in
vivo. Moreover, miRNA-148a was also reported to function as a
tumor metastasis suppressor to suppress cancer cell migration,
invasion and lung metastasis in gastric cancer (22). Taken together, our results suggest
that miRNA-148a functions as a negative regulator for lung cancer
metastasis.
In our study, the mechanism by which miRNA-148a
inhibited the migratory and invasive abilities of NSCLC cells was
further investigated. We found that levels of DNMT1 were
significantly inhibited by miRNA-148a in lung cancer cells.
Supporting our result, DNMT1 was recently confirmed as a direct
target of miRNA-148a (19,26). DNMT1 is essential for maintaining
DNA methylation patterns of cells, which causes inactivation of
methylation-sensitive genes including metastatic repressor gene
E-cadherin, while the depletion of DNMT1 is sufficient to result in
gene overexpression via promoter demethylation (25,32,33).
There is increasing evidence to support the correlation between
reduced E-cadherin expression and gene promoter methylation in lung
cancer (25,34). Moreover the loss of E-cadherin
function was found in several invasive cancers including lung
cancer and was associated with metastasis and invasion. The present
study showed that enforced miRNA-148a expression restored
E-cadherin gene expression by demethylating its promoter. Similar
to our results, inhibition of miRNA-152 increased global DNA
methylation in liver cell lines including the methylation levels of
E-cadherin by targeting DNMT1 (33). Hence, we speculated that the
inhibition of DNMT1 induced by miRNA-148a leads to hypomethylation
of E-cadherin, which results in re-expression of E-cadherin in lung
cancer cells. This in turn leads to inhibition of cell invasion in
human lung cancers.
In conclusion, we describe miRNA-148a as an
important anti-metastatic miRNA which was downregulated in NSCLC.
Downregulated levels of miRNA-148a due to DNA methylation were
correlated with aggressive tumor behavior and poor prognosis in
patients with NSCLC. Our study also provides an important rationale
for developing epigenetic therapies targeting miRNA-148a to
re-express the methylation-silenced tumor-suppressor gene
E-cadherin in lung cancer. miRNA-148a may have the potential to
assist with preoperative assessment and provide additional
information to complement clinical decisions.
References
|
1
|
Jemal A, Tiwari RC, Murray T, et al:
Cancer statistics, 2004. CA Cancer J Clin. 54:8–29. 2004.
View Article : Google Scholar
|
|
2
|
Minna JD, Roth JA and Gazdar AF: Focus on
lung cancer. Cancer Cell. 1:49–52. 2002. View Article : Google Scholar
|
|
3
|
Dykxhoorn DM: MicroRNAs and metastasis:
little RNAs go a long way. Cancer Res. 70:6401–6406. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Yanaihara N, Caplen N, Bowman E, et al:
Unique microRNA molecular profiles in lung cancer diagnosis and
prognosis. Cancer Cell. 9:189–198. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Farazi TA, Spitzer JI, Morozov P and
Tuschl T: miRNAs in human cancer. J Pathol. 223:102–115. 2011.
View Article : Google Scholar
|
|
6
|
Garzon R, Fabbri M, Cimmino A, Calin GA
and Croce CM: MicroRNA expression and function in cancer. Trends
Mol Med. 12:580–587. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhang B, Pan X, Cobb GP and Anderson TA:
microRNAs as oncogenes and tumor suppressors. Dev Biol. 302:1–12.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Schetter AJ, Leung SY, Sohn JJ, et al:
MicroRNA expression profiles associated with prognosis and
therapeutic outcome in colon adenocarcinoma. JAMA. 299:425–436.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hummel R, Hussey DJ, Michael MZ, et al:
MiRNAs and their association with locoregional staging and survival
following surgery for esophageal carcinoma. Ann Surg Oncol.
18:253–260. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Langevin SM, Stone RA, Bunker CH, et al:
MicroRNA-137 promoter methylation is associated with poorer overall
survival in patients with squamous cell carcinoma of the head and
neck. Cancer. 117:1454–1462. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Watanabe K, Emoto N, Hamano E, et al:
Genome structure-based screening identified epigenetically silenced
microRNA associated with invasiveness in non-small-cell lung
cancer. Int J Cancer. 130:2580–2590. 2012. View Article : Google Scholar
|
|
12
|
Visone R, Rassenti LZ, Veronese A, et al:
Karyotype-specific microRNA signature in chronic lymphocytic
leukemia. Blood. 114:3872–3879. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Magrelli A, Azzalin G, Salvatore M, et al:
Altered microRNA expression patterns in hepatoblastoma patients.
Transl Oncol. 2:157–163. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Xu Q, Jiang Y, Yin Y, et al: A regulatory
circuit of miR-148a/152 and DNMT1 in modulating cell transformation
and tumor angiogenesis through IGF-IR and IRS1. J Mol Cell Biol.
5:3–13. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ueda T, Volinia S, Okumura H, et al:
Relation between microRNA expression and progression and prognosis
of gastric cancer: a microRNA expression analysis. Lancet Oncol.
11:136–146. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Katada T, Ishiguro H, Kuwabara Y, et al:
microRNA expression profile in undifferentiated gastric cancer. Int
J Oncol. 34:537–542. 2009.PubMed/NCBI
|
|
17
|
Chen Y, Song Y, Wang Z, et al: Altered
expression of MiR-148a and MiR-152 in gastrointestinal cancers and
its clinical significance. J Gastrointest Surg. 14:1170–1179. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhang H, Li Y, Huang Q, et al: MiR-148a
promotes apoptosis by targeting Bcl-2 in colorectal cancer. Cell
Death Differ. 18:1702–1710. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhu A, Xia J, Zuo J, et al: MicroRNA-148a
is silenced by hypermethylation and interacts with DNA
methyltransferase 1 in gastric cancer. Med Oncol. 29:2701–2709.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Liffers ST, Munding JB, Vogt M, et al:
MicroRNA-148a is down-regulated in human pancreatic ductal
adenocarcinomas and regulates cell survival by targeting CDC25B.
Lab Invest. 91:1472–1479. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Lujambio A, Calin GA, Villanueva A, et al:
A microRNA DNA methylation signature for human cancer metastasis.
Proc Natl Acad Sci USA. 105:13556–13561. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zheng B, Liang L, Wang C, et al:
MicroRNA-148a suppresses tumor cell invasion and metastasis by
downregulating ROCK1 in gastric cancer. Clin Cancer Res.
17:7574–7583. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Reddel RR, Ke Y, Gerwin BI, et al:
Transformation of human bronchial epithelial cells by infection
with SV40 or adenovirus-12 SV40 hybrid virus, or transfection via
strontium phosphate coprecipitation with a plasmid containing SV40
early region genes. Cancer Res. 48:1904–1909. 1988.
|
|
24
|
Hanoun N, Delpu Y, Suriawinata AA, et al:
The silencing of microRNA 148a production by DNA hypermethylation
is an early event in pancreatic carcinogenesis. Clin Chem.
56:1107–1118. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang G, Hu X, Lu C, Su C, Luo S and Luo
ZW: Promoter-hypermethylation associated defective expression of
E-cadherin in primary non-small cell lung cancer. Lung Cancer.
62:162–172. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Pan W, Zhu S, Yuan M, et al: MicroRNA-21
and microRNA-148a contribute to DNA hypomethylation in lupus
CD4+ T cells by directly and indirectly targeting DNA
methyltransferase 1. J Immunol. 184:6773–6781. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Xi Y, Nakajima G, Gavin E, et al:
Systematic analysis of microRNA expression of RNA extracted from
fresh frozen and formalin-fixed paraffin-embedded samples. RNA.
13:1668–1674. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhang X, Chen J, Radcliffe T, Lebrun DP,
Tron VA and Feilotter H: An array-based analysis of microRNA
expression comparing matched frozen and formalin-fixed
paraffin-embedded human tissue samples. J Mol Diagn. 10:513–519.
2008. View Article : Google Scholar
|
|
29
|
Leite KR, Canavez JM, Reis ST, et al:
miRNA analysis of prostate cancer by quantitative real time PCR:
comparison between formalin-fixed paraffin embedded and
fresh-frozen tissue. Urol Oncol. 29:533–537. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Esquela-Kerscher A and Slack FJ: Oncomirs
- microRNAs with a role in cancer. Nat Rev Cancer. 6:259–269. 2006.
View Article : Google Scholar
|
|
31
|
Calin GA and Croce CM: MicroRNA-cancer
connection: the beginning of a new tale. Cancer Res. 66:7390–7394.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Qiu X, Qiao F, Su X, Zhao Z and Fan H:
Epigenetic activation of E-cadherin is a candidate therapeutic
target in human hepatocellular carcinoma. Exp Ther Med. 1:519–523.
2010.PubMed/NCBI
|
|
33
|
Huang J, Wang Y, Guo Y and Sun S:
Down-regulated microRNA-152 induces aberrant DNA methylation in
hepatitis B virus-related hepatocellular carcinoma by targeting DNA
methyltransferase 1. Hepatology. 52:60–70. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Shimamoto T, Ohyashiki JH, Hirano T, Kato
H and Ohyashiki K: Hypermethylation of E-cadherin gene is frequent
and independent of p16INK4A methylation in non-small
cell lung cancer: Potential prognostic implication. Oncol Rep.
12:389–395. 2004.PubMed/NCBI
|