1. Introduction
HUGE is a database for human unidentified
gene-encoded proteins larger than 50 kDa, summarising the results
from the sequence analysis of human novel large cDNAs (>4 kb)
identified in the Kazusa cDNA sequencing project (1). All genes newly characterised are
conventionally identified by KIAA plus a 4-digit number. Until
2004, there were more than 2,400 entries in HUGE starting with
KIAA0001, and each entry has its own gene/protein characteristic
table (2). KIAA1199 was identified
in 1999 (3).
KIAA1199 (Hs.459088, TMEM2L) is located on
chromosome band 15q25.1, where a brain tumour-suppressor gene has
been mapped (4). It encodes a
150-kDa protein and was originally described as an inner
ear-specific protein (5).
Structurally, the KIAA1199 protein has three domains and an
N-terminal secretion signal, or signal peptide, according to
recently published research. The GG domain is composed of seven
β-strands and two α-helices and is ~100 amino acid residues in
size. KIAA1199 has two GG domains, and the phylogenetic tree
indicates that these two GG domains originated from separate
combination events, instead of intragenic duplication. In the two
GG domains, the N-terminal one is more homologous to the phage gp35
proteins and Dictyostelium proteins (6). Another domain presented in the
N-terminus of KIAA1199 is called G8, containing eight conserved
glycine residues and five repeated β-strand pairs. Most
G8-containing proteins are predicted to be integral membrane
proteins with signal peptides and/or transmembrane segments. Based
on the structural and functions of G8-associated domains and
proteins, it is reasonable to predict that G8 may be involved in
extracellular ligand binding and catalysis processing (7).
The gene expression of KIAA1199 is tightly
controlled by both genetic and epigenetic regulatory mechanisms
(8). A site-direct mutagenesis
study involving a series of truncated KIAA1199 promoters
combined with EMSA and ChIP demonstrated the existence of
cis-acting elements, specifically AP-1 and NF-κB sites,
within the KIAA1199 basic promoter. The basic promoter
activity of KIAA1199, however, is dependent on the DNA
methylation status with the effective region in the first intron
area of the predicted 1.9-kb long CpG island (−444 to +1509).
Further analysis showed that there are two subregions in the CpG
island based on their relatively higher GC content with respect to
the rest of the island. The first subregion was identified between
−444 and +280 mainly in the proximal part, and the second one was
identified between +525 and +1059 in the first intron. Furthermore,
the link between hypomethylation and upregulation of KIAA1199 in
human breast cancer has been established. Thus, KIAA1199 is
coordinately regulated through genetic and epigenetic mechanisms to
control its gene expression.
KIAA1199 is an endonuclear protein and can be
secreted into the extracellular environment (9). In colon cancer, strong expression of
the KIAA1199 protein is localised in the cytoplasm (10), and perinuclear space (probably the
ER), and the cell membrane of adenocarcinomas (11) which is different compared with
gastric cancer (12) and cochlea
(13) where the protein is
exclusively localised in the cytoplasm.
The first publication noting KIAA1199 was a study by
Abe et al (14) showing that
KIAA1199 was a preferentially expressed gene in the inner ear with
a Cy3/Cy5 ratio of >10 and it was localised at 15q24 (14). Early studies have laid the
foundation for more than 20 publications on human disease and the
role of KIAA1199 in cell signalling, adhesion, migration and
proliferation in human types of cancer.
2. KIAA1199 regulation and signalling
During the 10 years following the first publication
relating KIAA1199 and human disease, possible signal transduction
cascades involved in KIAA1199 signalling have been reported.
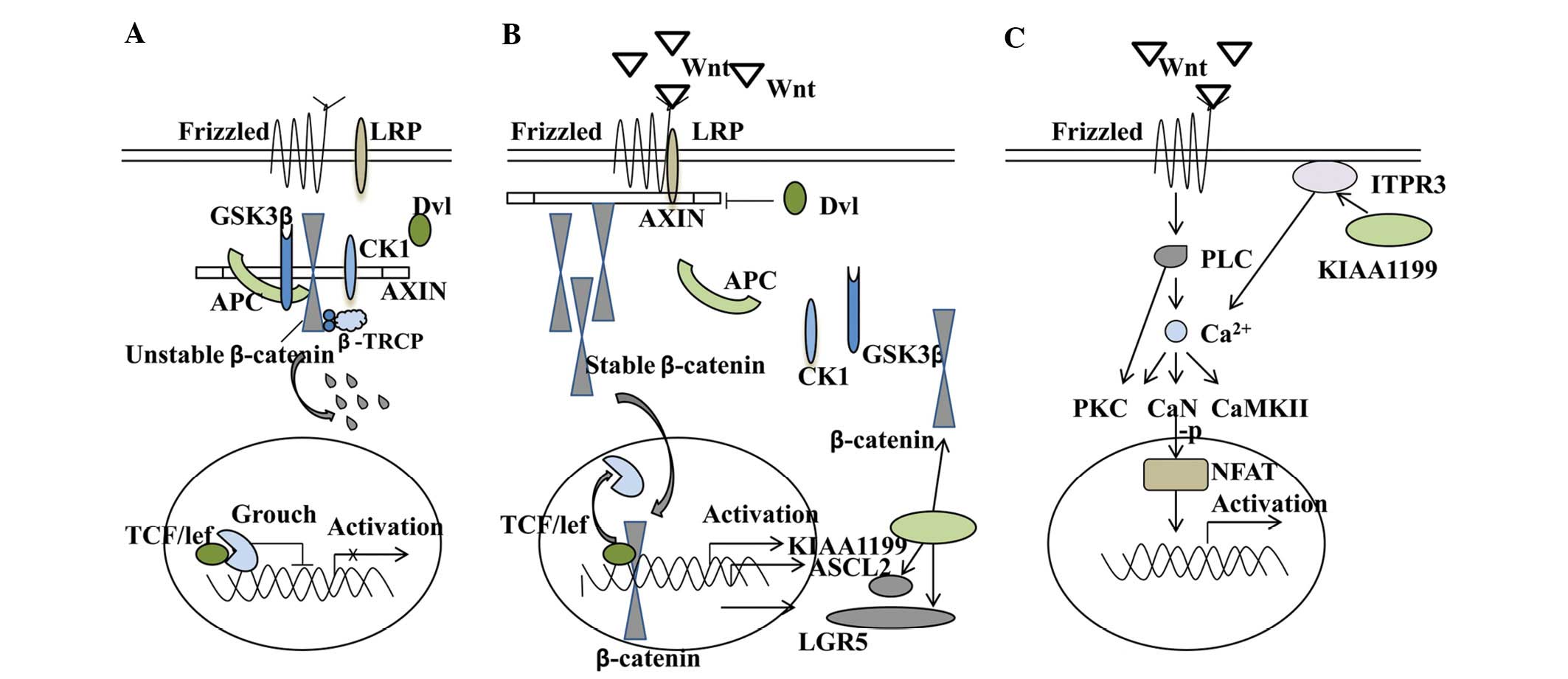
Wnt/β-catenin signalling pathway
Wnt/β-catenin signalling is a branch of an extensive
functional network that developed around a class of proteins and is
involved in a broad range of biological systems, including stem
cell biology, developmental biology and the adult organ system
(15). Wnt proteins bind to the
cell surface receptors Frizzled (Fzd) and LRP5/6, thereby
inhibiting glycogen synthase kinase (GSK-3β) and casein kinase 1
(CK1). Inhibition of these enzymes allows the stabilization of
cytoplasmic β-catenin and its eventual translocation to the
nucleus, where it interacts with TCF/LEF transcription factors and
induces transcription of a select subset of proteins, including
PPARδ, c-myc, c-jun, cyclin D1 and ASCL2. Most colorectal cancers
are caused by aberrant Wnt signalling, in which 70–80% of cases are
rooted in mutational inactivation of the tumour-suppressor gene
APC. This leads to stabilisation and nuclear translocation of the
proto-oncogene β-catenin. Perhaps the best studied pathways of
KIAA1199 is Wnt signalling in colorectal cancer.
Induction of NH2-terminal-deleted TCF4
(dominant-negative TCF4) proteins or siRNA-mediated knockdown of
β-catenin in LS174 colon cancer cells is able to decrease KIAA1199
expression. The TCF4 protein is a transcription factor involved in
the Wnt/β-catenin signalling pathway, and findings from the
genome-wide TCF4 ChIP-on-chip analyses have indicated that the
KIAA1199 locus is surrounded by four TCF4-binding regions.
Furthermore, KIAA1199 mRNA and protein are both confined to the
proliferative compartment of normal intestinal crypts, where Wnt
signalling is normally active. KIAA1199 and Wnt are highly
expressed in colorectal adenomas and carcinomas, in which this
pathway is almost always aberrantly activated. These features of
KIAA1199 expression are also compatible with its putative role as a
Wnt target gene, suggesting that KIAA1199 is a novel positively
regulated target gene of the Wnt/β-catenin signalling pathway and a
putative marker of colorectal adenomatous transformation (9). The same research group, using
immunoprecipitation experiments, further confirmed the interaction
of KIAA1199 with the ER receptor ITPR3, a key player in
Ca2+ signalling. Intracellular Ca2+ release
upon ligand binding is mediated by trimeric G-proteins, which are
involved in all three components of the Wnt network, i.e., the
canonical Wnt/β-catenin pathway, the Wnt/calcium pathway and the
Wnt/Jun N-terminal kinase pathway (16–18).
By modulating Ca2+ signalling, KIAA1199 is able to
regulate different aspects of the Wnt and exert a negative feedback
on this signalling (11). Notably,
the ingenuity pathway analysis of colorectal cell line SW480
revealed that KIAA1199 knockdown affected the expression of 67
genes involved in Wnt/β-catenin signalling; most of the genes being
downregulated upon KIAA1199 knockdown. This impact was further
substantiated by immunofluorescence microscopy and western
blotting, showing a decreased protein expression of β-catenin and
the Wnt-related stem cell marker ASCL2 (10). Together, this suggests that there
may be a dual-regulation between KIAA1199 and the Wnt/β-catenin
signalling pathway. Possible crosstalks of KIAA1199 are summarised
in Fig. 1.
Interactions between large KIAA
proteins
To detect the protein-protein interactions between
large KIAA proteins, a study based on yeast two-hybrid screening
suggested that KIAA1199 may interact with KIAA0463 (plexin 2
precursor) (19). The transmembrane
plexins interact with transmembrane semaphorins on nearby cells,
providing ‘stop’ and ‘go’ signals that are crucial for remodelling
of the cytoskeleton, cell motility and invasive growth (20). KIAA1199/plexin 2 interaction may
thus play important roles in tumourigenesis and the invasiveness of
cells.
COX2 inhibitor NS398 attenuates KIAA1199
expression
Elevated COX2 levels may lead to tumour development
and progression through activation of the EGFR and Tcf/Lef signal
transduction pathways.
N-(2-cyclohexyloxy-4-nitrophenyl)-methanesulfonamide (NS398), a
selective COX2 inhibitor, exerts its anticarcinogenic effect by
inducing apoptosis (21),
inhibiting cell cycle progression, angiogenesis (22) metastasis (23) and enhancing the effect of antitumour
agents (24). Dose-dependent
treatment of NS398 had a reverse effect on the expression of genes
increased in the colorectal normal-adenoma sequence in HT29 colon
adenocarcinoma cells, including KIAA1199 (25), although the mechanism is not yet
clear.
3. KIAA1199 and cellular motility,
invasiveness, proliferation and adhesion
KIAA1199 has been shown to be involved in several
human diseases or conditions that are marked by aberrant cell
migration and proliferation. Cell culture studies demonstrated a
potential biological role for KIAA1199 in the regulation of cell
motility, invasiveness, proliferation and adhesion.
Motility, invasiveness and
metastasis
Cell migration is a critical determinant that
precedes proliferation in the pathogenesis of a number of diseases,
including cancer, atherosclerosis, asthma and many others. The role
of KIAA1199 in promoting cell motility, invasiveness and metastasis
has been demonstrated in a number of different cell types.
In colorectal cancer SW480 cells, KIAA1199 knockdown
was found to reduce the actin filament or microtubule-based
cellular movement involving genes such as MYO1E, KIF1B or TUBE1 as
well as the maintenance of cytoskeleton signalling involving genes
such as MTSS1, ARHGAP26, DIAPH2, ABLIM1 or DAAM1. Real-time cell
analysis (RTCA)-based cellular migration analysis demonstrated
decreased migration upon KIAA1199 knockdown. Moreover, ingenuity
pathway analysis showed that molecules modulating cell-cell
contacts were altered upon KIAA1199 knockdown (10). In breast cancer MDA-MB-435 cells,
silencing of KIAA1199 resulted in mesenchymal-epithelial transition
(MET) that reduced cell migratory ability in vitro and
decreased metastasis in vivo. Gain-of-function assays
further demonstrated the role of KIAA1199 in promoting cell
migration. KIAA1199-enhanced cell migration was found to require
endoplasmic reticulum (ER) localization, where it forms a stable
complex with the chaperone binding immunoglobulin protein (BiP). A
novel ER-retention motif within KIAA1199 was identified and is
known to be required for its ER localization, interaction with ER
glucose-regulated protein 78/binding immunoglobulin protein
(GRP-78/BiP) and enhancement of cellular migration. The
KIAA1199-mediated cellular migration was hypothesised to be due to
KIAA1199-mediated ER calcium leakage and the resultant increase in
cytosolic calcium ultimately leading to protein kinase C α
activation and subsequent cell migration (26).
Proliferation
Proliferation analysis using RTCA and MTT
demonstrated that knockdown using three different constructs
reduced the proliferation rate of colorectal cancer cells by ~50%
as well as a decrease in the proliferation marker Ki-67 and
phospho-Rb (10). Knockdown of
KIAA1199 decreased the proliferation of gastric cancer cell lines
AGS and HGC27 (27).
Adhesion
Using colorectal cancer SW480 cells, RTCA
demonstrated that knockdown of KIAA1199 led to a 50% reduced
cellular adhesion. Decreased adhesion was confirmed by a
colorimetric assay on fibronectin-coated plates (10). In colorectal cancer, mutational
inactivation of the APC led to the stabilisation and nuclear
translocation of the proto-oncogene β-catenin, which has a crucial
role in cell-cell adhesion. KIAA1199 knockdown was found to
attenuate the effects of Wnt/β-catenin signalling, thus attenuating
the adhesion of SW480 cells.
4. KIAA1199 and human diseases
KIAA1199 is expressed in a wide range of normal
human tissues, with the highest level of expression noted in brain.
Mutated KIAA1199 was shown to be linked to systematic hearing loss.
Dysregulated expression of mature KIAA1199 has been frequently
detected in cancers and senescence-related diseases. In addition,
immature KIAA1199 with the N-terminal signal peptide was found to
be involved in the metabolism of hyaluronan (HA), although the
basic function of KIAA1199 remains unknown.
Hereditary hearing loss
Hereditary hearing loss is a highly heterogeneous
sensory disorder in genetic terms. By cDNA microarray analysis,
KIAA1199 was found to be highly expressed in the inner ear
(14). In situ hybridisation
findings found that KIAA1199 was expressed in Deiters’ cells and/or
the spiral ligament and has three possible non-syndromic hearing
loss-causing point mutations of which an R187C mutation in one
family (located at the N-terminal GG domain), an R187H mutation in
two unrelated families, and an H783Y mutation in one sporadic case
of non-syndromic hearing loss were noted. Although the R187C and
R187H mutations did not appear to affect cytoplasmic localisation
of the gene product in vitro, the H783Y mutation showed an
irregular and worm-eaten pattern, that may underlie the molecular
mechanism of hearing impairment (5). However, Usami et al found that
in a Japanese population, mutations in GJB2, SLC26A4 and CDH23, and
the mitochondrial 12S rRNA, are the major causes of hearing loss.
Mutations in KIAA1199 and several other genes have been
noted in independent autosomal dominant families (28). Furthermore, Usami et al
demonstrated the distribution of KIAA1199 in Deiters’ cells, as
well as in various supporting cells in the organ of Corti including
border, inner phalangeal, and inner and outer pillar cells,
although the detailed function of KIAA1199 protein in these cells
remains unknown (13).
Keratoconus with cataract
To investigate the fine mapping of the keratoconus
with cataract locus on chromosome 15q and the mutational analysis
of positional candidate genes, Dash et al carried out the
genotyping of two novel microsatellite markers and a single
nucleotide polymorphism (SNP) in the critical region of linkage for
keratoconus with cataract on 15q (29). KIAA1199 was found to be one of the
positional candidate genes for mutation (29).
Gastric cancer
KIAA1199 was found to be highly expressed in gastric
cancer, and has been associated with prognosis and lymph node
metastasis in multivariate analyses (12). Chivou Economescu et al found
that KIAA1199 was one of the seven genes that was significantly
upregulated in gastric cancer and was associated with gastric
cancer progression (30). Jia et
al (27) found that high levels
of KIAA1199 expression in gastric cancer patients were noted in
advanced stages and poorer prognosis, thus indicating a potential
prognostic value.
Colorectal tumours
Expression of KIAA1199 has been extensively studied
in colorectal tumours with differing expression profiles. In normal
mucosa, KIAA1199 protein expression has been shown to be confined
to cells in the lower portion of intestinal crypts, where Wnt
signalling is physiologically active, but is markedly increased in
adenomas (9,25), colorectal cancer (11,31)
and stage I–IV adenocarcinomas (10). However, Galamb et al
discovered higher expression of KIAA1199 only in adenoma and not in
colorectal cancer, when compared to that in normal colorectal
tissue, suggesting a possible early role of KIAA1199 in the
oncogenesis of colorectal cancer. Array-based methylation analysis
and comparison with transcript expression data has provided
additional evidence that KIAA1199 expression may be shut down in
normal colon mucosa by promotion or methylation (10). KIAA1199 mRNA was detected in the
plasma of approximately 80% of individuals with colorectal adenomas
or cancers. Unfortunately, it was also identified in 30% of
tumour-free controls, thus the ultimate diagnostic value remains to
be determined (31).
Breast cancer
Kuscu et al found that KIAA1199 was
upregulated in invasive breast cancer specimens and invasive
MDA-MB-231 breast cancer cells whereas minimal expression was noted
in non-invasive MCF-7 breast cancer cells (8), indicating a role of KIAA1199 in the
invasive potential of cancer cells. In separate studies, two breast
cancer cell lines, T-47D and ZR-75-1, with low invasive potential,
expressed virtually a negative amount of KIAA1199 mRNA whereas the
highly invasive cell lines (MDAMB-435 and MDA-MB-231) expressed
higher levels of KIAA1199. This again suggests that the
upregulation of KIAA1199 may play a role in the development of
some, but not all, breast cancers. Another possibility is that
MDA-MB-435 and MDA-MB-231 cells have a mutation in the KIAA1199
gene (4,26).
Familial glioma
Importantly, the KIAA1199 gene is located on
chromosome 15q25, where putative tumour-suppressor genes of various
human cancers are located (e.g. brain tumours, ovarian and small
cell lung carcinomas) (32–34). Of particular interest is that the
KIAA1199 gene lies within the chromosomal region showing a strong
linkage to familial gliomas (32).
Given that the brain is the organ with the highest expression of
KIAA1199, investigation of whether KIAA1199 plays a role in the
development of familial and sporadic gliomas is warranted.
Oral squamous cell carcinoma (OSCC)
Using two-dimensional (2D) SDS-PAGE electrophoresis
accompanied by mass spectrometry, 10 pairs of OSCCs and adjacent
non-tumour tissues from 5 cases of early-stage and 5 cases of
late-stage OSCCs were examined to identify differentially expressed
genes. KIAA1199 was found to be highly expressed in both early- and
late-stage OSCCs, indicating that KIAA1199 may be a novel marker
and may be involved in the oncogenesis of OSCC (35).
Uterine leiomyosarcomas (ULMS)
A genome-wide array-based comparative genomic
hybridisation (array-CGH) analysis method was used to analyse 15
cases of ULMS in order to identify novel genes following genomic
DNA copy number changes. KIAA1199 was one of the representative
frequently gained BAC clones (36),
which suggests a new regulation mode for KIAA1199 expression.
Prostate cancer
Minimal expression of KIAA1199 was found in normal
prostate epithelial PrEC cells (4)
and in non-invasive LNCaP prostate cancer cells. However, high
levels of KIAA1199 were observed in invasive DU145 prostate cancer
cells (8), again indicating that
KIAA1199 may be correlated with the progression and invasiveness of
prostate cancer cells.
Tumour-related senescence
In a study concerning the mutual paracrine
interactions of human mesenchymal stem cells (hMSCs) and
glioblastoma multiforme (GBM) cells under in vitro
co-culture conditions compared with their monocultures, Motaln
et al (37) showed that
hMSCs were responsible for the impairment of GBM cell invasion and
growth, possibly via induction of their senescence. They found
CCL2/MCP-1 and other deregulated chemokines may account senescence
for the altered cocultured cell phenotype by affecting genes
associated with proliferation, invasion and senescence including
KIAA1199 and SerpinB2.
To identify the genes involved in chromosome
3-induced cellular mortality, Michishita et al (4) performed a cDNA subtraction experiment
using immortal renal cell carcinoma cells (RCC23) and the mortal
counterpart with the transferred chromosome 3 (RCC23+3). A striking
upregulation of KIAA1199 mRNA in mortal RCC23+3 was found. Taking
the DNA localisation into account, where a brain tumour-suppressor
gene has been mapped, the findings suggest that the KIAA1199 gene
may play a role in cellular mortality, and replicative senescence
of normal human cells, which counter cell immortalisation and
carcinogenesis.
In addition, KIAA1199 was also highly expressed in
neurally cultured human skeletal muscle cells when compared with
that in tissue biopsies (38).
Cultured muscle cells displayed reductive metabolic and
muscle-system transcriptome adaptations as observed in muscle
atrophy, and they activated tissue-remodelling and
senescence/apoptosis processes. Based on the role of KIAA1199 in
cellular mortality, this suggests that cultured muscle cells had
activated a senescence process that was not established in the
muscle tissues. Further IPA analysis revealing apoptosis as a
regulated signalling pathway supported this point.
Hyaluronan metabolism
Hyaluronan (HA) has an extraordinarily high output
in tissues such as skin, cartilage and other connective tissues,
providing structural and functional integrity to organs. HA
degradation is accelerated in inflammatory diseases and cancers.
CD44 (39) and two hyaluronidases
(HYAL1 and HYAL2) (40) are thought
to be responsible for HA binding and degradation. In a study by
Yoshida et al, using glycosaminoglycan-binding assays,
KIAA1199 was found to play a central role in HA binding and
depolymerisation that was independent of CD44 and HYAL enzymes
(41). Knockdown of KIAA1199
attenuated HA degradation by human skin fibroblasts, and
transfection of KIAA1199 cDNA into cells conferred the ability to
catabolise HA in an endo-β-N-acetylglucosaminidase-dependent manner
via the clathrin-coated pit pathway. Increased degradation of HA in
synovial fibroblasts from patients with osteoarthritis or
rheumatoid arthritis was correlated with increased levels of
KIAA1199 and was abrogated by the knockdown of KIAA1199. The level
of KIAA1199 in uninflamed synovium was less than that in
osteoarthritic or rheumatoid synovium. These data suggest that
KIAA1199 is a unique hyaladherin with an important role in HA
catabolism in the dermis of the skin and arthritic synovium.
Recently, Yoshida et al disclosed that a murine homologue of
human KIAA1199 (mKiaa1199) selectively catabolised HA via the
clathrin-coated pit pathway. A glycosaminoglycan-binding assay
demonstrated the specific binding of mKiaa1199 to HA, although
slight differences were found in the peak sizes of the minimum
degradates of HA (42). Together,
it is suggested that, similar to hKIAA1199, mKiaa1199 is also a
hyaladherin, leading to HA depolymerisation. Yoshida et al
carried out further research on KIAA1199 and hyaluronan metabolism.
They found that cleavage of N-terminal 30 amino acids occurs in
functionally matured KIAA1199 resulting in altered intracellular
trafficking of the molecule and loss of cellular HA
depolymerisation. This suggests that the N-terminal portion of
KIAA1199 functions as a cleavable signal sequence required for
proper KIAA1199 translocation and KIAA1199-mediated HA
depolymerisation. Notably, the secreted mature-form of KIAA1199
showed no HA degrading activity, together supporting the idea that
KIAA1199-mediated HA depolymerisation occurred through rapid
vesicle endocytosis (41,43).
5. Conclusion
KIAA1199 is a newly found gene with significance in
several research fields, including hereditary hearing loss,
hyaluronan metabolism, senescence and cancer. KIAA1199 accelerates
senescence/apoptosis in certain situations. KIAA1199 generally
behaves as an oncogene that is highly expressed in different types
of cancers and has been correlated with the progression, distant
metastasis and poor prognosis of cancer patients. In vitro,
KIAA1199 stimulates proliferation, adhesion, motility, invasiveness
and epithelial-to-mesenchymal transition of cancer cells. Pathway
signalling analysis indicates that it is a target gene of canonical
Wnt/β-catenin signalling and by regulating Ca2+,
KIAA1199 can feedback-control all of the three components of
Wnt/β-catenin signalling, thus providing a new and promising target
for broad-spectrum cancer therapy.
Acknowledgements
The authors wish to thank the Cancer Research Wales
and the Albert Hung Foundation for supporting the present study.
This study was also supported by the 2013 National Natural Science
Foundation of China (no. 81260363).
References
|
1
|
Suyama M, Nagase T and Ohara O: HUGE: a
database for human large proteins identified by Kazusa cDNA
sequencing project. Nucleic Acids Res. 27:338–339. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kikuno R, Nagase T, Nakayama M, et al:
HUGE: a database for human KIAA proteins, a 2004 update integrating
HUGEppi and ROUGE. Nucleic Acids Res. 32:D502–D504. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Nagase T, Ishikawa K, Kikuno R, Hirosawa
M, Nomura N and Ohara O: Prediction of the coding sequences of
unidentified human genes. XV The complete sequences of 100 new cDNA
clones from brain which code for large proteins in vitro. DNA Res.
6:337–345. 1999. View Article : Google Scholar
|
|
4
|
Michishita E, Garcés G, Barrett JC and
Horikawa I: Upregulation of the KIAA1199 gene is associated with
cellular mortality. Cancer Lett. 239:71–77. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Abe S, Usami S and Nakamura Y: Mutations
in the gene encoding KIAA1199 protein, an inner-ear protein
expressed in Deiters’ cells and the fibrocytes, as the cause of
nonsyndromic hearing loss. J Hum Genet. 48:564–570. 2003.PubMed/NCBI
|
|
6
|
Guo J, Cheng H, Zhao S and Yu L: GG: a
domain involved in phage LTF apparatus and implicated in human MEB
and non-syndromic hearing loss diseases. FEBS Lett. 580:581–584.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
He QY, Liu XH, Li Q, Studholme DJ, Li XW
and Liang SP: G8: a novel domain associated with polycystic kidney
disease and non-syndromic hearing loss. Bioinformatics.
22:2189–2191. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kuscu C, Evensen N, Kim D, Hu YJ, Zucker S
and Cao J: Transcriptional and epigenetic regulation of
KIAA1199 gene expression in human breast cancer. PLoS One.
7:e446612012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Sabates-Bellver J, Van der Flier LG, de
Palo M, et al: Transcriptome profile of human colorectal adenomas.
Mol Cancer Res. 5:1263–1275. 2007. View Article : Google Scholar
|
|
10
|
Birkenkamp-Demtroder K, Maghnouj A,
Mansilla F, et al: Repression of KIAA1199 attenuates Wnt-signalling
and decreases the proliferation of colon cancer cells. Br J Cancer.
105:552–561. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Tiwari A, Schneider M, Fiorino A, et al:
Early insights into the function of KIAA1199, a markedly
overexpressed protein in human colorectal tumors. PLoS One.
8:e694732013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Matsuzaki S, Tanaka F, Mimori K, Tahara K,
Inoue H and Mori M: Clinicopathologic significance of
KIAA1199 overexpression in human gastric cancer. Ann Surg
Oncol. 16:2042–2051. 2009.
|
|
13
|
Usami S, Takumi Y, Suzuki N, et al: The
localization of proteins encoded by CRYM, KIAA1199,
UBA52, COL9A3, and COL9A1, genes highly
expressed in the cochlea. Neuroscience. 154:22–28. 2008.
|
|
14
|
Abe S, Katagiri T, Saito-Hisaminato A, et
al: Identification of CRYM as a candidate responsible for
nonsyndromic deafness, through cDNA microarray analysis of human
cochlear and vestibular tissues. Am J Hum Genet. 72:73–82.
2003.
|
|
15
|
Voronkov A and Krauss S: Wnt/beta-catenin
signaling and small molecule inhibitors. Curr Pharm Des.
19:633–664. 2013. View Article : Google Scholar
|
|
16
|
Katanaev VL, Ponzielli R, Sémériva M and
Tomlinson A: Trimeric G protein-dependent frizzled signaling in
Drosophila. Cell. 120:111–122. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ma L and Wang HY: Suppression of cyclic
GMP-dependent protein kinase is essential to the
Wnt/cGMP/Ca2+ pathway. J Biol Chem. 281:30990–31001.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kestler HA and Kühl M: From individual Wnt
pathways towards a Wnt signalling network. Philos Trans R Soc Lond
B Biol Sci. 363:1333–1347. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Nakayama M, Kikuno R and Ohara O:
Protein-protein interactions between large proteins: two-hybrid
screening using a functionally classified library composed of long
cDNAs. Genome Res. 12:1773–1784. 2002. View Article : Google Scholar
|
|
20
|
Zachary IC, Frankel P, Evans IM and
Pellet-Many C: The role of neuropilins in cell signalling. Biochem
Soc Trans. 37:1171–1178. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ramer R, Walther U, Borchert P, Laufer S,
Linnebacher M and Hinz B: Induction but not inhibition of COX-2
confers human lung cancer cell apoptosis by celecoxib. J Lipid Res.
54:3116–3129. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Abdelrahim M and Safe S: Cyclooxygenase-2
inhibitors decrease vascular endothelial growth factor expression
in colon cancer cells by enhanced degradation of Sp1 and Sp4
proteins. Mol Pharmacol. 68:317–329. 2005.
|
|
23
|
Zhou L, Wang DS, Li QJ, Sun W, Zhang Y and
Dou KF: The down-regulation of Notch1 inhibits the invasion and
migration of hepatocellular carcinoma cells by inactivating the
cyclooxygenase-2/Snail/E-cadherin pathway in vitro. Dig Dis Sci.
58:1016–1025. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Que W, Li S and Chen J: NS-398 enhances
the efficacy of bortezomib against RPMI8226 human multiple myeloma
cells. Mol Med Rep. 7:1641–1645. 2013.PubMed/NCBI
|
|
25
|
Galamb O, Spisák S, Sipos F, et al:
Reversal of gene expression changes in the colorectal
normal-adenoma pathway by NS398 selective COX2 inhibitor. Br J
Cancer. 102:765–773. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Evensen NA, Kuscu C, Nguyen HL, et al:
Unraveling the role of KIAA1199, a novel endoplasmic reticulum
protein, in cancer cell migration. J Natl Cancer Inst.
105:1402–1416. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Jia S, Ji J and Jiang WG: KIAA1199
knockdown attenuate cell growth of gastric cancer cells and its
over-expression is associated with disease progression in patient
with gastric cancer. Eur J Cancer. 45(Suppl 2): S5752013.
|
|
28
|
Usami S, Wagatsuma M, Fukuoka H, et al:
The responsible genes in Japanese deafness patients and clinical
application using Invader assay. Acta Otolaryngol. 128:446–454.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Dash DP, Silvestri G and Hughes AE: Fine
mapping of the keratoconus with cataract locus on chromosome 15q
and candidate gene analysis. Mol Vis. 12:499–505. 2006.PubMed/NCBI
|
|
30
|
Chivu Economescu M, Necula LG, Dragu D, et
al: Identification of potential biomarkers for early and advanced
gastric adenocarcinoma detection. Hepatogastroenterology.
57:1453–1464. 2010.PubMed/NCBI
|
|
31
|
LaPointe LC, Pedersen SK, Dunne R, et al:
Discovery and validation of molecular biomarkers for colorectal
adenomas and cancer with application to blood testing. PLoS One.
7:e290592012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Paunu N, Lahermo P, Onkamo P, et al: A
novel low-penetrance locus for familial glioma at 15q23–q26.3.
Cancer Res. 62:3798–3802. 2002.PubMed/NCBI
|
|
33
|
Zweemer RP, Ryan A, Snijders AM, et al:
Comparative genomic hybridization of microdissected familial
ovarian carcinoma: two deleted regions on chromosome 15q not
previously identified in sporadic ovarian carcinoma. Lab Invest.
81:1363–1370. 2001. View Article : Google Scholar
|
|
34
|
Stanton SE, Shin SW, Johnson BE and
Meyerson M: Recurrent allelic deletions of chromosome arms 15q and
16q in human small cell lung carcinomas. Genes Chromosomes Cancer.
27:323–331. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Chanthammachat P, Promwikorn W,
Pruegsanusak K, et al: Comparative proteomic analysis of oral
squamous cell carcinoma and adjacent non-tumour tissue from
Thailand. Arch Oral Biol. 58:1677–1685. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Raish M, Khurshid M, Ansari MA, et al:
Analysis of molecular cytogenetic alterations in uterine
leiomyosarcoma by array-based comparative genomic hybridization. J
Cancer Res Clin Oncol. 138:1173–1186. 2012. View Article : Google Scholar
|
|
37
|
Motaln H, Gruden K, Hren M, et al: Human
mesenchymal stem cells exploit the immune response mediating
chemokines to impact the phenotype of glioblastoma. Cell
Transplant. 21:1529–1545. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Raymond F, Métairon S, Kussmann M, et al:
Comparative gene expression profiling between human cultured
myotubes and skeletal muscle tissue. BMC Genomics. 11:1252010.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Harada H and Takahashi M: CD44-dependent
intracellular and extracellular catabolism of hyaluronic acid by
hyaluronidase-1 and -2. J Biol Chem. 282:5597–5607. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Csoka AB, Frost GI and Stern R: The six
hyaluronidase-like genes in the human and mouse genomes. Matrix
Biol. 20:499–508. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Yoshida H, Nagaoka A, Kusaka-Kikushima A,
et al: KIAA1199, a deafness gene of unknown function, is a new
hyaluronan binding protein involved in hyaluronan depolymerization.
Proc Natl Acad Sci USA. 110:5612–5617. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Yoshida H, Nagaoka A, Nakamura S, Sugiyama
Y, Okada Y and Inoue S: Murine homologue of the human KIAA1199 is
implicated in hyaluronan binding and depolymerization. FEBS Open
Bio. 3:352–356. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Yoshida H, Nagaoka A, Nakamura S, Tobiishi
M, Sugiyama Y and Inoue S: N-terminal signal sequence is required
for cellular trafficking and hyaluronan-depolymerization of
KIAA1199. FEBS Lett. 588:111–116. 2013. View Article : Google Scholar : PubMed/NCBI
|