Introduction
In 2013, cancer of the prostate (CaP) was determine
to be the most common type of tumor in males in the United States
(1,2) and worldwide (3). Early diagnostis of CaP is important
due to the increase in treatment success leading to elimination of
metastatic expansion. Currently, there is no complex test available
for CaP diagnosis and evaluation of prostate cancer stage (4). The testing process usually used for
the diagnosis of CaP includes digital rectal examination,
determination of prostate-specific antigen (PSA) (5), transrectal sonography with biopsy of
the prostate (6), magnetic
resonance imaging (7) and positron
emission tomography (8). PSA, first
described in 1977 (9), is the most
widely used biomarker of CaP to date. It is commonly used to
estimate the stage of disease and disease progression. Despite the
fact that the sensitivity (49–91%) and the specificity (68–80%) of
PSA are high, the prognosis estimate is unreliable, in early stages
in particular (10). In this
context, new biomarkers of CaP carcinoma are increasingly studied
with the prospect to serve as a useful tool for early diagnosis
without clinical examinations and/or invasive interventions
(11,12). Novel potential biomarkers with the
possibility to be determined in urine include
α-methylacyl-CoA-racemase (AMACR) (13), PCA3 (prostatic antigen 3) (14,15)
and Annexin A3 (13,16). Other reported potential biomarkers
detectable in serum include kallikrein 2, fibronectin 1,
urokinase-type plasminogen activator/urokinase-type plasminogen
activator receptor, pigment epithelium-derived factor (PEDF),
interleukin-6 and interleukin-6 receptor (17,18).
Nevertheless, the use of these markers in clinical practice and
their role in the active surveillance scenario require further
investigation. The amino acid sarcosine is currently the most
studied biomarker showing the capability to serve as a diagnostic
substance for the early stages of prostate carcinoma. Interest in
this molecule was increased in 2009 when Sreekumar and colleagues
(19) published their study
regarding metabolomic profiles of urine obtained from CaP patients.
Even though the linkage of sarcosine with prostate carcinoma
development was reported (20,21) as
well as its potential in the diagnosis of early-stage tumors
(4,22), its use as a marker is still under
discussion (23). Hence, it is
necessary to study the functions of sarcosine and other amino
acids, acting as the intermediate products of metabolism influenced
by tumor growth. Therefore, the aim of the present study was to
determine the amino acid profile of urine samples obtained from
patients suffering from CaP and to compare them with control
subjects. Biochemical analyses of samples were also carried out,
and the sarcosine content was determined. Further statistical
analysis was performed to reveal correlation between the parameters
obtained.
Materials and methods
Biological samples
Urine samples from patients suffering from cancer of
the prostate (n=32), obtained from the Department of Urology, St.
Anne’s University Hospital, Brno were used. The average age of the
patients was 68.45 years. All cases were diagnosed with different
types of acinar adenocarcinoma. Detailed information concerning the
patients is documented in Table I.
For a control measurement, urine samples from volunteers (n=32)
with an average age of 24.69 years were used. Enrollment of
patients into the clinical study was approved by the Ethics
Committee of the Faculty of Medicine, Masaryk University, Brno,
Czech Republic.
 | Table IOverview of the information of the
CaP patients, from whom urine samples were obtained. |
Table I
Overview of the information of the
CaP patients, from whom urine samples were obtained.
| Patient | Age (years) | Histology | Stage | Comorbidities | Smoker |
|---|
| 86 | 80 | Low differentiated
acinar adenocarcinoma (GS 4+5) | pT2c cN0cM0 | Polyneuropathy,
hypothyreose, glaucoma | No |
| 87 | 65 | Acinar
adenocarcinoma (GS 3+3) | pT2c cN0cM0 | HHD, HLP | No |
| 88 | 71 | Acinar
adenocarcinoma (GS 3+4) | pT2c cN0cM0 | HHD, DM II,
AFL | Stop-smoker |
| 89 | 62 | Medium
differentiated acinar adenocarcinoma (GS 3+2) | pT2c cN0cM0 | HLP, VAS | No |
| 90 | 73 | Medium
differentiated acinar adenocarcinoma (GS 3+3) | pT2c cN0cM0 | HHD | No |
| 91 | 61 | Medium
differentiated acinar adenocarcinoma (GS 3+3) | pT2a cN0cM0 | HHD, CMP | No |
| 92 | 76 | Low differentiated
acinar adenocarcinoma (GS 3+4) | pT3b cN0cMx | GIST | No |
| 93 | 64 | Medium
differentiated acinar adenocarcinoma (GS 2+3) | pT2c cN0cM0 | HHD, thyropathy,
PAOD | Yes |
| 94 | 77 | Low differentiated
acinar adenocarcinoma (GS 5+4) | pT3b cN0cM0 | HHD, DM II | Yes |
| 95 | 61 | Low differentiated
acinar adenocarcinoma (GS 3+4) | pT3b cN0cM0 | - | Yes |
| 96 | 67 | Acinar
adenocarcinoma (GS 3+4) | pT2c cN0cM0 | - | Yes |
| 97 | 65 | Medium
differentiated acinar adenocarcinoma (GS 4+3) | pT3b cN0cM0 | HLP, arthritis | No |
| 98 | 78 | Acinar
adenocarcinoma (GS 3+3) | pT1c cN0cM0 | HHD, COPD | Yes |
| 99 | 66 | Low to medium
differentiated acinar adenocarcinoma (GS 3+4) | pT2c cN0cM0 | HHD | No |
| 100 | 65 | Medium
differentiated acinar adenocarcinoma (GS 3+3) | pT2c cN0cM0 | - | No |
| 101 | 62 | Low differentiated
acinar adenocarcinoma (GS 3+4) | pT2c cN0cM0 | HHD | No |
| 102 | 66 | High grade acinar
adenocarcinoma (GS 4+5) | pT3b cN0cM0 | DM II, HHD, HLP,
hepatopathy | No |
| 103 | 63 | Acinar
adenocarcinoma (GS 3+4) | pT2a cN0cM0 | HHD, HLP, COPD | No |
| 104 | 62 | Differentiated
acinar adenocarcinoma (GS 2+3) | pT2a cN0cM0 | HHD, HLP, COPD | Yes |
| 105 | 60 | Medium
differentiated acinar adenocarcinoma (GS 3+3) | pT2c cN0cM0 | HHD, HLP, AB | No |
| 106 | 68 | Acinar
adenocarcinoma (GS 3+4) | pT2c cN0cM0 | HHD, A-Fib | No |
| 107 | 72 | Low differentiated
acinar adenocarcinoma (GS 5+4) | pT3b cN0cM0 | - | Yes |
| 108 | 72 | Medium
differentiated microacinar adenocarcinoma (GS 3+3) | pT2c cN0cM0 | HHD, ICHS, HLP | No |
| 109 | 71 | Acinar
adenocarcinoma (GS 3+3) | pT2c cN0cM0 | DM II, depressive
disorder | No |
| 110 | 67 | Acinar
adenocarcinoma (GS 3+3) | pT3a cN0cM0 | HLP | No |
| 111 | 84 | Acinar
adenocarcinoma (GS 4+5) | cT3–4 cN0cM1 | Hypertension,
vertigo | No |
| 112 | 65 | Acinar
adenocarcinoma (GS 3+4) | pT3a cN0cM0 | IHD, HHD, DM II.,
HLP | No |
| 113 | 70 | Acinar
adenocarcinoma (GS 3+4) | pT3a cN0cM0 | Hypertension,
vertigo | No |
| 114 | 84 | Acinar
adenocarcinoma (GS 5+3) | cT3–4 cN1cM1 | CKD -
hemodialyzed | No |
| 115 | 65 | Acinar
adenocarcinoma (GS 3+3) | pT1c cN0cM0 | CKD, HHD, PUD,
anemia | Stop-smoker |
| 116 | 68 | High grade acinar
adenocarcinoma (GS 5+4) | pT3b cN0cM0 | Hypertension,
overactive bladder | No |
| 117 | 72 | Acinar
adenocarcinoma (GS/+/) | pT3b cN0cM0 | Hypertension | No |
Chemicals and pH measurement
All chemicals were obtained from Sigma-Aldrich (St.
Louis, MO, USA) at ACS-specified purity unless noted otherwise. As
a buffer for ion-exchange liquid chromatographic sample preparation
sodium dilution buffer composed of 0.10 g of N3Na, 11.5
g of NaCl, 14 g of citric acid all diluted in 1 liter of water was
used. Chemicals used as a part of the kits for urine biochemical
parameters were glucose, pyrogallol red, creatinine and uric acid
(Medesa s.r.o. Policka, Czech Republic) and urea.
4-Methylumbelliferyl phosphate was obtained from Tosoh Bioscience
(Tokyo, Japan). As a derivatization agent used for ion-exchange
chromatographic analyses, ninhydrin with methyl Cellosolve (Ingos,
Prague, Czech Republic) and SnCl2 as a reduction agent
were used. Washing solutions were prepared in Milli-Q water
obtained using reverse osmosis equipment Aqual 25 (Aqual s.r.o.,
Brno, Czech Republic). The deionized water was further purified by
using apparatus Direct-Q 3 UV Water Purification system equipped
with a UV lamp from Millipore (Billerica, MA, USA). The resistance
was established at 18 MΩ·cm−1. The pH was measured using
the pH meter WTW inoLab (WTW, Weilheim, Germany).
Sample preparation for the determination
of the urine amino acid profile
The urine sample (500 μl) was pipetted into
mineralization vials and mixed with 500 μl of 35% HCl and
mineralized using the microwave equipment MW 3000 (Anton Paar,
Graz, Austria) using parameters: Power 80; Ramp, 15 min; Hold, 90
min; Max, pressure 25 bar, Rotor XF100-6. The mineralized sample
(100 μl) was diluted with 900 μl of dilution buffer and centrifuged
using Centrifuge 5417R (Eppendorf, Hamburg, Germany) under the
following conditions: temperature 4°C, 25,000 × g for 20 min.
Subsequently, 500 μl of the sample was diluted in 500 μl of 0.6 M
NaOH prior to analysis by ion-exchange chromatography.
Sample preparation for determination of
sarcosine
The urine sample (500 μl) was pipetted into a
96-well evaporation plate (Deepwell plate 96; Eppendorf AG) and
evaporated by the nitrogen blow-down evaporator Ultravap 96 with
spiral needles (Porvair Sciences Ltd., Leatherhead, UK). After this
procedure, the sample was diluted with 500 μl of dilution buffer
and was subsequently used for analysis by ion-exchange
chromatography.
Ion-exchange liquid chromatography
For determination of sarcosine, an ion-exchange
liquid chromatography (Model AAA-400; Ingos) with post column
derivatization by ninhydrin and an absorbance detector in visible
light range (VIS) was used. A glass column with an inner diameter
of 3.7 and length of 350 mm was filled manually with strong cation
exchanger (Ostion LG ANB; Ingos) in sodium cycle with ~12 μm
particles and 8% porosity. The column was thermostated at 60°C.
Double channel VIS detector with an inner cell of 5 μl was set to
two wavelengths: 440 and 570 nm. Prepared solution of ninhydrin was
stored under nitrogen atmosphere in the dark at 4°C. Elution of
amino acids was carried out by buffer containing 10.0 g of citric
acid, 5.6 g of sodium citrate, and 8.36 g of natrium chloride per
liter of solution (pH 3.0). The flow rate was 0.25
ml·min−1. The reactor temperature was set to 120°C.
Spectrophotometric analysis
For determination of all biochemical parameters a
BS-400 automated spectrophotometer (Mindray, Shenzhen, China) was
used. It is composed of cuvette space tempered to 37°C, reagent
space with a carousel for reagents (tempered to 4°C), sample space
with a carousel for preparation of samples and an optical detector.
The cuvette contents are mixed by an automatic mixer including a
stirrer immediately after addition of reagents or samples.
Contamination is reduced due to its rinsing system. For detection
itself, the following range of wavelengths were used: 340, 380,
412, 450, 505, 546, 570, 605, 660, 700, 740 and 800 nm.
Total protein was determined using the SKALAB CBT
600T kit (Skalab, Svitavy, Czech Republic), glucose was determined
using a glucose assay (Greiner, Stuttgart, Germany), creatinine
using a creatinine kit (Greiner), uric acid using a uric acid kit
(Greiner), and urea using the Urea UV 5+1 assay (Greiner) according
to the manufacturer’s instructions.
Immunoenzymometric assay (IEMA)
For analysis of PSA and fPSA in the sample of urine,
IEMA was used. Measurement was carried out using the automated
analyzer AIA 600 II (Tosoh Bioscience). Seventy microliters of
urine sample was pipetted into the testing cup ST AIA-PACK PSAII
obtained by Tosoh Bioscience containing lyophilized reagent
(magnetic microbeads with murine anti-PSA and mouse anti-PSA
conjugated with bovine alkaline phosphatase). Subsequently, the
sample was incubated at 37°C for 10 min. Non-bound antibodies were
removed by washing solution (Tosoh Bioscience). Finally fluorogenic
substrate (4-methylumbelliferyl phosphate) was added, and the
intensity of the fluorescence for determination of the activity of
the enzyme was measured.
Statistical analysis
The statistical analysis was carried out using
several tests. All values in the present study are expressed as
means ± SD. Firstly, data were checked for normality using
Shapiro-Wilk test. t-tests were used to analyze differences between
cases and controls. To outline dependencies between variables,
hierarchical clustering on normalized data was used. A P-value
<0.05 was considered to indicate a statistically significant
result. Statistica Software 10 (StatSoft, Inc., Tulsa, OK USA) was
used for analyses.
Results and Discussion
Amino acid determination in the urine
samples
The purpose of the first part of the present study
was to investigate the amino acid content in the urine of patients
suffering from prostate carcinoma and to compare the results with
the urine samples of the controls obtained from healthy
individuals. Previously, it was reported that metabolism of amino
acids is perturbed in tumor cells (24,25),
and urine amino acid profiles are consistently altered during tumor
development (20,26,27).
Proline
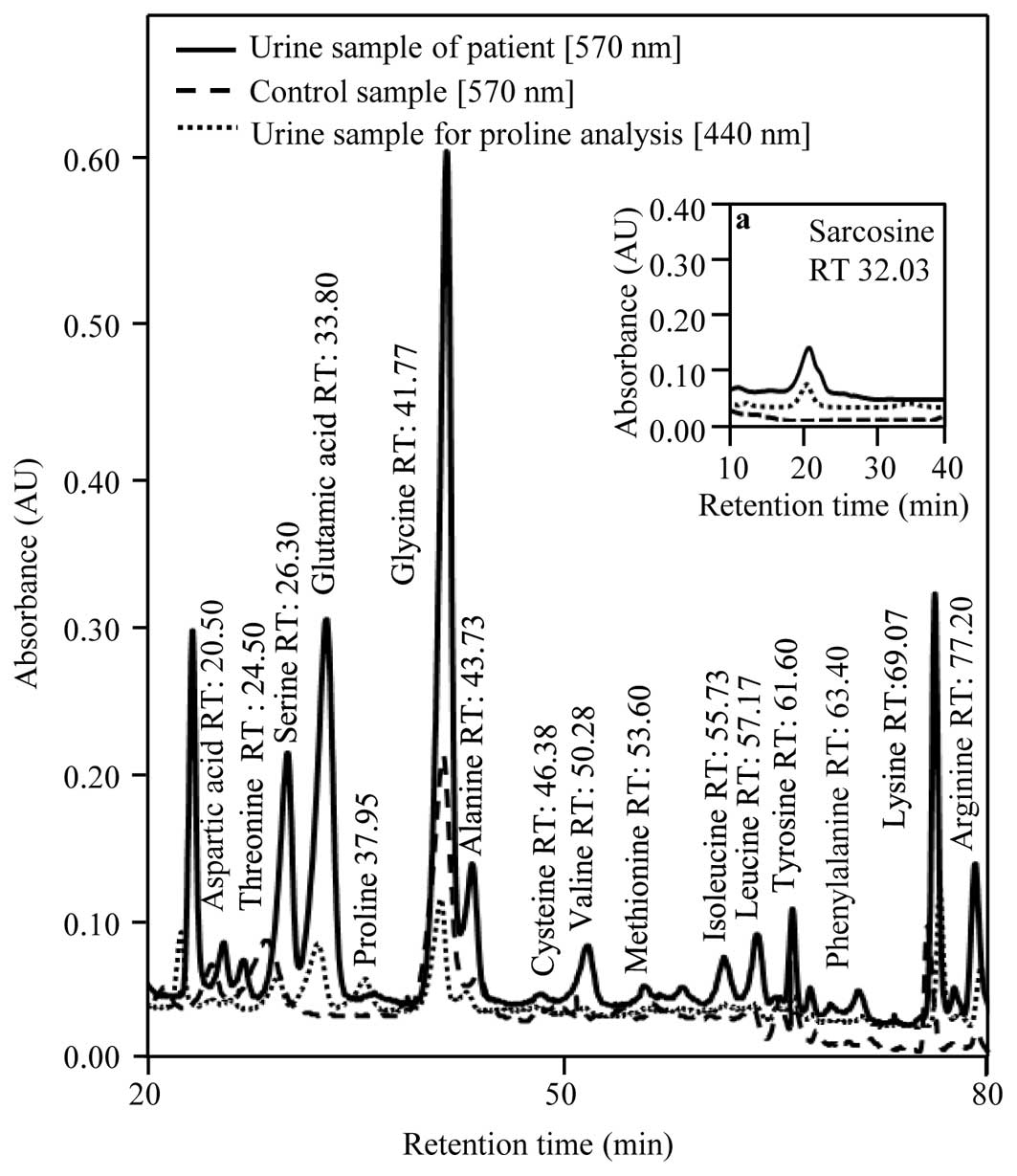
As shown in Fig. 1,
the content of most of the analyzed amino acids in the urine
obtained from the CaP patients was considerably increased. Proline
was absent in the control samples while found in all patient
samples at relatively high amounts. Proline is the only secondary
amino acid incorporated into protein. It functions with its own
distinct metabolic system, responsive to special metabolic
requirements (28). POX/PRODH, the
first enzyme in proline catabolism, is induced by genotoxic (p53),
inflammatory (PPARγ) (29) and
nutrient stress (glucose deprivation) (30). Polyak and colleagues (31) used adenoviral-p53 expression with
subsequent, extensive analysis of gene expression. They found that
14 out of 7,202 genes were induced more than 7-fold. Proline
oxidase was one of them and was marked as p53-induced gene-6
(PIG6). Proline catabolism catalyzed by POX produces
proline-dependent radical oxygen species (ROS) specifically
superoxides, resulting in proline-dependent apoptosis with
potential to serve as a novel mitochondrial tumor suppressor
(32,33). In addition, Liu et al
(34) showed that transcription
factor MYC inhibits POX/PRODH expression and, thus, inhibits its
function. microRNA miR-23b* an inhibitor of POX is highly expressed
in different types of tumors (35,36).
Due to the lack of POX, the conversion of proline to
pyrroline-5-carboxylate (P5C) is altered (37), and subsequently the amount of
proline in urine is increased. Based on these facts and the results
showing increased levels of proline compared to controls, proline
appears to be a biomolecule with the potential to enlarge the
spectrum of diagnostic tools for CaP.
Sarcosine
The role of sarcosine as a potential biomarker of
prostate carcinoma was confirmed. Sarcosine was determined in all
patient urine samples (Fig. 1a). An
elevated amount of sarcosine was probably caused by the
overexpression of glycine N-methyl transferase (GNMT),
cleaving glycine to sarcosine (23). Overexpression of GNMT, encoded by
the GNMT gene, was previously observed in patients suffering
from CaP (38). The expression of
GNMT induced in this manner leads to elevated synthesis of
GNMT that subsequently contributes to the regulation of the levels
of S-adenosylmethionine (SAM), subsequently affecting the gene
expression by influencing DNA methylation (38). The role of SAM is to transfer the
methyl groups and to use them for formation of many essential
compounds as creatine or phosphatidylcholine. It has been
previously reported that the increased conflux of GNMT results in
the elevated formation of sarcosine through increased utilization
of SAM (39). The absence of
sarcosine in control samples indicates that it is applicable for
diagnosis, due to the reduction in false-positive or negative
results (23) similar to
proline.
Total amino acid content
Furthermore, the basic statistical comparison of
amino acid content in the cases and controls was carried out, and
the results are summarized in Table
II. Values measured were recalculated to urinary creatinine
concentration. From these means, standard deviations and P-values
were calculated. All amino acids were significantly increased
except for phenylalanine amounts.
 | Table IIOverview of the amino acid content in
the urine of 32 prostate cancer patients and 32 controls. |
Table II
Overview of the amino acid content in
the urine of 32 prostate cancer patients and 32 controls.
| Cancer
patients | Healthy
controls | |
|---|
|
|
| |
|---|
| Amino acid | Mean
(μmol/mmol) | SD | Mean
(μmol/mmol) | SD | P-value |
|---|
| ASP | 3.07 | 2.69 | 0.74 | 2.28 | 0.000 |
| THR | 1.72 | 2.12 | 0.08 | 2.92 | 0.013 |
| SER | 3.43 | 0.72 | 3.28 | 1.66 | 0.640 |
| GLU | 0.92 | 3.55 | 0.50 | 3.02 | 0.605 |
| PRO | 4.79 | 1.96 | 0.31 | 0.55 | 0.000 |
| GLY | 4.08 | 1.54 | 3.61 | 1.76 | 0.260 |
| ALA | 2.65 | 1.15 | 2.36 | 1.72 | 0.433 |
| CYS | 1.43 | 1.81 | 1.02 | 1.47 | 0.324 |
| VAL | 1.14 | 1.61 | 0.72 | 1.45 | 0.280 |
| MET | 0.75 | 1.51 | 0.02 | 1.17 | 0.025 |
| ILE | 0.85 | 2.27 | 0.23 | 1.73 | 0.005 |
| LEU | 1.58 | 2.11 | 0.17 | 1.54 | 0.000 |
| TYR | 24.11 | 34.93 | 9.87 | 15.44 | 0.039 |
| PHE | 9.16 | 25.50 | 20.70 | 83.89 | 0.459 |
| HIS | 68.17 | 70.99 | 49.72 | 68.92 | 0.296 |
| LYS | 32.82 | 67.23 | 18.62 | 34.28 | 0.291 |
| ARG | 24.80 | 33.82 | 8.17 | 16.01 | 0.015 |
| SAR | 23.49 | 18.96 | 0.00 | 0.00 | 0.000 |
Statistical significant results were observed for
aspartic acid, threonine, methionine, isoleucine, leucine, tyrosine
and arginine. Levels of sarcosine and proline in the controls were
negligible (mean 0.31 μmol/mmol of creatinine for proline) or at
zero (absent or below the limit of detection) for sarcosine. Levels
of these biomolecules found in samples obtained from the patients
included proline (4.79 μmol/mmol of creatinine) and sarcosine
(23.49 μmol/mmol of creatinine) (Table
II). These findings support possible utilization of these
biomolecules for diagnosis. Relative standard deviation of proline
(1.96%) indicated relatively similar values in all cases. In
contrast, the relative standard deviations for sarcosine showed
higher scatter (18.96%). This was probably caused by the different
stages of carcinomas diagnosed in the patients. Levels of other
amino acids were altered when compared to the control samples,
supporting the general theory concerning the perturbation of tumor
cell metabolism (24,25).
Biochemical parameters of the urine
samples
Using various spectrophotometric methods, the
concentrations of K+, Na+, Cl−,
uric acid, urea, PSA, glucose, total proteins (pyrogallol method),
fPSA, creatinine and pH were measured simultaneously with the amino
acids (Table III). All parameters
were related to creatinine content and subjected to basic
statistical analysis. Statistically significant differences between
patients and controls were observed for levels of K+
ions, uric acid, urea and creatinine. Serum PSA is currently the
most widely used method for CaP diagnosis (40–42).
Detecting cancer using low PSA values risks excessive unnecessary
biopsies and the detection of clinically insignificant disease.
Although PSA has high diagnostic value in the early diagnosis of
CaP, there is a considerable overlap of PSA values between various
stages of prostate cancer, and decreasing levels cannot be used to
evaluate treatment efficacy in all patients (43). Based on this fact, it has been
suggested that the ratio of fPSA/tPSA may improve the specificity
of PSA for the diagnosis of CaP (44). As shown in Table III, the levels of both PSA and
fPSA were below the detection limits in the controls. In patients,
the levels were 4.93 μmol/mmol of creatinine and 17.46 μmol/mmol of
creatinine for PSA and fPSA, respectively, with relative standard
deviations of 7.52% for PSA and 2.12% for fPSA.
 | Table IIIOverview of the biochemical
parameters and levels of ions and pH in urine samples of 32
prostate cancer patients and 32 controls. |
Table III
Overview of the biochemical
parameters and levels of ions and pH in urine samples of 32
prostate cancer patients and 32 controls.
| Cancer
patients | Healthy
controls | |
|---|
|
|
| |
|---|
| Parameter | Mean
(μmol/mmol) | SD | Mean
(μmol/mmol) | SD | P-value |
|---|
| K+ | 5.73 | 3.84 | 8.03 | 4.08 | 0.042 |
| Na+ | 13.18 | 8.12 | 10.62 | 4.38 | 0.186 |
| Cl− | 9.12 | 6.98 | 9.98 | 4.14 | 0.607 |
| Uric acid | 15.44 | 57.95 | 0.26 | 0.10 | 0.026 |
| Urea | 38.66 | 18.42 | 23.79 | 12.26 | 0.002 |
| PSA | 4.93 | 7.52 | 0.00 | 0.00 | 0.000 |
| Glucose | 0.05 | 0.13 | 0.02 | 0.02 | 0.341 |
| Pyrogallol | 0.08 | 0.21 | 0.00 | 0.00 | 0.081 |
| fPSA | 17.46 | 2.12 | 0.00 | 0.00 | 0.000 |
| pH | 6.05 | 0.75 | 6.49 | 0.32 | 0.058 |
| Creatinine | 15.04 | 4.74 | 9.57 | 18.94 | 0.019 |
Androgen deprivation therapy (ADT) was found to
delay disease progression in the management of advanced CaP.
Nevertheless, the suppression of testosterone associated with ADT
may often lead to hypogonadal conditions with harmful effects on
renal function leading to acute kidney injury (45,46).
Deteriorated kidney tissue loses its capability to maintain its
naturally functions resulting in disturbances in urine electrolytes
(Table III). We found 5.73
μmol/mmol of creatinine of K+ ions, and their
concentrations in the controls were established at 8.03 μmol/mmol
of creatinine. The downward trend observed in K+ ions
was also noted in Cl− ions, but at a much lower level
(mean 9.12 μmol/mmol of creatinine for patients compared to 9.98
μmol/mmol of creatinine measured in the controls). An opposite
upward trend was observed in Na+ ions (mean 13.18
μmol/mmol of creatinine in patients and 10.62 μmol/mmol of
creatinine in controls). Levels of different ions in the urine of
prostate cancer patients are not well understood, mainly due to
problems regarding other associated health complications,
significantly affecting the urine electrolyte composition, such as
inflammation. Hence, these factors prevent their utilization as
auxiliary diagnostic markers of CaP.
Uric acid is an important antioxidant and free
radical scavenger formed in the body as a product of purine
degradation. Several studies have reported that the uric acid level
is depleted during tumor development (47,48).
In contrast, uric acid may be increased due to cancer therapy, such
as by chemotherapy or irradiation during treatment. Kolonel et
al (50) carried out
comprehensive analysis of different types of cancer (prostate,
stomach, colorectal, lung, urinary bladder and leukemia). There
were no significant associations between the type of cancer and
uric acid level except for prostate cancer. In prostate carcinoma a
positive association was found. Similarly we observed distinct
differences in the content of uric acid (mean 15.44 μmol/mmol of
creatinine in patients and 0.26 μmol/mmol of creatinine in
controls; Table III). Our results
confirmed the implication of uric acid in cancer pathogenesis and
indicates good accessibility of uric acid as a possible additional
diagnostic marker of CaP. This compound can be measured simply and
at low costs, and with the possibility of method automation. In
patients suffering from CaP, higher levels of urea are observed.
Values of 38.66 μmol/mmol of creatinine for patients compared to
controls (mean 23.79 μmol/mmol of creatinine; Table III) again indicate impaired
functioning of the kidneys. Higher levels of urea excreted in the
urine may potentially serve as a marker of acute kidney injury
(AKI), commonly observed in patients undergoing anticancer therapy.
Koyner et al (51) showed
that fractional excretion of urea is not able to be used to detect
a difference in AKI course, severity, and outcomes, but on the
other hand may serve as an early detection marker of kidney injury,
a frequent complication in CaP patients. It clearly follows from
the results obtained that patients were affected by kidney function
deterioration. For this reason we also noted higher levels of
creatinine, a marker of the correct glomerular filtration rate of
the kidney (52), ordinarily used
for standardization of urine waste substances (mean 15.04 μmol/l in
cases compared to 9.57 μmol/l in control samples; Table III).
For glucose content, only minimal differences with
no statistical significance were found as well as in total proteins
determined using pyrogallol red (Table III). Higher levels of proteins in
urine (mean 0.08 μmol/mmol of creatinine compared to 0.00 μmol/mmol
of creatinine in control samples) were probably caused by the
presence of chronic inflammation accompanying prostate cancer.
Inflammation has been proposed as one of the potential carcinogens
for CaP. It was shown that inflammation may be found in prostate
biopsy tissues, prostatectomy specimens, and chips from
transurethral resection of the prostate (53). Minimal differences were observed
also in the pH of the urine, but generally urine samples from
patients with prostate carcinoma showed lower pH values (mean 6.05)
when compared with the control samples (mean 6.49). Slightly acidic
pH corresponds with higher levels of proteins and uric acid;
nevertheless, this value is still within the physiologic range
(54).
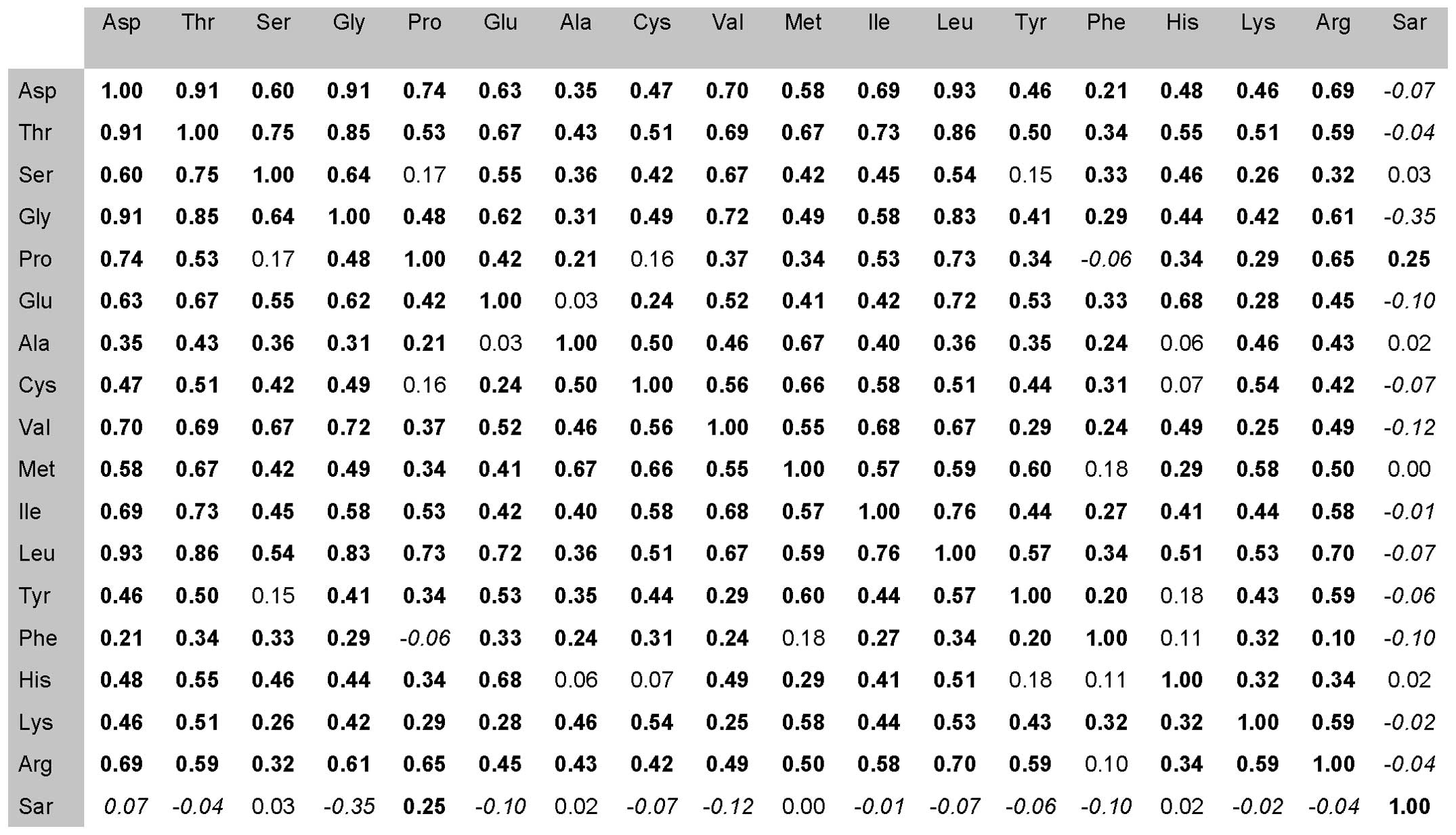
Furthermore, we carried out a correlation analysis
for each amino acid determined in the urine samples from the CaP
patients. From the values obtained from previous measurements, the
correlation coefficients were prepared and subsequently compared to
each other (Fig. 2). A positive
correlation is highlighted in bold print, negative in italics, no
correlation in normal font. As shown in Fig. 2, for sarcosine, instead of serine,
alanine, methionine and histidine, negative correlations were
observed. In contrast, when comparing sarcosine with proline, a
positive correlation was noted. Negative correlations indicate an
increased sarcosine amount in urine in comparison with other amino
acids. The only negative correlation with statistical significance
was observed in the case of glycine. On the biochemical basis of
sarcosine formation during prostate carcinoma development this
phenomenon is caused due to glycine degradation at the expense of
sarcosine creation. Montrose et al (55) previously confirmed that within tumor
tissue, sarcosine dehydrogenase (SARDH), the enzyme which converts
sarcosine to glycine, is dysregulated. On the other hand, enzymes
generating sarcosine from glycine, glycine N-methyl
transferase (GNMT) and dimethylglycine dehydrogenase (DMGDH) are
elevated in CaP patients (56).
This process may explain the increases in sarcosine levels in
urine. A high positive correlation of proline was probably random
and caused by high increases in proline levels in the patients when
compared to sarcosine.
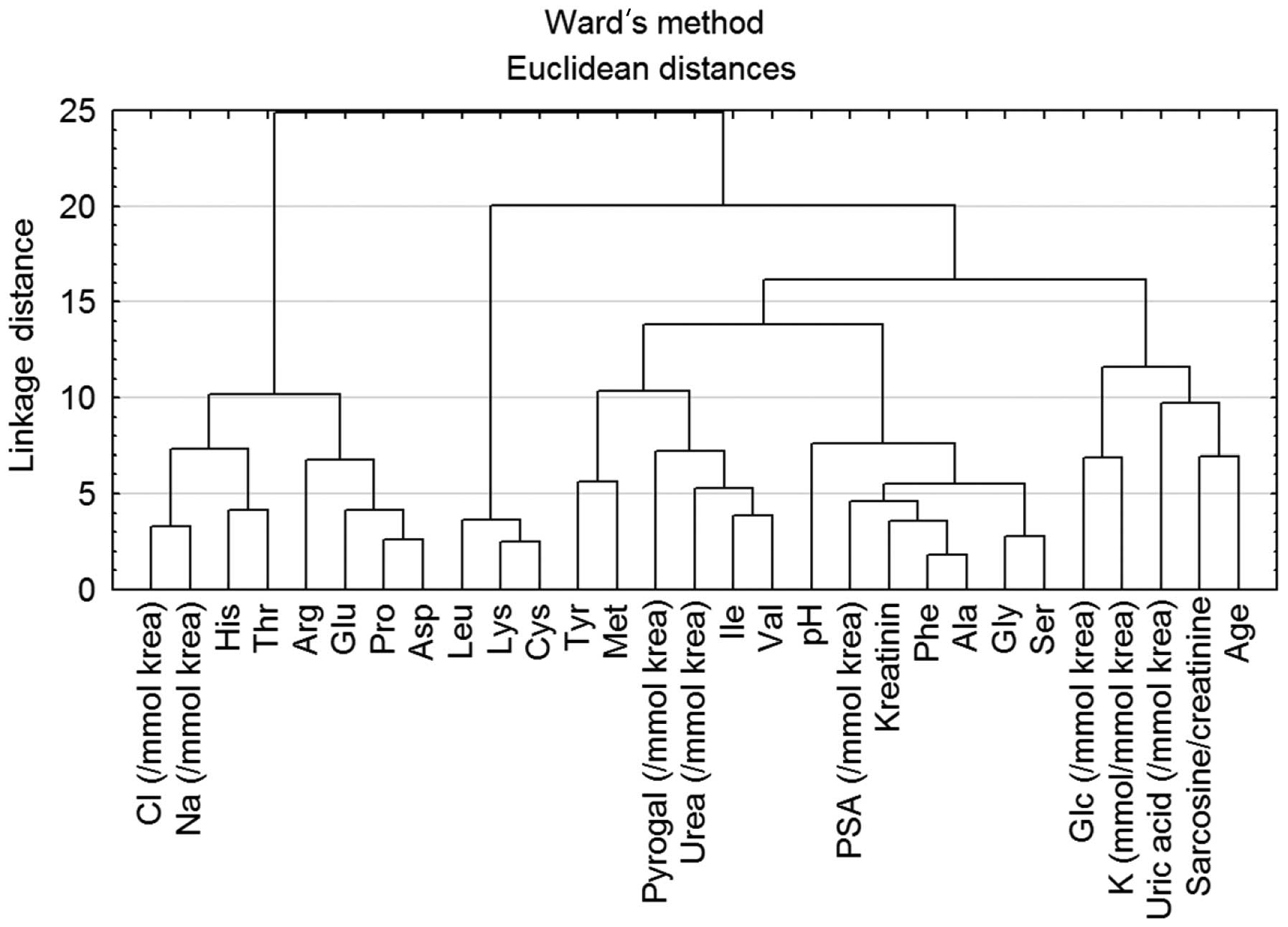
Cluster analysis of all measured urine
parameters
For revealing the correlation between all
parameters, Ward’s method of hierarchical cluster analysis was
carried out (Fig. 3). Because of
the different units for each parameter, the data were standardized
to average zero and standard deviation 1. Due to this fact the
dimensionless y-axis - linkage distance was used. From the
dendrogram, two main dependencies may be observed. First one is the
dependence of Na+ and Cl− ions. As mentioned
above, the concentration of ions in urine electrolytes is highly
influenced by kidney conditions and associated diseases. Due to
this fact, the correlation of these two parameters was difficult to
evaluate. The dependence of sarcosine on age was more significant
(Fig. 3). Although the significant
dependence was evident, in order to obtain more detailed insight
into this correlation it was necessary to perform a correlation
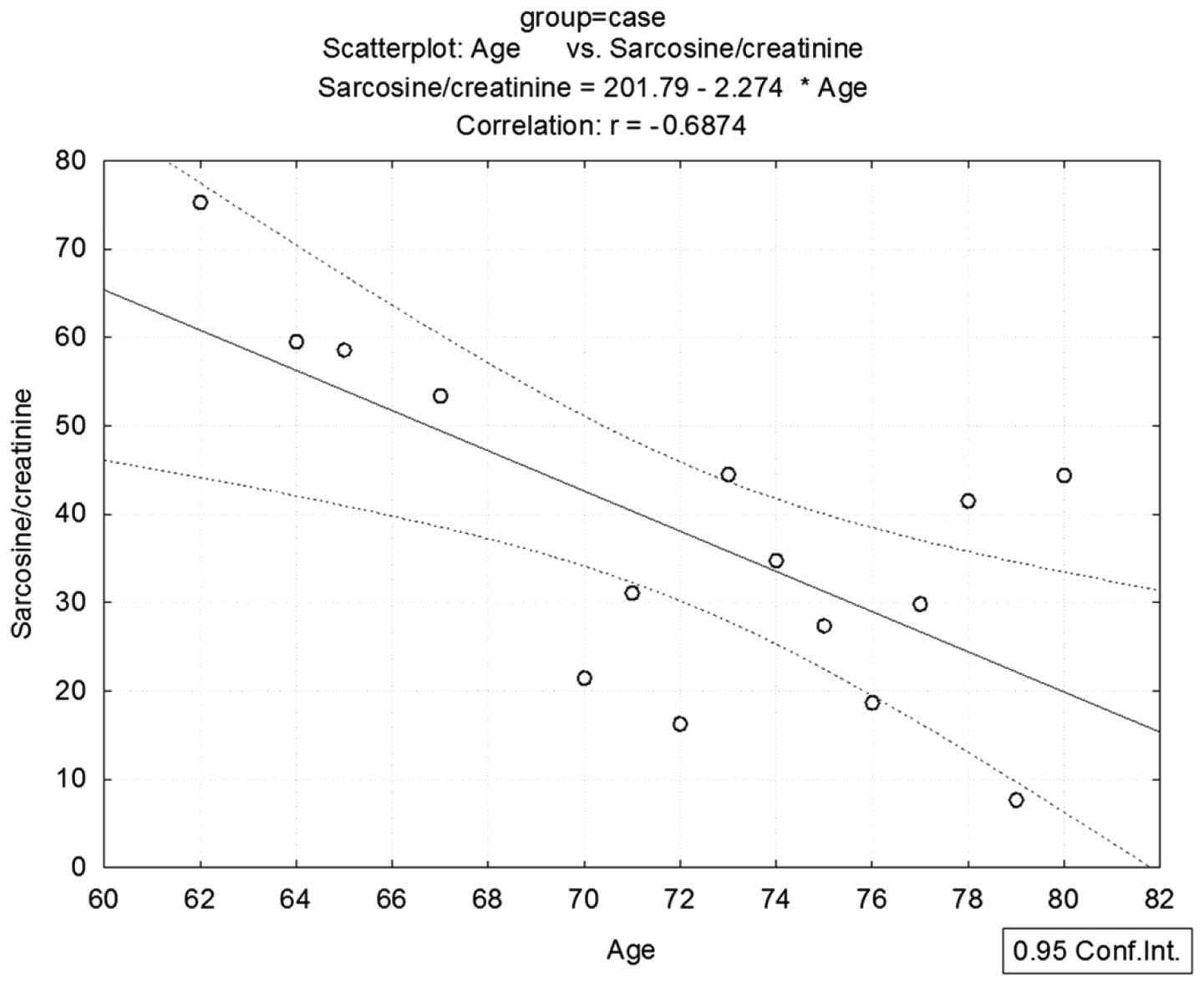
test. Statistical analysis revealed a negative dependence between
sarcosine and age (r=−0.068) (Fig.
4). Sarcosine was originally proven to be a mechanistic
biomarker of mainly aggressive prostate cancer (19). In accordance with this fact, levels
of sarcosine are decreased in elderly patients who have a higher
probability to suffer from non-aggressive prostate carcinoma with a
lower ability to produce sarcosine due to dysregulation of the
enzymes producing and catabolizing sarcosine (57). The current hypothesis is that most
aged men have prostate cancer, and they have cells in their
prostates that if observed on a needle biopsy would be diagnosed as
prostate cancer (58). This fact is
supported by the study of Powell et al (59) showing that prostate carcinoma can be
found in 50% of males 50 years of age, and 70–80% of men over the
age of 70 years, dying from non-prostate cancer-related causes.
These lesions are called ‘microscopic’ or ‘latent’ foci of prostate
cancer, typical of their small size, non-progressiveness,
clinically insignificance and rare detection through routine
prostate cancer screening. These properties are fundamentally
different from aggressive tumors in most cases detected in younger
men producing more sarcosine when compared to the non-aggressive
forms of the tumors.
In conclusion, in the present study various urine
parameters were compared between patients suffering from prostate
carcinoma and healthy individuals. Our results indicate that urine
sarcosine, proline, uric acid and PSA may serve as a set of
non-invasive, rapid, screening panel for CaP examination. Searching
for new non-invasive markers of prostate carcinoma is still a great
challenge for researchers. PSA achieves relatively excellent
results, but specificity could be enhanced. For this reason we
searched for correlations between well-known substances with
potential for routine analysis in urine samples. We found one
profile that may be utilized with relatively high meaningful
evaluation from urine samples. This profile includes analysis of
the widely studied amino acid sarcosine, amino acid proline, PSA
and uric acid. Higher levels of these substances were not found in
control samples obtained from health individuals, but were greatly
increased in the samples from CaP patients. Inclusion of these
analytes to a test panel could increase the specificity of prostate
carcinoma diagnosis. Other parameters such as urea, K+
ions or other amino acids were also altered, but their presence in
control samples hindered their utilization for diagnosis. Moreover,
their increased levels may rather indicate kidney injury following
treatment. Analysis of the above mentioned substances with
potential to serve as non-invasive biomarkers can be achieved at a
relatively low cost, but the utilization in clinical practice
requires examination of a larger cohort of patients.
Acknowledgements
The present study was financially supported by the
CEITEC CZ.1.05/1.1.00/02.0068 and the project for conceptual
development of research organization 00064203.
References
|
1
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2013. CA Cancer J Clin. 63:11–30. 2013. View Article : Google Scholar
|
|
2
|
Ding ZH, Wu CJ, Chu GC, et al:
SMAD4-dependent barrier constrains prostate cancer growth and
metastatic progression. Nature. 470:269–273. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Boyle P and Ferlay J: Cancer incidence and
mortality in Europe, 2004. Ann Oncol. 16:481–488. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Armstrong AJ, Eisenberger MA, Halabi S, et
al: Biomarkers in the management and treatment of men with
metastatic castration-resistant prostate cancer. Eur Urol.
61:549–559. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Prensner JR, Rubin MA, Wei JT and
Chinnaiyan AM: Beyond PSA: the next generation of prostate cancer
biomarkers. Sci Transl Med. 4:1–12. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lattanzi J, McNeely S, Hanlon A, Das I,
Schultheiss TE and Hanks GE: Daily CT localization for correcting
portal errors in the treatment of prostate cancer. Int J Radiat
Oncol Biol Phys. 41:1079–1086. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
van Vugt HA, Roobol MJ, Busstra M, et al:
Compliance with biopsy recommendations of a prostate cancer risk
calculator. BJU Int. 109:1480–1488. 2012.PubMed/NCBI
|
|
8
|
Schoder H and Larson SM: Positron emission
tomography for prostate, bladder, and renal cancer. Semin Nucl Med.
34:274–292. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Fukushima K, Satoh T, Baba S and Yamashita
K: α 1,2-Fucosylated and β-N-acetylgalactosaminylated
prostate-specific antigen as an efficient marker of prostatic
cancer. Glycobiology. 20:452–460. 2010.
|
|
10
|
Page ST, Hirano L, Gilchriest J, et al:
Dutasteride reduces prostate size and prostate-specific antigen in
older hypogonadal men with benign prostatic hyperplasia undergoing
testosterone replacement therapy. J Urol. 186:191–197. 2011.
View Article : Google Scholar
|
|
11
|
Zitka O, Cernei N, Heger Z, et al:
Microfluidic chip coupled with modified paramagnetic particles for
sarcosine isolation in urine. Electrophoresis. 34:2639–2647. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Berger MF, Lawrence MS, Demichelis F, et
al: The genomic complexity of primary human prostate cancer.
Nature. 470:214–220. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Cao DL, Ye DW, Zhang HL, Zhu Y, Wang YX
and Yao XD: A multiplex model of combining gene-based,
protein-based, and metabolite-based with positive and negative
markers in urine for the early diagnosis of prostate cancer.
Prostate. 71:700–710. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Rigau M, Morote J, Mir MC, et al: PSGR and
PCA3 as biomarkers for the detection of prostate cancer in urine.
Prostate. 70:1760–1767. 2010.PubMed/NCBI
|
|
15
|
Crawford ED, Rove KO, Trabulsi EJ, et al:
Diagnostic performance of PCA3 to detect prostate cancer in men
with increased prostate specific antigen: a prospective study of
1,962 cases. J Urol. 188:1726–1731. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Jamaspishvili T, Kral M, Khomeriki I,
Student V, Kolar Z and Bouchal J: Urine markers in monitoring for
prostate cancer. Prostate Cancer Prostatic Dis. 13:12–19. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Pin E, Fredolini C and Petricoin EF: The
role of proteomics in prostate cancer research: biomarker discovery
and validation. Clin Biochem. 46:524–538. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Vesprini D, Liu S and Nam R: Predicting
high risk disease using serum and DNA biomarkers. Curr Opin Urol.
23:252–260. 2013.PubMed/NCBI
|
|
19
|
Sreekumar A, Poisson LM, Rajendiran TM, et
al: Metabolomic profiles delineate potential role for sarcosine in
prostate cancer progression. Nature. 457:910–914. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Cavaliere B, Macchione B, Monteleone M,
Naccarato A, Sindona G and Tagarelli A: Sarcosine as a marker in
prostate cancer progression: a rapid and simple method for its
quantification in human urine by solid-phase microextraction-gas
chromatography-triple quadrupole mass spectrometry. Anal Bioanal
Chem. 400:2903–2912. 2011. View Article : Google Scholar
|
|
21
|
Petersen LF, Brockton NT, Bakkar A, et al:
Elevated physiological levels of folic acid can increase in
vitro growth and invasiveness of prostate cancer cells. BJU
Int. 109:788–795. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lucarelli G, Fanelli M, Larocca AMV, et
al: Serum sarcosine increases the accuracy of prostate cancer
detection in patients with total serum PSA less than 4.0 ng/ml.
Prostate. 72:1611–1621. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Cernei N, Heger Z, Gumulec J, et al:
Sarcosine as a potential prostate cancer biomarker: a review. Int J
Mol Sci. 14:13893–13908. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Chen JL, Tang HQ, Hu JD, Fan J, Hong J and
Gu JZ: Metabolomics of gastric cancer metastasis detected by gas
chromatography and mass spectrometry. World J Gastroenterol.
16:5874–5880. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Hirayama A, Kami K, Sugimoto M, et al:
Quantitative metabolome profiling of colon and stomach cancer
microenvironment by capillary electrophoresis time-of-flight mass
spectrometry. Cancer Res. 69:4918–4925. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Slupsky CM, Steed H, Wells TH, et al:
Urine metabolite analysis offers potential early diagnosis of
ovarian and breast cancers. Clin Cancer Res. 16:5835–5841. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kim K, Taylor SL, Ganti S, Guo LN, Osier
MV and Weiss RH: Urine metabolomic analysis identifies potential
biomarkers and pathogenic pathways in kidney cancer. OMICS.
15:293–303. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Phang JM: The regulatory functions of
proline and pyrroline-5-carboxylic acid. Curr Top Cell Regul.
25:91–132. 1985. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Pandhare J, Cooper SK and Phang JM:
Proline oxidase, a proapoptotic gene, is induced by troglitazone:
evidence for both peroxisome proliferator-activated receptor
γ-dependent and -independent mechanisms. J Biol Chem.
281:2044–2052. 2006.PubMed/NCBI
|
|
30
|
Pandhare J, Donald SP, Cooper SK and Phang
JM: Regulation and function of proline oxidase under nutrient
stress. J Cell Biochem. 107:759–768. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Polyak K, Xia Y, Zweier K, Kinzler W and
Vogelstein B: A model for p53-induced apoptosis. Nature.
389:300–305. 1997. View
Article : Google Scholar : PubMed/NCBI
|
|
32
|
Donald SP, Sun XY, Hu CAA, et al: Proline
oxidase, encoded by p53-induced gene-6, catalyzes the generation of
proline-dependent reactive oxygen species. Cancer Res.
61:1810–1815. 2001.PubMed/NCBI
|
|
33
|
Liu Y, Borchert GL, Surazynski A, Hu CA
and Phang JM: Proline oxidase activates both intrinsic and
extrinsic pathways for apoptosis: the role of ROS/superoxides, NFAT
and MEK/ERK signaling. Oncogene. 25:5640–5647. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Liu W, Le A, Hancock C, et al:
Reprogramming of proline and glutamine metabolism contributes to
the proliferative and metabolic responses regulated by oncogenic
transcription factor c-MYC. Proc Natl Acad Sci USA. 109:8983–8988.
2012. View Article : Google Scholar
|
|
35
|
Phang JM and Liu W: Proline metabolism and
cancer. Front Biosci. 17:1835–1845. 2012. View Article : Google Scholar
|
|
36
|
Liu W, Zabirnyk O, Wang H, et al: miR-23b*
targets proline oxidase, a novel tumor suppressor protein in renal
cancer. Oncogene. 29:4914–4924. 2010.
|
|
37
|
Liu Y, Borchert GL, Surazynski A and Phang
JM: Proline oxidase, a p53-induced gene, targets
COX-2/PGE2 signaling to induce apoptosis and inhibit
tumor growth in colorectal cancers. Oncogene. 27:6729–6737. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Ianni M, Porcellini E, Carbone I, et al:
Genetic factors regulating inflammation and DNA methylation
associated with prostate cancer. Prostate Cancer Prostatic Dis.
16:56–60. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Luka Z, Cerone R, Phillips JA, Mudd SH and
Wagner C: Mutations in human glycine N-methyltransferase give
insights into its role in methionine metabolism. Hum Genet.
110:68–74. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Pelzer AE, Volgger H, Bektic J, et al: The
effect of percentage free prostate-specific antigen (PSA) level on
the prostate cancer detection rate in a screening population with
low PSA levels. BJU Int. 96:995–998. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Sirovich BE, Schwartz LM and Woloshin S:
Screening men for prostate and colorectal cancer in the United
States: does practice reflect the evidence? JAMA. 289:1414–1420.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Seo HS and Lee NK: Predictors of PSA
screening among men over 40 years of age who had ever heard about
PSA. Korean J Urol. 51:391–397. 2010.PubMed/NCBI
|
|
43
|
Demers LM, Costa L, Chinchilli VM, Gaydos
L, Curley E and Lipton A: Biochemical markers of bone turnover in
patients with metastatic bone disease. Clin Chem. 41:1489–1494.
1995.PubMed/NCBI
|
|
44
|
Catalona WJ, Partin AW, Slawin KM, et al:
Use of the percentage of free prostate-specific antigen to enhance
differentiation of prostate cancer from benign prostatic disease: a
prospective multicenter clinical trial. JAMA. 279:1542–1547. 1998.
View Article : Google Scholar
|
|
45
|
Lapi F, Azoulay L, Niazi T, Yin H,
Benayoun S and Suissa S: Androgen deprivation therapy and risk of
acute kidney injury in patients with prostate cancer. JAMA.
310:289–296. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Nariculam J, Murphy DG, Jenner C, et al:
Nephrostomy insertion for patients with bilateral ureteric
obstruction caused by prostate cancer. Br J Radiol. 82:571–576.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Panis C, Victorino VJ, Herrera ACSA, et
al: Differential oxidative status and immune characterization of
the early and advanced stages of human breast cancer. Breast Cancer
Res Treat. 133:881–888. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Xu XL, Rao GS, Groh V, et al: Major
histocompatibility complex class I-related chain A/B (MICA/B)
expression in tumor tissue and serum of pancreatic cancer: role of
uric acid accumulation in gemcitabine-induced MICA/B expression.
BMC Cancer. 11:1942011. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Burgaz S, Torun M, Yardim S, Sargin H,
Orman MN and Ozdamar NY: Serum carotenoids and uric acid levels in
relation to cancer. J Clin Pharm Ther. 21:331–336. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Kolonel LN, Yoshizawa C, Nomura AMY and
Stemmermann GN: Relationship of serum uric-acid to cancer
occurrence in a prospective male cohort. Cancer Epidemiol
Biomarkers Prev. 3:225–228. 1994.PubMed/NCBI
|
|
51
|
Koyner JL, Vaidya VS, Bennett MR, et al:
Urinary biomarkers in the clinical prognosis and early detection of
acute kidney injury. Clin J Am Soc Nephrol. 5:2154–2165. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Chen CH and Lin MS: A novel structural
specific creatinine sensing scheme for the determination of the
urine creatinine. Biosens Bioelectron. 31:90–94. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Elsberger B, Lankston L, McMillan DC,
Underwood MA and Edwards J: Presence of tumoural C-reactive protein
correlates with progressive prostate cancer. Prostate Cancer
Prostatic Dis. 14:122–128. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Grases F, Costa-Bauza A and Prieto RM:
Renal lithiasis and nutrition. Nutr J. 5:1–7. 2006. View Article : Google Scholar
|
|
55
|
Montrose DC, Zhou XK, Kopelovich L, et al:
Metabolic profiling, a noninvasive approach for the detection of
experimental colorectal neoplasia. Cancer Prev Res. 5:1358–1367.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Green T, Chen XF, Ryan S, Asch AS and
Ruiz-Echevarria MJ: TMEFF2 and SARDH cooperate to modulate
one-carbon metabolism and invasion of prostate cancer cells.
Prostate. 73:1561–1575. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Khan AP, Rajendiran TM, Ateeq B, et al:
The role of sarcosine metabolism in prostate cancer progression.
Neoplasia. 15:491–501. 2013.PubMed/NCBI
|
|
58
|
Isaacs WB: Inherited susceptibility for
aggressive prostate cancer. Asian J Androl. 14:415–418. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Powell IJ, Bock CH, Ruterbusch JJ and Sakr
W: Evidence supports a faster growth rate and/or earlier
transformation to clinically significant prostate cancer in black
than in white American men, and influences racial progression and
mortality disparity. J Urol. 183:1792–1796. 2010. View Article : Google Scholar
|