Introduction
Gliomas are the most malignant and prevalent brain
tumors of glial origin and are divided into four clinical grades on
the basis of their histology and prognosis. Grade IV GBM is the
most malignant of all brain tumors. Despite recent advances in
diagnostics and treatments, the prognosis of advanced patients
suffering from GBM remains poor (1). One of the most important reasons for
this dismal outcome is that the migration and invasion of GBM cells
eventually lead to tumor recurrence and patient death. However, the
factors that mediate GBM migration and invasion are still poorly
understood.
MicroRNAs are a class of 20–25 nucleotide long
non-coding RNAs that modulate gene expression
post-transcriptionally by regulating mRNA translation or stability
(2). The human miR-127 gene is
located within a cluster on chromosome 14q32.31. The hsa-miR-127
precursor can express two mature miRNAs, miR-127-3p and miR-127-5p.
Changes in the expression of miR-127 were previously observed in
esophageal squamous cell carcinoma (3) and medullary thyroid carcinoma
(4), and miR-127 was proposed to be
a tumor suppressor in human bladder cancer cells (5). However, the role of miR-127 in GBM
development has not been well documented and little is known in
regards to its target genes.
We decided to investigate the roles of miR-127-3p in
GBM. We found that miR-127-3p promoted cell migration and invasion
using in vitro cell lines and in vivo mouse models.
We further demonstrated that SEPT7 is a direct target of miR-127-3p
and partially mediated the process of cell migration and invasion
initiated by miR-127-3p. In addition, we performed microarray
analysis after miR-127-3p overexpression was induced in a GBM cell
line and found that the expression of many migration and
invasion-related genes was significantly altered. Finally,
immunofluorescent cell staining showed that miR-127-3p affected the
remodeling of the actin cytoskeleton by regulating SEPT7 in GBM
cells.
Materials and methods
Cell culture and reagents
Human GBM cell lines, LN229, T98G, A172, U87 and
U251, were obtained from ATCC (http://www.atcc.org/) and grown in Dulbecco’s Modified
Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum
(FBS). Cells were incubated at 37°C and supplemented with 5%
CO2. Antibodies specific to SEPT7 and GAPDH were
obtained from Santa Cruz Biotechnology (Santa Cruz, CA, USA).
Horseradish peroxidase-coupled secondary antibodies were purchased
from MultiSciences Biotech Co., Ltd. (Hangzhou, China). miR-127-3p
inhibitors, which are antisense sequences of miR-127-3p with
2′-O-methyl modification for in vitro transfection, and
their respective negative controls were obtained from Guangzhou
RiboBio Co., Ltd. (Guangzhou, China).
Plasmid construction and establishment of
stable miR-127-3p-overexpressing cells
The genomic sequence of the human miR-127-3p gene
was PCR amplified using the primer
5′-GGAAGATCTGTAGTCCTGTCTGTTGGTCAG-3′ and
5′-CCCAAGCTTCCTGAAGAACTGCTTCCGCC-3′ from LN229 cells and cloned
into pSUPER.neo (Oligo Engine, Seattle, WA, USA). It was designated
as pSUPER.neo-miR-127-3p. Lipofectamine 2000 (Invitrogen, Carlsbad,
CA, USA) was used for the transfection of the DNA plasmid according
to the manufacturer’s protocol. Positive cells selected with 1
mg/ml G418 were confirmed by stem-loop RT-PCR. The full-length
3′UTR of SEPT7 was subcloned into the pmirGLO vector (Promega,
Madison, WI, USA) to generate pmirGLO-SEPT7-3′UTR. Mutant construct
of SEPT7-3′UTR, named pmirGLO-SEPT7-3′UTR-mut, which carried a
substitution of 4 nucleotides within the core binding site of
SEPT7-3′UTR, was carried out using Takara MutanBEST kit. The coding
sequence of SEPT7 was synthesized and subcloned into the
pUC57-simple vector (GenScript, Piscataway, NJ, USA), and then was
amplified by PCR and subcloned into the pcDNA3.1 vector, named
pcDNA3.1-SEPT7. The primers for SEPT7-3′UTR were
5′-CCGCTCGAGCTCTCTATTGACCACCAGTTAACG-3′ and
5′-AGCTTTCCTGCAGGCAGTGCCAATATATGGAAAATATC-3′. The primers for
mutation were 5′-TAGGGTTTGACCAATTTGCACCAGTTTTATCC-3′ and
5′-CCAACAAACACTGATGTCCAAGCTGGC-3′. All PCR products were verified
by DNA sequencing.
RNA quantification
Total RNA, containing miRNA, was extracted with
TRIzol reagent (Invitrogen) following the manufacturer’s
instructions. For mRNA analysis, reverse transcription was
performed by using M-MLV reverse transcriptase (Promega).
Amplification reactions were performed using the SYBR®
Premix Ex Taq™ (Takara, Dalian, China). Data were normalized to the
level of glyceraldehyde-3-phosphate dehydrogenase expression in
each sample. Expression of miR-127-3p was quantified using
stem-loop RT-PCR analysis as reported (6). The primers and other reagents for
stem-loop RT-PCR were obtained from Guangzhou RiboBio. U6 RNA was
used as an internal control. The 2−ΔΔCt method for
relative quantification of gene expression was used to determine
the expression levels of the transcripts.
Migration and invasion assays
For the migration assay, cells were plated onto
24-well Transwell chambers (Corning, Corning, NY, USA) with an 8-μm
pore polycarbonate membrane. For the invasion assay, cells were
plated on chambers precoated with ECM gel (Sigma, St. Louis, MO,
USA). In both assays, the cells were plated in medium without
serum, and medium containing 10% FBS in the lower chamber served as
a chemoattractant. After 24 h, the cells on the upper surface were
scraped and washed away, whereas the cells on the lower surface
were fixed and stained with 0.05% crystal violet. Finally, six
visual fields for each insert were randomly selected and counted
under a microscope. The cell numbers were normalized to the control
cell number, which was set to 100. The results were averaged from
three independent experiments.
Animal model
Five-week-old female nude mice were purchased from
Zhejiang Chinese Medical University (Zhejiang, China). All
experimental protocols were approved by the Ethics Committee for
Animal Experimentation of Zhejiang University. Exponentially
growing LN229 cells (2×106) with stable ectopic
expression of miR-127-3p or control vector were injected into nude
mice through the tail vein (n=5). Eight weeks after injection, the
animals were sacrificed. The liver and lung tissues were removed,
sectioned, and stained with hematoxylin and eosin (H&E) stain
to count the tumor loci by a pathologist.
Luciferase assay
LN229 cells were transiently transfected with the
luciferase reporter gene constructs and miR-127-3p-expressing
plasmid, and U251 cells were transiently transfected with the
luciferase reporter gene constructs and miR-127-3p inhibitors.
After 48 h, luciferase activities were measured using a dual
luciferase reporter assay system according to the manufacturer’s
protocol (Promega). For each plasmid construct, three independent
transfection experiments were performed in triplicate.
Western blot analysis
The cells were lysed by a standard procedure in
radioimmunoprecipitation assay buffer containing protease inhibitor
cocktail (Calbiochem, Darmstadt, Germany). Protein concentrations
of total cell lysates were measured using a BCA protein assay kit
(Pierce Biotechnology, Rockford, IL, USA). Protein was then
separated on polyacrylamide gels, transferred to a nitrocellulose
membrane, incubated with the relevant antibodies, and detected with
HRP-conjugated antibodies. Bands were visualized using ECL Western
blotting detection reagents (Thermo Fisher Scientific, Rockford,
IL, USA).
Microarray analysis
Two separate total RNA samples were extracted from
LN229 cells with miR-127-3p overexpression and corresponding
control cells. Genes regulated by miR-127-3p were identified by
Affymetrix U133 Plus 2 GeneChip (Affymetrix, Santa Clara, CA, USA).
The CEL files were imported into the Affymetrix’s Expression
Console (V1.1) program, and the data were normalized using RMA
normalization. Then the data were exported to the GeneSpring
Program (Agilent, Inc., Palo Alto, CA, USA) for further analysis.
In order to remove the low-expressed genes in the chips, probes
with intensities <100 in both chips were excluded. Genes with
absolute change of ≥2-fold and P<0.05 were considered
differentially regulated by miR-127-3p. The online High-Throughput
GoMiner program was used to analyze the Gene Ontology (GO) using
biological processes terms at level 4. U251 GBM cells naturally
present higher expression of miR-127-3p (as shown in Fig. 1), its array data (GSM803632) was
downloaded from the GEO database on NCI 60 cell lines (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE32474).
The same analysis pipeline was used to identify differentially
expressed genes between LN229 with miR-127-3p and U251.
Immunofluorescence cell staining
Cells were seeded on coverslips and incubated for 24
h and then fixed with formalin for 15 min, washed with PBS,
permeabilized for 3 min at room temperature with PBS containing
0.25% Triton X-100 and blocked for 30 min with PBS containing 1% of
BSA and 0.5% Tween-20. Slides were incubated overnight at 4°C in a
blocking solution containing anti-SEPT7 (Santa Cruz Biotechnology),
washed with PBS, and then incubated in a blocking solution
containing the secondary antibody conjugated with Cy3 (red)
(Millipore, Darmstadt, Germany) and phalloidin-FITC (Beyotime
Institute of Biotechnology, Haimen, China) for 1 h. After washing,
coverslips were attached to glass slides. Cells were imaged using a
fluorescence microscope.
Statistical analysis
Data are presented as means ± SD. Statistical
comparisons of the studies were analyzed by Student’s t test or
Chi-square test as appropriate. Correlations between groups were
calculated with Pearson’s correlation analysis. A P value <0.05
was defined as indicative of a statistically significant
result.
Results
miR-127-3p promotes human GBM cell
migration and invasion
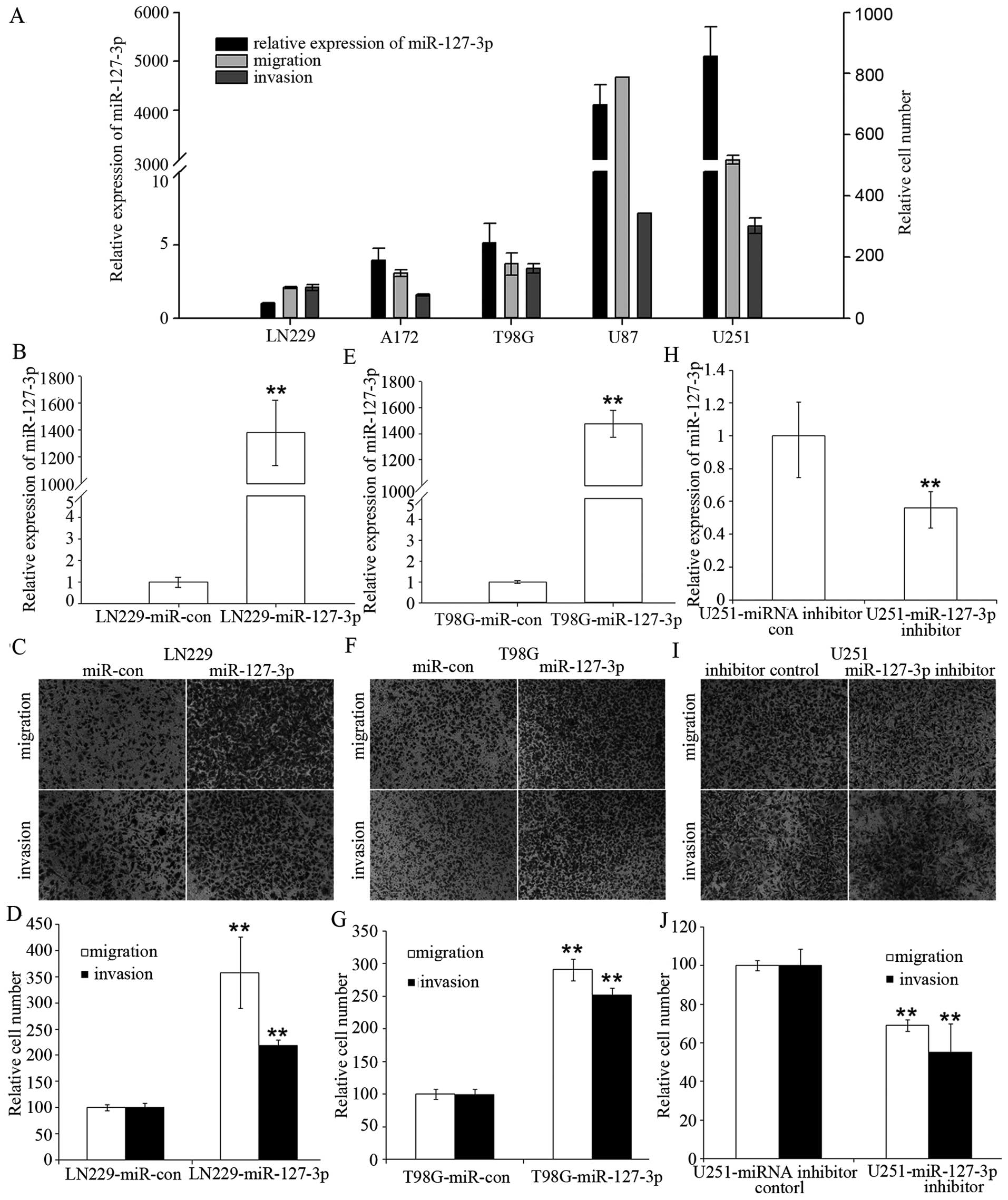
We initially analyzed the expression levels of
miR-127-3p in five human GBM cell lines and found that there was
great heterogeneity in miR-127-3p expression in the different GBM
cell lines. Furthermore, we observed a statistically significant
positive correlation between miR-127-3p levels and the cell
migration or the cell invasion potential of the five GBM cell lines
(r=0.89 and 0.93, respectively; P<0.05) (Fig. 1A). We observed a significant
difference between the miR-127-3p high-expressing cell lines (U251
and U87) and low-expressing cell lines (LN229, A172 and T98 cells)
in regards to migration and invasion potential (P=0.017 and 0.011,
respectively; Student’s t-test).
To determine whether miR-127-3p is merely correlated
or directly regulates human GBM cell migration and invasion, we
selected LN229 and T98G cells, which are two cell lines with low
endogenous miR-127-3p expression, and U251 cells, which have a high
endogenous miR-127-3p expression, for further analyses. Fig. 1B and 1E show that we were able to
induce miR-127-3p overexpression in the LN229 and T98G cells. We
observed a >2-fold increase in cell migration and invasion
potential in the LN229 and T98G cells with stable overexpression of
miR-127-3p compared to the control cells (Fig. 1C, D, F and G).
In addition, we performed a complementary analysis
by silencing miR-127-3p with an miR-127-3p inhibitor in U251 cells
with high endogenous miR-127-3p expression (Fig. 1H). Inhibition of miR-127-3p in U251
cells led to ~40–50% reduction in migration and invasion compared
with the control cells (Fig. 1I and
J).
miR-127-3p increases tumor formation in a
mouse model
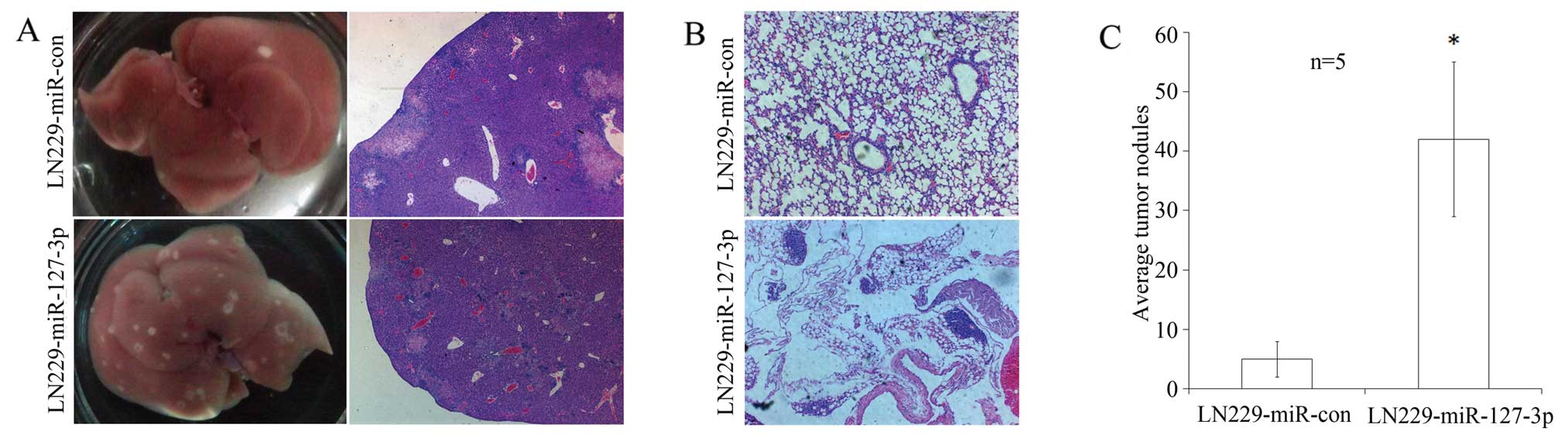
To determine the effects of miR-127-3p on tumor
invasive potential in vivo, LN229 cells either
overexpressing miR-127-3p or transfected with a control vector were
injected via the tail vein into nude mice. Eight weeks after
injection, the animals were sacrificed. The liver and lung tissues
were removed, sectioned, and stained with H&E to count the
tumor loci by a pathologist.
In the mice inoculated with
miR-127-3p-overexpressing cells, 5 of 5 cases had tumor formation
in the liver and 2 of 5 cases had tumor formation in the lung. In
contrast, 4 of 5 cases had tumor formation in the liver and none
had tumor formation in the lung in the control group, both
confirmed by pathohistological examinations (Fig. 2A and B). Although the number of
tumors formed was higher in the miR-127-3p-overexpressing tumor
group compared to the control cell group, the number was not
statistically significant (P=0.29 and 0.11, respectively). However,
as shown in Fig. 2C, the average
number of tumors formed in the liver in the
miR-127-3p-overexpressing group was 42, while it was 5 for the
vector control group (P=0.02).
miR-127-3p targets the SEPT7 gene and its
effects on cell migration and invasion are partially mediated
through SEPT7
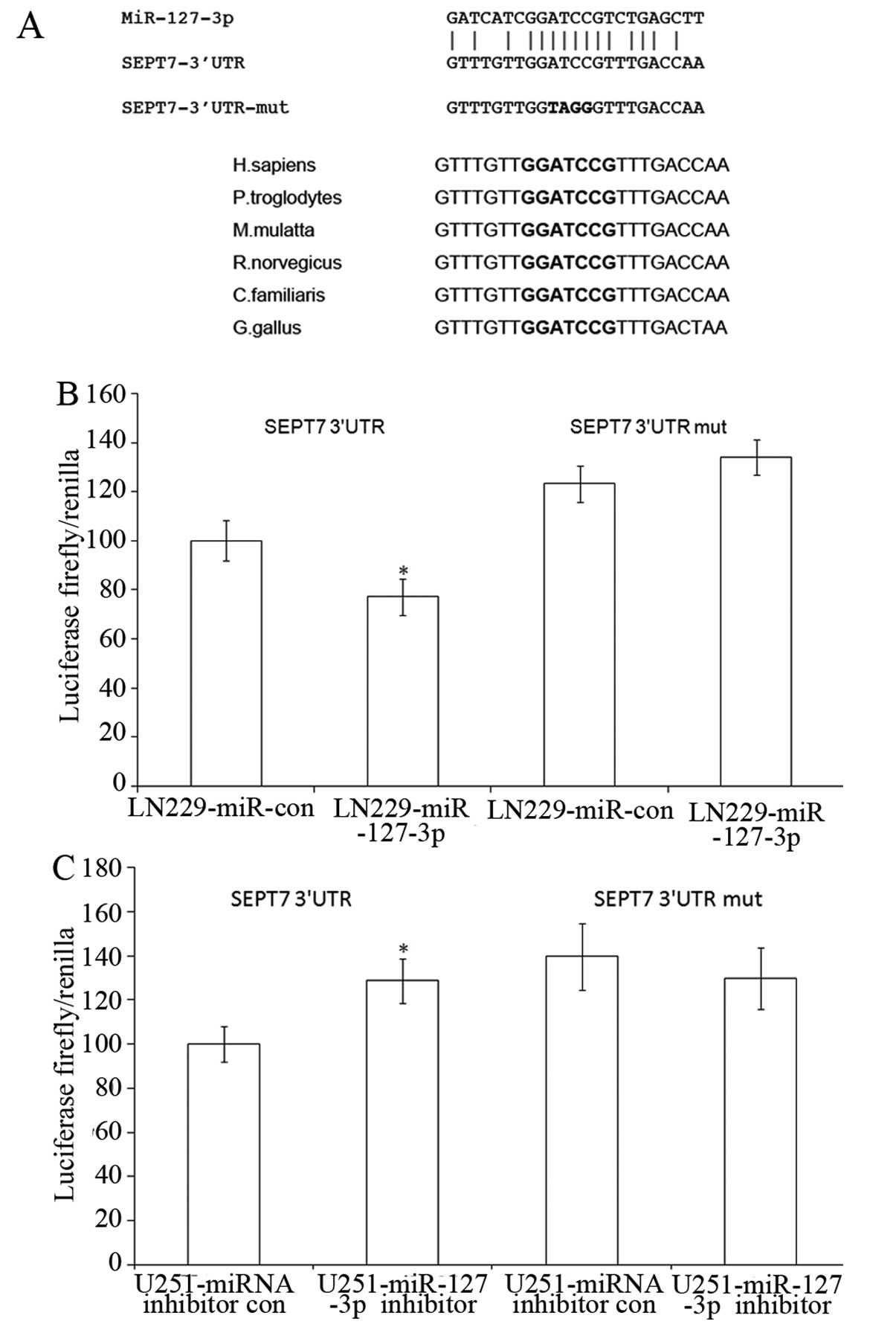
To explore the molecular mechanism of miR-127-3p
function, we used the EIMMo program (http://www.mirz.unibas.ch/ElMMo2), which combined mRNA
expression and targeted prediction algorithm, to predict targets of
miR-127-3p in the human. We used the whole brain mRNA expression
data set on the server to help predict miR-127-3p. One hundred and
fifty-eight predicted targets were found. We found that SEPT7 had
two entries (variant 1 and variant 2) that were among the top 10
predicted targets. Fig. 3A shows a
miR-127-3p core binding site at nucleotides 67–74 of the SEPT7
3′UTR. In addition, the miR-127-3p target sequence at nucleotides
67–74 of the SEPT7 3′UTR is highly conserved among different
species (Fig. 3A). We then decided
to conduct additional analysis to determine whether SEPT7 is a
target of miR-127-3p.
We constructed pmirGLO-SEPT7-3′UTR and
pmirGLO-SEPT7-3′UTR-mut the latter contains mutated targeted
sequences. Cotransfection of LN229 cells with pmirGLO-SEPT7-3′UTR
and pSUPER.neo-miR-127-3p caused a 22% decrease in the luciferase
activity compared with the negative control. Four-nucleotide
substitution in the core binding site (pmirGLO-SEPT7-3′UTR-mut)
abolished the suppressive effects (Fig.
3B). In addition, pmirGLO-SEPT7-3′UTR and
pmirGLO-SEPT7-3′UTR-mut were cotransfected with either miR-127-3p
inhibitor or the control oligonucleotide into U251 cells,
expressing high levels of endogenous miR-127-3p. The inhibition of
miR-127-3p by the inhibitors resulted in a significant increase in
luciferase activity of the pmirGLO-SEPT7-3′UTR but not that of the
pmirGLO-SEPT7-3′UTR-mut-transfected cells (Fig. 3C).
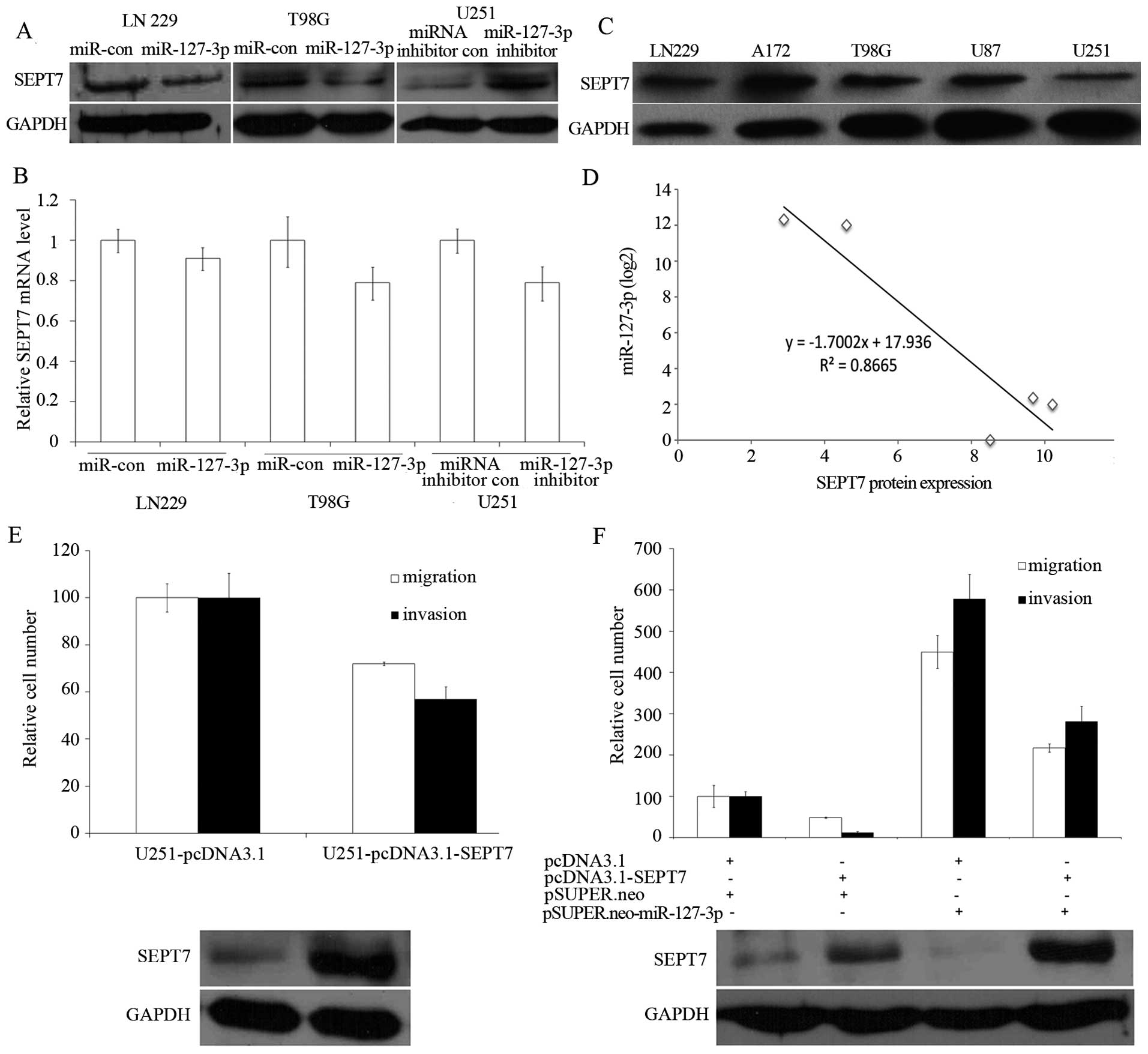
To test whether miR-127-3p expression affected
endogenous SEPT7 expression, we transfected pSUPER.neo-miR-127-3p
and the control plasmid into LN229 and T98G cells; a decrease in
SEPT7 protein level in LN229 and T98G cells was observed (Fig. 4A, the left and the middle panel).
Consistent with these results, silencing of miR-127-3p in U251
cells showed an increase in the SEPT7 protein level (Fig. 4A, the right panel). However, the
SEPT7 mRNA level was not significantly influenced by overexpression
or inhibition of miR-127-3p (Fig.
4B).
We then examined the SEPT7 protein levels in five
GBM cell lines (Fig. 4C) and
compared them with the miR-127-3p levels. In cell lines with high
endogenous miR-127-3p (for example, U87 and U251; Fig. 1A, lanes 4 and 5), a low amount of
SEPT7 protein at 49 kDa was observed (Fig. 4C, lanes 4 and 5), whereas cell lines
with low miR-127-3p (for example, LN229, A172 and T98G; Fig. 1A, lanes 1–3) showed high amounts of
SEPT7 protein (Fig. 4C, lanes 1–3).
Across all five cell lines tested, we found a significant inverse
correlation (P<0.05) between miR-127-3p and SEPT7 protein levels
(Fig. 4D). We compared the
quantified SEPT7 protein expression between the miR-127-3p high
expression cell lines (U251 and U87) and low expression cell lines
(LN229, A172 and T98 cells) and found that there existed a
significant difference between the two groups (P=0.008; Student’s
t-test).
We investigated whether EPT7 suppressed cell
migration and invasion in LN229 and U251 GBM cells. As shown in
Fig. 4E, overexpression of SEPT7 in
U251 cells resulted in a decreased cell migration and invasion. To
further establish a functional connection between miR-127-3p and
SEPT7, we tested whether SEPT7 deregulation was required for
miR-127-3p regulation of the migration and invasion of GBM cells.
pcDNA3.1-SEPT7, which carried the whole SEPT7 coding sequence
without 3′UTR and would thus not be suppressed by the miR-127-3p,
and pSUPER.neo-miR-127-3p, were cotransfected into LN229 cells. We
found that expression of SEPT7 partly abrogated the migration and
invasion initiated by miR-127-3p in the LN229 cells (Fig. 4F).
miR-127-3p regulates genes related to
migration and invasion
To identify the global effect of miR-127-3p on GBM
cells, we performed microarray analysis to compare the gene
expression profile of the miR-127-3p stably overexpressing LN229
cells with that of the LN229 control cells. Among 54,614 probe
sets, we identified 1320 (791 downregulated and 529 upregulated)
genes that were differentially expressed.
As the U251 GBM cells naturally present higher
expression of miR-127-3p (Fig. 1),
we compared its expression with LN229, and identified 5280 probe
sets with 2-fold changes (data not shown). We then intersected the
differentially expressed gene list for the comparison between
LN229-miR-127-3p and LN229 mock control with that for the
comparison between U251 and LN229 cells. We identified 855 probe
sets that showed differential expression in both comparisons.
Using GO analysis we determined biological process
gene categories that were significantly altered by miR-127-3p
overexpression (FDR <0.01). Biological processes including cell
proliferation, cell migration, cell motility, regulation of
locomotion, which are fundamental biological processes required to
achieve cell migration and invasion were found to be enriched
(Table I).
 | Table ImiR-127-3p overexpression results in
enriched GO categories related to migration and invasion. |
Table I
miR-127-3p overexpression results in
enriched GO categories related to migration and invasion.
| GO category | Total gene | Changed genes | Enrichments | Log10(p) | False discovery
rate |
|---|
|
GO:0032386_regulation_of_intracellular_transport | 60 | 13 | 5.11 | −6.02 | 0.0000 |
|
GO:0033157_regulation_of_intracellular_protein_transport | 53 | 12 | 5.34 | −5.81 | 0.0000 |
|
GO:0046822_regulation_of_nucleocytoplasmic_transport | 53 | 12 | 5.34 | −5.81 | 0.0000 |
|
GO:0008283_cell_proliferation | 680 | 54 | 1.87 | −5.43 | 0.0000 |
|
GO:0016477_cell_migration | 292 | 29 | 2.34 | −4.81 | 0.0000 |
|
GO:0046824_positive_regulation_of_nucleocytoplasmic_transport | 29 | 8 | 6.51 | −4.72 | 0.0000 |
|
GO:0090316_positive_regulation_of_intracellular_protein_transport | 29 | 8 | 6.51 | −4.72 | 0.0000 |
|
GO:0040012_regulation_of_locomotion | 140 | 18 | 3.03 | −4.63 | 0.0000 |
|
GO:0032388_positive_regulation_of_intracellular_transport | 30 | 8 | 6.29 | −4.61 | 0.0000 |
|
GO:0048870_cell_motility | 301 | 29 | 2.27 | −4.56 | 0.0000 |
|
GO:0051674_localization_of_cell | 301 | 29 | 2.27 | −4.56 | 0.0000 |
|
GO:0006928_cellular_component_movement | 387 | 34 | 2.07 | −4.42 | 0.0025 |
|
GO:0032879_regulation_of_localization | 473 | 39 | 1.94 | −4.37 | 0.0031 |
|
GO:0042127_regulation_of_cell_proliferation | 476 | 39 | 1.93 | −4.31 | 0.0029 |
|
GO:0002376_immune_system_process | 729 | 53 | 1.71 | −4.25 | 0.0053 |
|
GO:0042307_positive_regulation_of_protein_import_into_nucleus | 25 | 7 | 6.60 | −4.24 | 0.0050 |
|
GO:0034764_positive_regulation_of_transmembrane_transport | 26 | 7 | 6.35 | −4.12 | 0.0071 |
|
GO:0034762_regulation_of_transmembrane_transport | 65 | 11 | 3.99 | −4.11 | 0.0067 |
|
GO:0042306_regulation_of_protein_import_into_nucleus | 45 | 9 | 4.72 | −4.04 | 0.0074 |
|
GO:0048856_anatomical_structure_development | 1367 | 84 | 1.45 | −3.81 | 0.0085 |
|
GO:0008284_positive_regulation_of_cell_proliferation | 253 | 24 | 2.24 | −3.76 | 0.0100 |
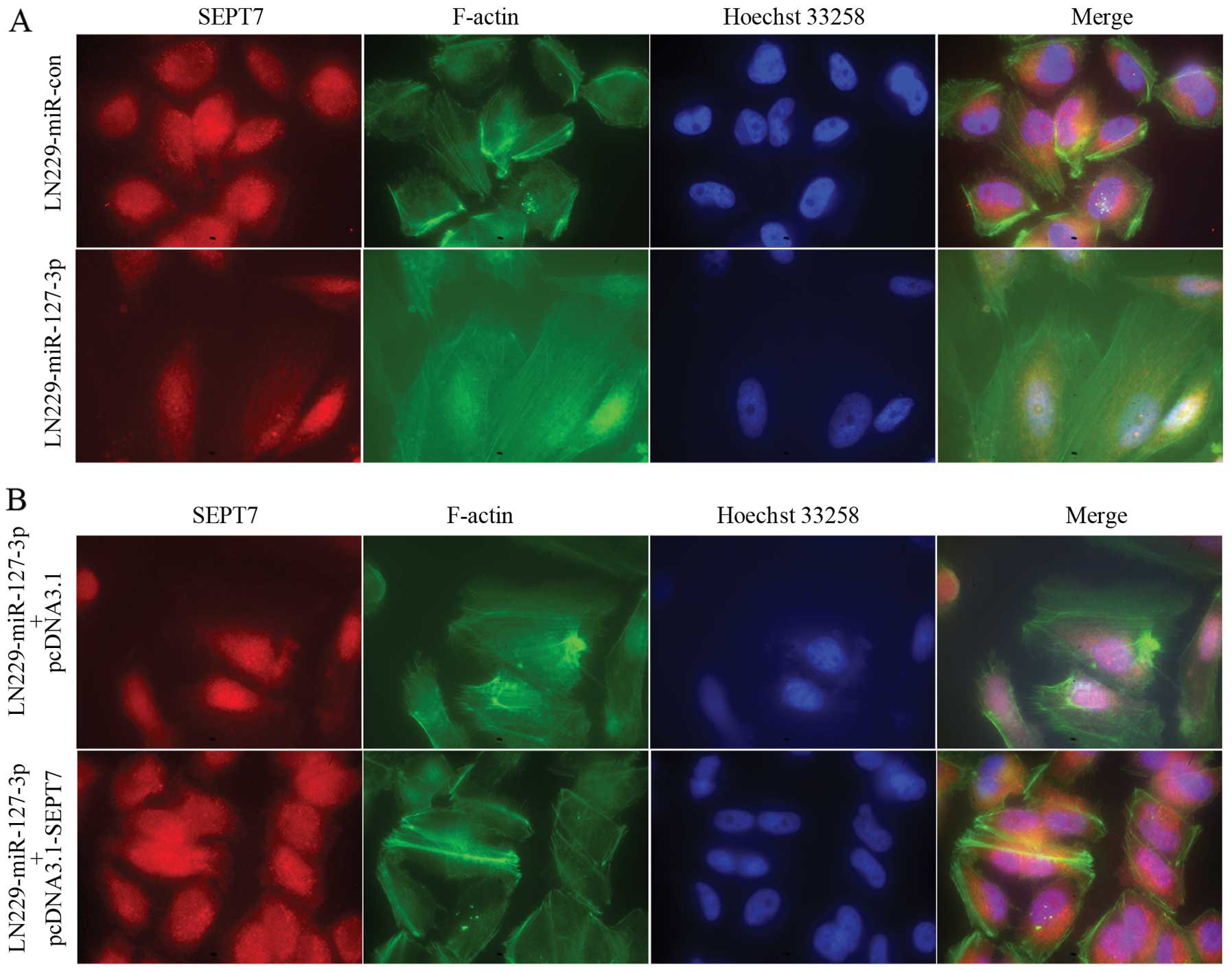
miR-127-3p affects remodeling of the
actin cytoskeleton by regulating SEPT7 in GBM cells
Dynamic regulation of the filamentous actin
(F-actin) cytoskeleton is critical to cell adhesion and migration.
We, therefore, examined changes in F-actin remodeling after
miR-127-3p expression with a fluorescence microscope. In the
control cells, the actin cytoskeleton formed a thick peripheral
cytoskeleton near the cell membranes and the cells showed more
rounded morphology (Fig. 5A, upper
panel), while in the miR-127-3p-overexpressing LN229 GBM cells, the
actin cytoskeleton formed thin stress fibers across the cytoplasm
and the cells showed spindle-shaped morphology (Fig. 5A, lower panel). We then examined the
F-actin remodeling of LN229 cells stably overexpressing miR-127-3p
transfected with pcDNA3.1 or pcDNA3.1-SEPT7. The observed changes
in cell morphology and the actin cytoskeleton structure in cells
overexpressing miR-127-3p were rescued by SEPT7 overexpression
(Fig. 5B).
Discussion
We investigated for the first time the role of
miR-127-3p in GBM. Previous studies suggested that miR-127-3p might
have tumor suppressing or promotive roles (3–5,7–10).
Our inadvertent observation that there existed a statistically
significant positive correlation between miR-127-3p expression and
the migration and invasion potential of GBM cell lines prompted us
to analyze the roles of miR-127-3p in GBM. We showed that
miR-127-3p promoted migration and invasion of human GBM cells by
in vitro cell analysis, and that miR-127-3p promoted
metastasis in in vivo mouse models. This result is
consistent with a demonstrated association of miR-127-3p with
increases in lymph node metastasis in cervical carcinoma (11). This suggests that miR-127-3p may
function as an oncomiR in GBM.
Accumulating evidence suggests that miRNAs play
important roles in gliomas including regulation of their migration
and invasion (12–14). As initial evidence for a role of
miRNA in promoting glioma invasion, Gabriely et al (12) found that miR-21 promotes glioma
invasion by targeting MMP inhibitors. Quintavalle et al
(15) showed that miR-221/222
overexpession in human GBM increased its invasiveness by targeting
the protein phosphate PTPμ. The inhibitory roles of miRNAs in
glioma migration and invasion have also been reported. For example,
miR-146b inhibits glioma cell migration and invasion by targeting
MMPs (16). Ectopic expression of
miR-26b, miR-124a and miR-34a in GBM cell lines resulted in
significant inhibition of migration and invasion (13,17,18).
miR-7 was downregulated in GBM and it was found to target insulin
receptor substrate 2 (IRS2), epidermal growth factor receptor
(EGFR) (19) and focal adhesion
kinase (FAK). In addition, overexpression of miR-7 resulted in
decreased migration and invasion, possibly mediated in part by
decreased levels of MMP-2 and MMP-9 (14).
We found that SEPT7 was targeted by miR-127-3p.
SEPT7 is a member of the septin family genes. Septins are a group
of highly-conserved GTP binding proteins located at the plasma
membranes in eukaryote cells (20),
and 13 members of septins have been identified in the human
(21). Septin members can act as
either tumor suppressors or promoters. For example, the expression
of ARTS (apoptosis-related protein in the TGF-β signaling pathway,
the short isoform splice variant SEPT4_v2) is often lost in human
leukemia (22). The loss of
Sept4 function in mice was found to promote spontaneous
leukemia or lymphoma (23).
However, SEPT2 was found to be upregulated in human hepatoma
carcinoma cells (HCC) and the phosphorylation of Ser218 of SEPT2 is
crucial to the proliferation of human hepatoma carcinoma cells
(24). SEPT1 plays an important
role in spreading in squamous cell carcinoma DJM-1 cells (25). SEPT9 is overexpressed in diverse
human tumors including breast, cenral nervous system (CNS),
endometrial, kidney, liver, lung, lymphoid, oesophageal, ovarian
and pancreatic cancer (26).
The SEPT7 gene is located on human chromosome
7p14.3, and alternative splicing of the gene results in the
transcription of three isoforms. Although SEPT7 is expressed in
most tissues, it is especially abundantly expressed in brain
tissues (26). SEPT7 is involved in
several human diseases including neoplasia (27,28)
and neurodegenerative disorders (29). Previous data suggest that SEPT7 is a
tumor suppressor. SEPT7 was reported to be downregulated in
low-grade diffuse astrocytomas (30) and in other CNS tumor tissues
compared with normal tissues (26,27).
In addition, the high level of SEPT7 expression in neuroblastoma
and glioma may be associated with favorable clinical and biological
characteristics (27,28). SEPT7 has been shown to inhibit the
migration and invasion of GBM cell lines (31). This observation indicates that SEPT7
might have a negative role in migration and invasion of GBM cells.
Our observation in GBM U251 cells confirmed the suppressive role of
SEPT7 on cell migration and invasion (Fig. 4E).
To establish the functional connection between
miR-127-3p and SEPT7, we overexpressed SEPT7 and miR-127-3p in
LN229 cells and found that SEPT7 protein could partly abrogate the
function of miR-127-3p (Fig. 4F).
Therefore, SEPT7 protein might be a mediator of the regulation of
cell migration and invasion by miR-127-3p. Nonetheless,
overexpression of SEPT7 did not completely abolish the effect of
miR-127-3p, suggesting that additional mediators might exist.
Additional experimentation is necessary to identify these
mediators.
Our expression profile analysis of
miR-127-3p-regulated genes provided a mechanistic link with its
role in promoting migration and invasion as we found that
biological processes such as cell migration, cell motility,
regulation of locomotion are enriched in miR-127-3p-regulated genes
(Table I).
Our data demonstrated that cells with high levels of
miR-127-3p exhibited a spindle-shaped morphology and the actin
cytoskeleton formed thin stress fibers across the cytoplasm, in
contrast with more rounded morphology and a thick peripheral actin
cytoskeleton near cell membranes shown in the control cells
(Fig. 5). Our data is consistent
with reported roles of SEPT7 in cytoskeleton remodeling, cell shape
and cell movement (32–35). Recently, SEPT7 was reported to
affect the actin cytoskeleton and cellular morphology via the
SOCS7/NCK signaling pathway (29).
There is also evidence showing that upregulation of SEPT7 inhibits
invasion of GBM cells by distribution of α-tubulin (31). Collectively, these data suggest that
miR-127-3p regulates cell morphology and the structure of the actin
cytoskeleton by targeting SEPT7. Our observation that
reintroduction of SEPT7 into LN229 cells overexpressing miR-127-3p
rescues the phenotype seems to confirm this hypothesis.
In conclusion, our findings revealed that miR-127-3p
promoted the migration and invasion of GBM cells partly by
targeting SEPT7. miR-127-3p may be explored as a novel biomarker
for predicting the migratory and invasive potential of glioma cells
or may be exploited as a therapeutic target for GBM.
Acknowledgements
This study was funded by grant 81072060 from the
National Natural Science Foundation of China; grants 2008DFA11320
and 2012AA022705 from the Ministry of Science and Technology,
China; grant 20110101120153 from the Ministry of Education, China;
grant 2012R10021 from the Zhejiang Provincial Government; grant
2011ZX09307-001-05 from National Science and Technology Major
Project, China.
References
|
1
|
Huse JT, Holland E and Deangelis LM:
Glioblastoma: molecular analysis and clinical implications. Annu
Rev Med. 64:59–70. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Filipowicz W, Bhattacharyya SN and
Sonenberg N: Mechanisms of post-transcriptional regulation by
microRNAs: are the answers in sight? Nat Rev Genet. 9:102–114.
2008. View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zhang C, Wang C, Chen X, et al: Expression
profile of microRNAs in serum: a fingerprint for esophageal
squamous cell carcinoma. Clin Chem. 56:1871–1879. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Mian C, Pennelli G, Fassan M, et al:
MicroRNA profiles in familial and sporadic medullary thyroid
carcinoma: preliminary relationships with RET status and outcome.
Thyroid. 22:890–896. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Saito Y, Liang G, Egger G, et al: Specific
activation of microRNA-127 with downregulation of the
proto-oncogene BCL6 by chromatin-modifying drugs in human cancer
cells. Cancer Cell. 9:435–443. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Chen C, Ridzon DA, Broomer AJ, et al:
Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic
Acids Res. 33:e1792005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Yan LX, Huang XF, Shao Q, et al: MicroRNA
miR-21 overexpression in human breast cancer is associated with
advanced clinical stage, lymph node metastasis and patient poor
prognosis. RNA. 14:2348–2360. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Tryndyak VP, Ross SA, Beland FA and
Pogribny IP: Down-regulation of the microRNAs miR-34a, miR-127, and
miR-200b in rat liver during hepatocarcinogenesis induced by a
methyl-deficient diet. Mol Carcinog. 48:479–487. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Pan C, Chen H, Wang L, et al:
Down-regulation of miR-127 facilitates hepatocyte proliferation
during rat liver regeneration. PLoS One. 7:e391512012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Dixon-McIver A, East P, Mein CA, et al:
Distinctive patterns of microRNA expression associated with
karyotype in acute myeloid leukaemia. PLoS One. 3:e21412008.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lee JW, Choi CH, Choi JJ, et al: Altered
microRNA expression in cervical carcinomas. Clin Cancer Res.
14:2535–2542. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Gabriely G, Wurdinger T, Kesari S, et al:
MicroRNA 21 promotes glioma invasion by targeting matrix
metalloproteinase regulators. Mol Cell Biol. 28:5369–5380. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Guessous F, Zhang Y, Kofman A, et al:
microRNA-34a is tumor suppressive in brain tumors and glioma stem
cells. Cell Cycle. 9:1031–1036. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wu DG, Wang YY, Fan LG, et al: MicroRNA-7
regulates glioblastoma cell invasion via targeting focal adhesion
kinase expression. Chin Med J (Engl). 124:2616–2621.
2011.PubMed/NCBI
|
|
15
|
Quintavalle C, Garofalo M, Zanca C, et al:
miR-221/222 overexpession in human glioblastoma increases
invasiveness by targeting the protein phosphate PTPmu. Oncogene.
31:858–868. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Xia H, Qi Y, Ng SS, et al: microRNA-146b
inhibits glioma cell migration and invasion by targeting MMPs.
Brain Res. 1269:158–165. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wu N, Zhao X, Liu M, et al: Role of
microRNA-26b in glioma development and its mediated regulation on
EphA2. PLoS One. 6:e162642011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Fowler A, Thomson D, Giles K, et al:
miR-124a is frequently down-regulated in glioblastoma and is
involved in migration and invasion. Eur J Cancer. 47:953–963. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kefas B, Godlewski J, Comeau L, et al:
microRNA-7 inhibits the epidermal growth factor receptor and the
Akt pathway and is down-regulated in glioblastoma. Cancer Res.
68:3566–3572. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Mostowy S and Cossart P: Septins: the
fourth component of the cytoskeleton. Nat Rev Mol Cell Biol.
13:183–194. 2012.PubMed/NCBI
|
|
21
|
Hall PA and Russell SE: The pathobiology
of the septin gene family. J Pathol. 204:489–505. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Elhasid R, Sahar D, Merling A, et al:
Mitochondrial pro-apoptotic ARTS protein is lost in the majority of
acute lymphoblastic leukemia patients. Oncogene. 23:5468–5475.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Garcia-Fernandez M, Kissel H, Brown S, et
al: Sept4/ARTS is required for stem cell apoptosis and tumor
suppression. Genes Dev. 24:2282–2293. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yu W, Ding X, Chen F, et al: The
phosphorylation of SEPT2 on Ser218 by casein kinase 2 is important
to hepatoma carcinoma cell proliferation. Mol Cell Biochem.
325:61–67. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Mizutani Y, Ito H, Iwamoto I, et al:
Possible role of a septin, SEPT1, in spreading in squamous cell
carcinoma DJM-1 cells. Biol Chem. 394:281–290. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Scott M, Hyland PL, McGregor G, Hillan KJ,
Russell SE and Hall PA: Multimodality expression profiling shows
SEPT9 to be overexpressed in a wide range of human tumours.
Oncogene. 24:4688–4700. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Jia ZF, Huang Q, Kang CS, et al:
Overexpression of septin 7 suppresses glioma cell growth. J
Neurooncol. 98:329–340. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Nagata T, Takahashi Y, Asai S, et al: The
high level of hCDC10 gene expression in neuroblastoma may be
associated with favorable characteristics of the tumor. J Surg Res.
92:267–275. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kremer BE, Adang LA and Macara IG: Septins
regulate actin organization and cell-cycle arrest through nuclear
accumulation of NCK mediated by SOCS7. Cell. 130:837–850. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Huang H, Colella S, Kurrer M, Yonekawa Y,
Kleihues P and Ohgaki H: Gene expression profiling of low-grade
diffuse astrocytomas by cDNA arrays. Cancer Res. 60:6868–6874.
2000.PubMed/NCBI
|
|
31
|
Xu S, Jia ZF, Kang C, et al: Upregulation
of SEPT7 gene inhibits invasion of human glioma cells. Cancer
Invest. 28:248–258. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Nagata K, Asano T, Nozawa Y and Inagaki M:
Biochemical and cell biological analyses of a mammalian septin
complex, Sept7/9b/11. J Biol Chem. 279:55895–55904. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Bowen JR, Hwang D, Bai X, Roy D and
Spiliotis ET: Septin GTPases spatially guide microtubule
organization and plus end dynamics in polarizing epithelia. J Cell
Biol. 194:187–197. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Kim SK, Shindo A, Park TJ, et al: Planar
cell polarity acts through septins to control collective cell
movement and ciliogenesis. Science. 329:1337–1340. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Tada T, Simonetta A, Batterton M,
Kinoshita M, Edbauer D and Sheng M: Role of Septin cytoskeleton in
spine morphogenesis and dendrite development in neurons. Curr Biol.
17:1752–1758. 2007. View Article : Google Scholar : PubMed/NCBI
|