Introduction
Liver cancer is one of the most common malignancies
and is the second and sixth most frequent cause of cancer-related
death in men and women, respectively (1). Among primary liver cancers,
hepatocellular carcinoma (HCC) represents the major histological
subtype, accounting for 70–85% of the total liver cancer burden
worldwide (2). Therefore,
identification of novel clinical biomarkers and molecules
contributing to tumor progression in HCC is critically
required.
One of the main functions of ribosomes is
translation of mRNA to protein, and cancer cells generally exhibit
increased ribosomal synthesis (3,4).
Ribosomes consist of ribosomal proteins and ribosomal RNA (rRNA)
molecules (5), and the biogenesis
of ribosomes requires the coordinated actions of all three
DNA-dependent RNA polymerases (Pol I, II and III) (6,7).
Transcription termination factor, RNA polymerase I (TTF1) is a
protein encoded by the TTF1 gene at 9q34.13; this protein
mediates rRNA gene activation or silencing (8) and is essential for ribosome biogenesis
(9). TTF1 can bind to multiple
sites both downstream and upstream of rRNA genes (10–12),
and TTF1 binding to the upstream proximal promoter sites of rRNA
genes activates their transcription, while TTF1 binding to sites
downstream of rRNA genes is related to termination of transcription
(13,14). Excessive ribosome biogenesis and
translation initiation lead to tumor progression in HCC (15). Since cancer cells exhibit increased
ribosomal activity and since TTF1 is closely associated with
ribosomal activity, TTF1 may have clinical or biological importance
in cancer. However, the significance of TTF1 in solid cancer has
not been fully investigated. In the present study, we investigated
the clinicopathological significance of aberrant TTF1
expression in HCC cases. In order to validate the clinical
significance of TTF1 in the public database, we also performed gene
set enrichment analysis (GSEA) and assessed whether the levels of
TTF1 expression were associated with the unfavorable
prognostic signature in HCC. Finally, we examined the capacity for
TTF1-dependent HCC cell proliferation to provide an explanation for
clinical data in vitro.
Materials and methods
Patients and sample collection
Sixty patients with HCC who underwent hepatic
resection at Kyushu University Beppu Hospital and Affiliated
Hospitals between 2001 and 2004 were enrolled in the present study.
Resected HCC tumor and paired adjacent liver tissues were
immediately frozen in liquid nitrogen and kept at −80°C until RNA
extraction. An intermittent follow-up was conducted after the
operation, and the average follow-up period for the 60 patients was
70.1 months (range, 1.7–138.8 months). All protocols were approved
by the Ethics and Indications Committee of Kyushu University.
Written informed consent was obtained from all the patients.
Cell lines
Human HCC cells (HuH-7 and HepG2) were provided by
the Cell Resource Center for Biomedical Research (Institute of
Development, Aging and Cancer, Tohoku University, Japan). Cell
lines were maintained in Dulbecco's modified Eagle's medium (DMEM;
Gibco, Carlsbad, CA, USA) supplemented with 10% fetal calf serum
and antibiotics. All cells were cultured at 37°C in a humidified
atmosphere containing 5% CO2.
Cloning of human TTF1 cDNA
To generate TTF1 expression lentiviral
vectors, we amplified the insert (full-length human TTF1;
NM_001205296.1) by polymerase chain reaction (PCR) from human
reference cDNA. Transient transfections were performed using
CSII-CMV-MCS (empty) plasmid DNAs (5′-BamHI and
3′-HpaI sites) and Lipofectamine 2,000 (Invitrogen,
Carlsbad, CA, USA) according to the manufacturer's protocol.
RNA preparation and reverse transcription
(RT)-PCR
Total RNA from frozen tissue specimens and HCC cell
lines was extracted using ISOGEN (Nippon Gene, Tokyo, Japan). The
quality assessment of extracted RNA was performed by measuring
absorbance, and we confirmed that all of them kept satisfactory
quality. cDNA was synthesized from 8 µg total RNA with M-MLV
reverse transcriptase (Invitrogen).
Quantitative real-time PCR (qRT-PCR)
Gene-specific oligonucleotide primers were designed
for PCR. The following primers were used: TTF1,
5′-GAAAGGTTGTATGAAATAA ATGTGGA-3′ (sense) and
5′-TTGAACGTAAGATGGAGGAACA-3′ (antisense);
glyceraldehyde-3-phosphate dehydrogenase (GAPDH),
5′-TTGGTATCGTGGAAGGACTCA-3′ (sense) and 5′-TGTCATCATATTTGGCAGGTT-3′
(antisense); 28S ribosomal RNA, 5′-TTACCCTACTGATGATGTGTTGTTG-3′
(sense) and 5′-CCTGCGGTTCCTCTCGTA-3′ (anti-sense). PCR
amplification was performed in a LightCycler 480 instrument using a
LightCycler 480 Probes Master kit (both from Roche Applied Science
Basel, Switzerland). mRNA amplification conditions consisted of
initial denaturation at 95°C for 10 min, followed by 40 cycles of
denaturation at 95°C for 10 sec, annealing at 62°C (60°C for some
genes) for 10 sec, and elongation at 67°C (65°C for some genes) for
10 sec. Melting curve analysis was performed to distinguish
specific products from non-specific products and primer dimers. The
relative expression levels of the gene were obtained by normalizing
the amount of mRNA to that of GAPDH mRNA as an endogenous
control in each sample.
Western blot analysis
Total protein was extracted from
TTF1-expressing and mock-transfected cell lines. Aliquots of
total protein (40 µg) were electrophoresed on 10%
polyacrylamide gels and then electroblotted on nitrocellulose
membranes using Trans-Blot Transfer Medium (Bio-Rad Laboratories,
Hercules, CA, USA) at 0.4 A for 120 min. TTF1 protein was detected
using rabbit polyclonal antibodies (ab87726; Abcam, Cambridge, UK)
diluted 1:2,000. TTF1 protein levels were normalized to the level
of β-actin protein, which was detected using monoclonal antibodies
(Cytoskeleton Inc., Denver, CO, USA) at a 1:1,000 dilution. Blots
were developed with horseradish peroxidase-linked anti-rabbit or
anti-mouse immunoglobulin (Promega, Madison, WI, USA) diluted at
1:1,000. Enhanced chemiluminescence detection reagents (Amersham
Biosciences, Piscataway, NJ, USA) were used to detect
antigen-antibody reactions.
Immunohistochemical analysis
HCC tissues were surgically removed, embedded in
paraffin and sectioned (5-µm sections). Immunohistochemical
analysis was applied to determine the localization of TTF1. A
polyclonal rabbit anti-TTF1 antibody (ab87726; 1:200; Abcam) was
used as the primary antibody following the manufacturer's
protocols.
Data extraction from public clinical
dataset, copy number analysis, and GSEA
We obtained expression data of 371 HCC cases and
copy number data of 377 cases from The Cancer Genome Atlas (TCGA)
from the Broad Institute's Firehose (http://gdac.broadinstitute.org/runs/stddata_2014_12_06/data/LIHC/2014_12_06/).
Expression profiles of TCGA dataset were analyzed by GSEA (16). Gene sets extracted from the Broad
Institute database and the Uniform Resource Locator of their source
are as follows. LEE_LIVER_CANCER_SURVIVAL_DN (http://www.broadinstitute.org/gsea/msigdb/cards/LEE_LIVER_CANCER_SURVIVAL_DN.html),
LEE_LIVER_CANCER_SURVIVAL_UP (http://www.broadinstitute.org/gsea/msigdb/cards/LEE_LIVER_CANCER_SURVIVAL_UP.html),
RIBONUCLEOPROTEIN_COMPLEX (http://www.broadinstitute.org/gsea/msigdb/cards/RIBONUCLEOPROTEIN_COMPLEX.html),
STRUCTURAL_CONSTITUENT_OF_RIBOSOME (http://www.broadinstitute.org/gsea/msigdb/cards/STRUCTURAL_CONSTITUENT_OF_RIBOSOME.html),
RIBONUCLEOPROTEIN_COMPLEX_BIOGENESIS_AND_ASSEMBLY (http://www.broadinstitute.org/gsea/msigdb/cards/RIBONUCLEOPROTEIN_COMPLEX_BIOGENESIS_AND_ASSEMBLY.html),
PROTEIN_RNA_COMPLEX_ASSEMBLY (http://www.broadinstitute.org/gsea/msigdb/cards/PROTEIN_RNA_COMPLEX_ASSEMBLY.html).
We also acquired the expression profiles of TTF1 and the
survival information in Gene Expression Omnibus database (accession
code GSE14520) (17).
Cell proliferation assay
Twenty-four hours before the assay, cells were
transfected with TTF1 cDNA or empty cDNA.
3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT)
assays were conducted to evaluate cell proliferation. First, 10
µl of MTT-labeling reagent (final concentration, 0.5 mg/ml)
was added to each well, and the plate was incubated for 4 h in a
humidified atmosphere. Next, solubilization solution (100
µl) was added to each well, and the plate was incubated for
12 h in a humidified atmosphere. After confirming that the purple
formazan crystals were completely solubilized, the absorbance of
each well was measured using a Model 550 series microplate reader
(Bio-Rad Laboratories) at a wavelength of 570 nm corrected to 655
nm. The assay was performed using six replicates.
Statistical analysis
For continuous variables, data are expressed as
means ± standard deviations, and statistical analyses were
performed using paired t-tests in comparisons between corresponding
tumor and normal tissues. Other analyses were performed using
Welch's t-tests. Categorical variables were compared using the
Chi-square or Fisher's exact tests. Overall survival (OS) was
estimated using the Kaplan-Meier method, and survival curves were
compared using the log-rank test. Univariate and multivariate
analysis was performed using the Cox regression model to identify
independent variables predictive of OS. P-values of <0.05 were
considered to indicate a statistically significant result. Data
analyses of clinicopathological factors were performed using JMP 9
software (SAS Institute, Cary, NC, USA), and other analyses were
performed using R version 3.1.1 (The R Foundation for Statistical
Computing, Vienna, Austria).
Results
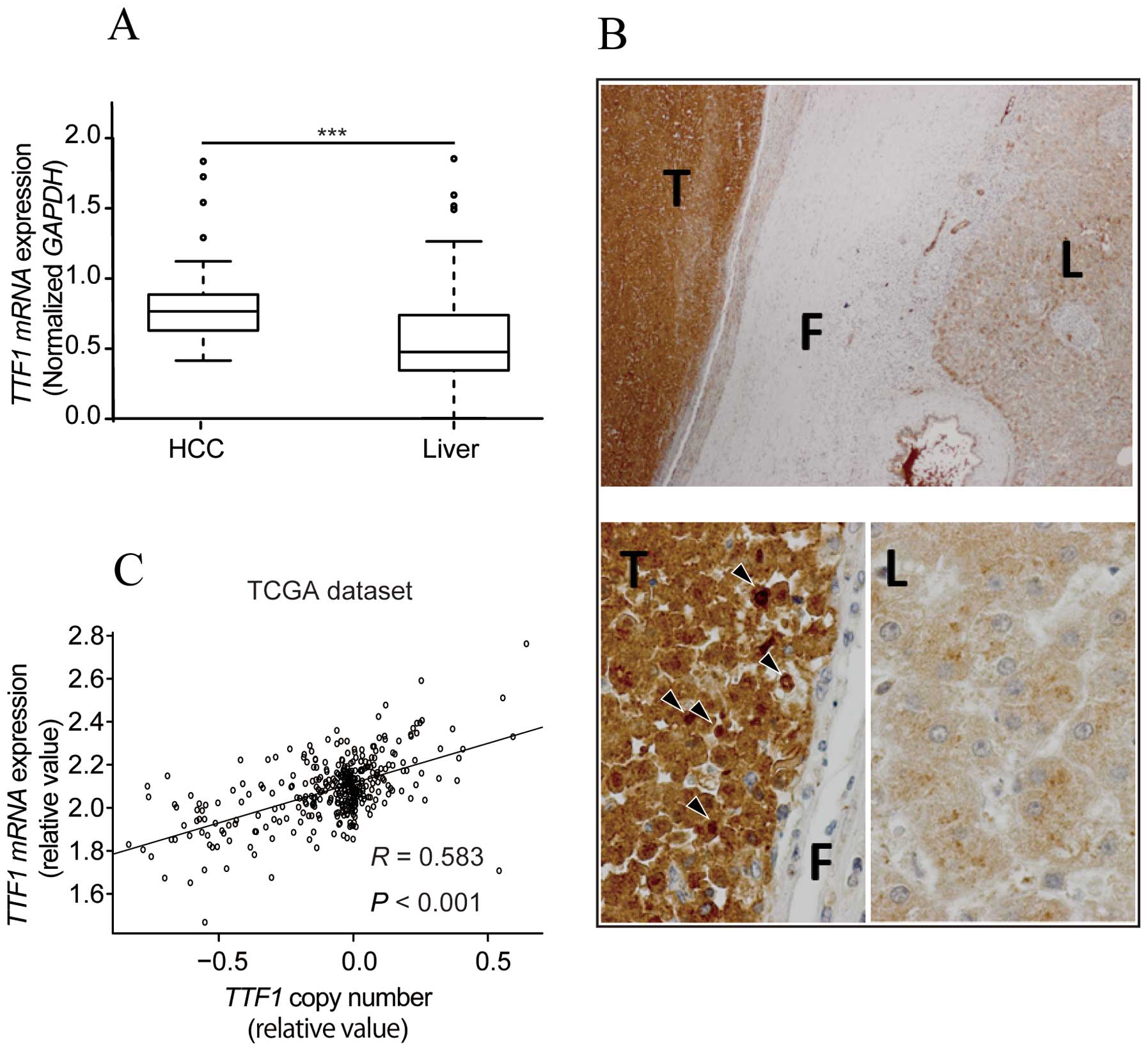
TTF1 mRNA and protein expression are
upregulated in HCC tissues
To identify whether TTF1 was critical for HCC
progression, we used qRT-PCR to examine TTF1 mRNA expression
in 58 clinical paired tumor and normal tissues. TTF1 mRNA
expression in cancerous tissues was significantly upregulated
compared with that in the matched normal tissues (P<0.001;
Fig. 1A). Moreover,
immunohistochemical analysis revealed that TTF1 protein levels were
markedly higher in HCC tissues than in corresponding liver tissues
(Fig. 1B). Staining in cytoplasm
was observed in both HCC and liver tissues. In contrast, staining
in nuclei was characteristically observed in tumor tissues.
TTF1 gene copy number analysis using the
TCGA HCC dataset
To assess whether TTF1 gene copy number
variations affected TTF1 gene expression, we extracted copy
number data from the TCGA dataset. A strong correlation between
copy number and TTF1 expression was observed in tumor
tissues (R=0.583, P<0.001; Fig.
1C).
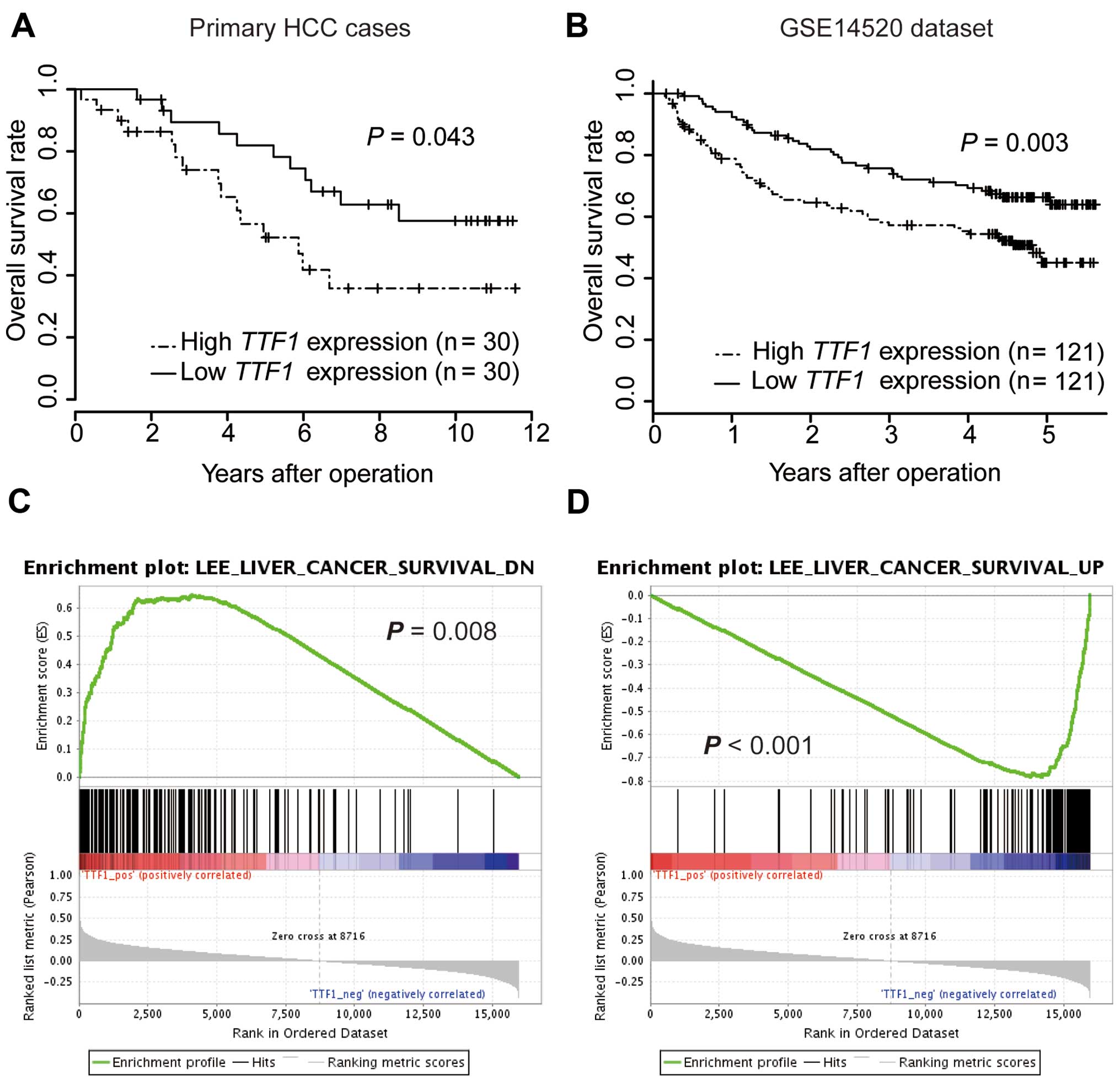
Increased expression of TTF1 is
associated with poor prognosis in patients with HCC
To estimate the clinical significance of TTF1
expression in HCC, TTF1 expression was analyzed by qRT-PCR
in 60 samples from patients who underwent resection of primary HCC.
According to TTF1 expression level, cases were divided into
two groups. Cut-off values were set at the median value of
TTF1 mRNA expression. There were no significant differences
in clinicopathological parameters between high and low TTF1
expression groups (data not shown). On univariate analysis, the OS
rate of the high TTF1 expression group was significantly
lower than that of the low TTF1 expression group (P=0.027;
Fig. 2A). Furthermore, multivariate
analysis showed that high TTF1 expression was an independent
prognostic factor for poorer OS (Table
I; P=0.020). We further analyzed OS for a public dataset of 242
HCC cases from GSE14520 (17) to
confirm the results of our series. In accordance with our data, the
OS of the high TTF1 expression group was significantly
poorer than that of the low TTF1 expression group (Fig. 2B; P=0.003).
 | Table IUnivariate and multivariate analyses
of clinicopathological factors affecting overall survival in
primary HCC cases (n=60). |
Table I
Univariate and multivariate analyses
of clinicopathological factors affecting overall survival in
primary HCC cases (n=60).
| Variable | No. of
patients | Univariate analysis
| Multivariate
analysis
|
|---|
| OS (%) | P-value | Hazard ratio (95%
CI) | P-value |
|---|
| Age (years) | | | 0.795 | | |
| >70 | 28 | 57.1 | | | |
| ≤70 | 32 | 53.1 | | | |
| Gender | | | 0.896 | | |
| Male | 39 | 53.8 | | | |
| Female | 21 | 57.1 | | | |
| HBV | | | 0.483 | | |
| (+) | 12 | 75.0 | | | |
| (−) | 48 | 50.0 | | | |
| HCV | | | 0.051 | | |
| (+) | 44 | 44.5 | | | |
| (−) | 16 | 81.3 | | | |
| Child-Pugh | | | 0.350 | | |
| A | 50 | 58.0 | | | |
| B or C | 10 | 40.0 | | | |
| Tumor size
(cm) | | | 0.320 | | |
| >3 | 29 | 48.4 | | | |
| ≤3 | 31 | 62.1 | | | |
| No. of tumors | | | 0.006 | 3.06
(1.40–6.52) | 0.002 |
| Single | 42 | 64.3 | | | |
| Multiple | 18 | 33.3 | | | |
| fc | | | 0.744 | | |
| (+) | 43 | 53.5 | | | |
| (−) | 17 | 58.8 | | | |
| vp | | | 0.890 | | |
| (+) | 33 | 54.5 | | | |
| (−) | 27 | 55.6 | | | |
| vva | | | 0.170 | | |
| (+) | 3 | 0.0 | | | |
| (−) | 56 | 57.1 | | | |
| b | | | 0.019 | 60.5
(2.39–1528) | 0.015 |
| (+) | 1 | 0.0 | | | |
| (−) | 59 | 55.9 | | | |
| TTF1
expression | | | 0.030 | 2.93
(1.02–10.5) | 0.020 |
| High | 30 | 46.6 | | | |
| Low | 30 | 63.3 | | | |
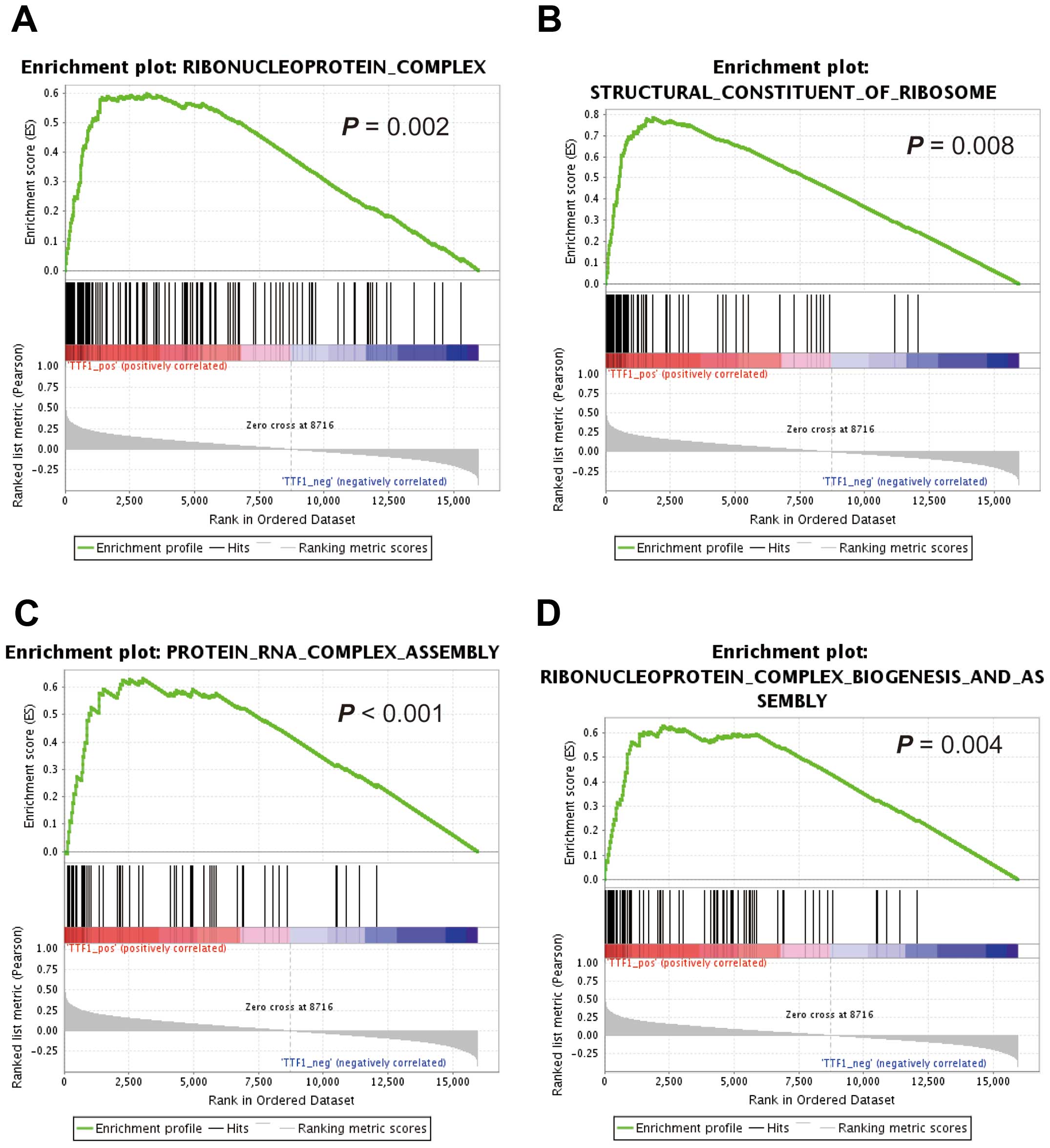
GSEA using reference datasets
To investigate whether the expression levels of
TTF1 were associated with known gene signatures, we applied
GSEA to HCC cases from TCGA datasets. GSEA revealed that
TTF1 expression levels positively correlated with an
unfavorable prognostic gene signature (Fig. 2C) and negatively correlated with a
signature representing favorable outcomes (Fig. 2D). Moreover, GSEA showed that
TTF1 expression levels were significantly correlated with
activity of gene sets which were involved in ribosomal function
(Fig. 3A–D).
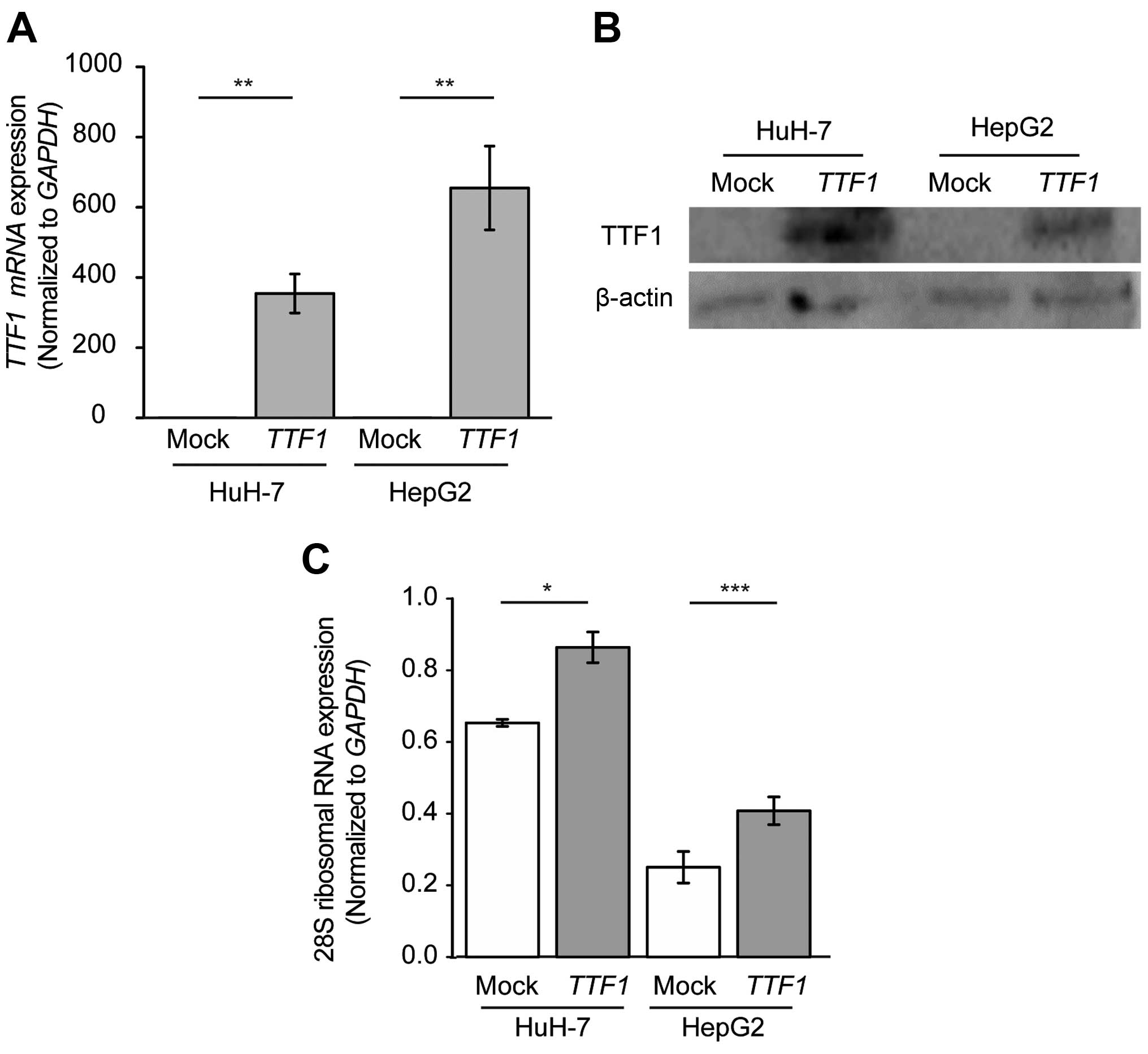
Overexpression of TTF1 enhances the
transcription of rRNA in human HCC cells
To explore the biological role of TTF1 in
HCC, we performed overexpression experiments in HuH-7 and HepG2
cells. We confirmed that TTF1 mRNA and TTF1 protein
expression was significantly higher in cells transfected with
TTF1 cDNA than in cells transfected with empty plasmid
(Fig. 4A and B). Next, to confirm
that upregulated expression of TTF1 contributed to the
increased synthesis of rRNA in HCC cells, we performed quantitative
analysis with qRT-PCR of 28S rRNA, which was one of the degradation
products of 45S rRNA and was reported to reflect the quantity of
transcribed rRNA (18). As a
result, overexpression of TTF1 led to the increased
expression of 28S rRNA, HuH-7 and HepG2 cells in comparison with
mock-transfected cells (Fig. 4C;
P=0.011 and P<0.001, respectively).
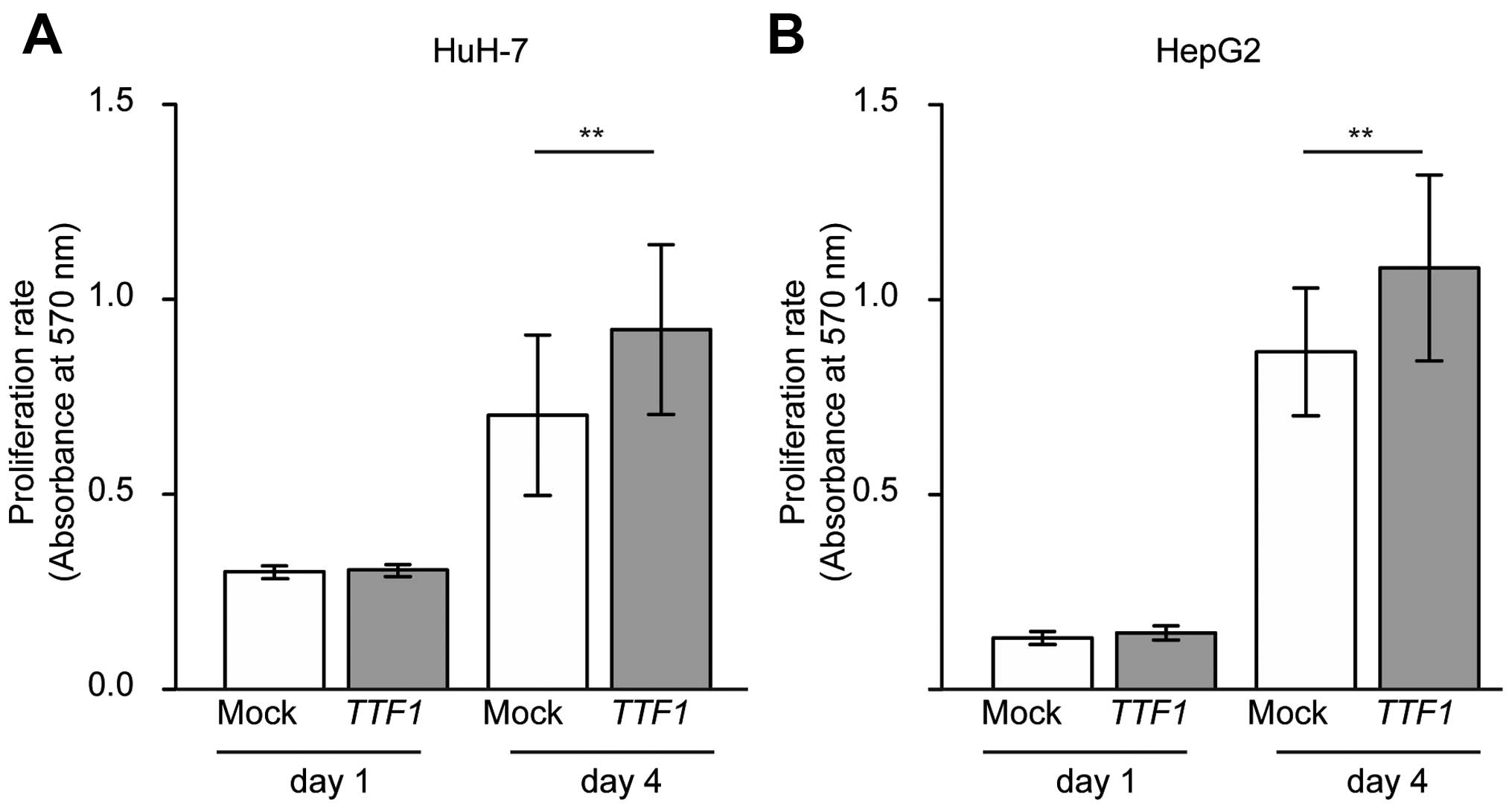
Ectopic TTF1 enhances the growth of HCC
cell lines
MTT assays were used to examine whether cell growth
rates were altered in cancer cells transfected with TTF1
cDNA. As a result, overexpression of TTF1 promoted the
proliferation of HuH-7 and HepG2 cells in comparison with
mock-transfected cells (Fig. 5A and
B; P=0.007 and P=0.018, respectively).
Discussion
It is well known that cancer cells frequently
exhibit relatively high nucleolar activity and increased ribosome
biogenesis, which contributes to cancer progression (19). TTF1 has been reported to mediate
ribosomal activity through regulating the activation/silencing of
rRNA (13,14). However, the clinicopathological
significance of TTF1 in solid cancers is not fully understood. In
the present study, we showed the involvement of TTF1 in tumor
progression of HCC.
First, we found that TTF1 mRNA and protein
expression was high in HCC tissues as compared to that in
corresponding normal tissues. Unlike adjacent liver tissues,
localization of TTF1 protein in nuclei was observed in HCC tissues.
This corresponds with previous studies that the nucleolus is the
site of biosynthesis of ribosomal RNA (20), and that increase in number of
nucleolus is observed in cancer cells (3). In cancer, copy number variation has
been shown to influence gene expression (21). Consistent with this, we observed a
strong correlation between copy number and TTF1 expression
in tumor tissues. Hence, the intertumoral differences in
TTF1 expression levels may be explained by copy number
variations.
Next, we showed that higher expression of
TTF1 predicted poor prognosis and was an independent
prognostic factor for HCC. To provide an adequate explanation for
clinical significance, we interrogated the public databases. GSEA
indicated that TTF1 mRNA expression positively correlated
with the expression levels of gene sets involved in ribosomal
function and with gene signatures predicting poor prognoses, which
suggested that upregulation of TTF1 affected the prognosis
of patients with HCC in accordance with elevated ribosomal
activity. This is consistent with previous reports showing that
increased ribosomal biogenesis predicts high-grade malignancy and
poor prognosis (22,23). Analysis of the GSEA database also
suggested that there was a correlation between TTF1
expression and intrahepatic metastatic or recurrent signatures,
which could explain the association with poor prognoses.
Finally, we found that TTF1 overexpression
resulted in upregulation of rRNA and promoted the proliferation of
HCC cells in vitro. Activation of ribosome synthesis has
been reported to be associated with tumor cell proliferation
(24), and cancer cell
proliferation affects clinical outcomes in patients with HCC
(25). Although it should be noted
that there exists many factors which are involved in the regulation
of ribosomal activity (26) and it
is necessary to examine the relation between TTF1 and such factors,
our findings suggested that upregulation of TTF1 may have
contributed to cancer progression through elevated ribosomal
activity.
In conclusion, the present study showed that TTF1
expression was upregulated in tumor tissues from patients with HCC
and that TTF1 mRNA levels were significantly associated with
poor prognoses as an independent prognostic factor. Moreover, we
showed that upregulation of TTF1 promoted the proliferation of HCC
cells in vitro. The function of TTF1 was not fully
elucidated, and further investigation would be needed to precisely
determine the significance of TTF1, and thus it would be premature
to predicate that TTF1 plays critical role in HCC. However, our
results suggested that TTF1 may be a novel biomarker and may be a
potential therapeutic target in HCC.
Acknowledgments
The present study used the super-computing resource
provided by the Human Genome Center at the Institute of Medical
Science, University of Tokyo (http://sc.hgc.jp/shirokane.html). Clinical samples and
corresponding clinical information were provided by the Oita Red
Cross Hospital (Oita, Japan), the Hiroshima Red Cross Hospital and
Atomic-bomb Survivors Hospital (Hiroshima, Japan), and the Iizuka
Hospital (Fukuoka, Japan). We thank K. Oda, M. Kasagi, S. Kono, M.
Aoyagi and T. Kawano for their excellent technical assistance. The
present study was supported in part by the Funding Program for Next
Generation World-Leading Researchers (LS094): Japan Society for the
Promotion of Science Grant-in-Aid for Scientific Research (grant
no. 24592005).
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Perz JF, Armstrong GL, Farrington LA,
Hutin YJ and Bell BP: The contributions of hepatitis B virus and
hepatitis C virus infections to cirrhosis and primary liver cancer
worldwide. J Hepatol. 45:529–538. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Busch H, Byvoet P and Smetana K: The
nucleolus of the cancer cell: A review. Cancer Res. 23:313–339.
1963.PubMed/NCBI
|
|
4
|
Nguyen XT, Chan SM, Ngo TD, Raval A, Kim
KK, Majeti R and Mitchell BS: Interaction of TIF-90 and filamin A
in the regulation of rRNA synthesis in leukemic cells. Blood.
124:579–589. 2014. View Article : Google Scholar
|
|
5
|
Noller HF: Structure of ribosomal RNA.
Annu Rev Biochem. 53:119–162. 1984. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Arabi A, Wu S, Ridderstråle K, Bierhoff H,
Shiue C, Fatyol K, Fahlén S, Hydbring P, Söderberg O, Grummt I, et
al: c-Myc associates with ribosomal DNA and activates RNA
polymerase I transcription. Nat Cell Biol. 7:303–310. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Grandori C, Gomez-Roman N, Felton-Edkins
ZA, Ngouenet C, Galloway DA, Eisenman RN and White RJ: c-Myc binds
to human ribosomal DNA and stimulates transcription of rRNA genes
by RNA polymerase I. Nat Cell Biol. 7:311–318. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Grummt I and Pikaard CS: Epigenetic
silencing of RNA polymerase I transcription. Nat Rev Mol Cell Biol.
4:641–649. 2003. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lessard F, Morin F, Ivanchuk S, Langlois
F, Stefanovsky V, Rutka J and Moss T: The ARF tumor suppressor
controls ribosome biogenesis by regulating the RNA polymerase I
transcription factor TTF-I. Mol Cell. 38:539–550. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Sander EE, Mason SW, Munz C and Grummt I:
The amino-terminal domain of the transcription termination factor
TTF-I causes protein oligomerization and inhibition of DNA binding.
Nucleic Acids Res. 24:3677–3684. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Evers R, Smid A, Rudloff U, Lottspeich F
and Grummt I: Different domains of the murine RNA polymerase
I-specific termination factor mTTF-I serve distinct functions in
transcription termination. EMBO J. 14:1248–1256. 1995.PubMed/NCBI
|
|
12
|
Kuhn A, Bartsch I and Grummt I: Specific
interaction of the murine transcription termination factor TTF I
with class-I RNA polymerases. Nature. 344:559–562. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Längst G, Blank TA, Becker PB and Grummt
I: RNA polymerase I transcription on nucleosomal templates: The
transcription termination factor TTF-I induces chromatin remodeling
and relieves transcriptional repression. EMBO J. 16:760–768. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lessard F, Stefanovsky V, Tremblay MG and
Moss T: The cellular abundance of the essential transcription
termination factor TTF-I regulates ribosome biogenesis and is
determined by MDM2 ubiquitinylation. Nucleic Acids Res.
40:5357–5367. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ruan Y, Sun L, Hao Y, Wang L, Xu J, Zhang
W, Xie J, Guo L, Zhou L, Yun X, et al: Ribosomal RACK1 promotes
chemoresistance and growth in human hepatocellular carcinoma. J
Clin Invest. 122:2554–2566. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
16
|
Subramanian A, Tamayo P, Mootha VK,
Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub
TR, Lander ES, et al: Gene set enrichment analysis: A
knowledge-based approach for interpreting genome-wide expression
profiles. Proc Natl Acad Sci USA. 102:15545–15550. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Roessler S, Jia HL, Budhu A, Forgues M, Ye
QH, Lee JS, Thorgeirsson SS, Sun Z, Tang ZY, Qin LX, et al: A
unique metastasis gene signature enables prediction of tumor
relapse in early-stage hepatocellular carcinoma patients. Cancer
Res. 70:10202–10212. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wen F, Zhou R, Shen A, Choi A, Uribe D and
Shi J: The tumor suppressive role of eIF3f and its function in
translation inhibition and rRNA degradation. PLoS One.
7:e341942012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Takada H and Kurisaki A: Emerging roles of
nucleolar and ribosomal proteins in cancer, development, and aging.
Cell Mol Life Sci. 72:4015–4025. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Nakamura T, Rapp F and Busch H: Common
features of the base composition of rapidly labeled RNA of nucleoli
in a number of experimental tumors. Cancer Res. 27:1084–1091.
1967.PubMed/NCBI
|
|
21
|
Albertson DG: Gene amplification in
cancer. Trends Genet. 22:447–455. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Sirri V, Roussel P, Trerè D, Derenzini M
and Hernandez-Verdun D: Amount variability of total and individual
Ag-NOR proteins in cells stimulated to proliferate. J Histochem
Cytochem. 43:887–893. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Tannapfel A, Geissler F, Köckerling F,
Katalinic A, Hauss J and Wittekind C: Apoptosis and proliferation
in relation to histopathological variables and prognosis in
hepatocellular carcinoma. J Pathol. 187:439–445. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Torres-Montaner A, Bolivar J, Ortiz M and
Valdivia MM: Immunohistochemical detection of ribosomal
transcription factor UBF: Diagnostic value in malignant specimens.
J Pathol. 184:77–82. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ito Y, Matsuura N, Sakon M, Takeda T,
Umeshita K, Nagano H, Nakamori S, Dono K, Tsujimoto M, Nakahara M,
et al: Both cell proliferation and apoptosis significantly predict
shortened disease-free survival in hepatocellular carcinoma. Br J
Cancer. 81:747–751. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kusnadi EP, Hannan KM, Hicks RJ, Hannan
RD, Pearson RB and Kang J: Regulation of rDNA transcription in
response to growth factors, nutrients and energy. Gene. 556:27–34.
2015. View Article : Google Scholar
|