Introduction
Head and neck squamous cell carcinoma (HNSCC) is one
of the aggressive malignancies and a majority of the HNSCC patients
belong to advanced stage with less than 50% of long-term survival
(1). Treatment of advanced-stage
HNSCC has undergone substantial changes in recent years. Combined
modality treatments including radical surgery and various regimens
of concomitant chemoradiotherapy (CCRT) have been used to improve
locoregional control and distant metastases of the cancer (1). However, even though CCRT has recently
become a mainstay of a primary treatment modality in advanced
HNSCC, some of the patients may experience cancer recurrence and
CCRT protocols can lead to some serious complications such as
swallowing dysfunction resulting in permanent gastrostomy tube
dependence (2). Therefore, if we
can distinguish the patients who would benefit from CCRT after the
surgery, from those that will not, before applying it, we would
reduce unnecessary CCRT thus avoiding risk of CCRT-related
complication.
Several molecular-genetic factors have been
suggested to be potentially contributing to the resistance to CCRT.
For example, CCRT-resistant HNSCCs showed a high rate of p53
aberrations and increased expression of MDM2, which confers tumor
cell survival after CCRT-induced cytotoxic stress (3,4).
Expression levels of hMLH1 and RKIP were also reported to be
predictive of responsiveness to CCRT in head and neck cancers
(5,6). Clinical factors such as the serum LDH
and hemoglobin levels have been suggested as prognostic factors
after CCRT (7,8).
Chromosomal copy number alteration (CNA), both
amplification and deletion, is one of the hallmarks of cancer
including HNSCC. Amplifications of the genes involved in
detoxification of chemotherapeutic agents or deletions of the genes
involved in DNA repair have been reported to be associated with
CCRT-resistance (9). More
specifically, gains of 3q11-q13, 3q21-q26.1 and 6q22-q27, and
losses of 3p11-pter and 4p11-pter were reported to be significantly
associated with CCRT resistance in HNSCC by comparative genomic
hybridization (CGH) analysis (10).
However, due to its limited resolution, the information from CGH
analyses has not been detailed enough to identify the causative
genes. Two genome-wide analyses, microarray based CGH (array-CGH)
and next generation sequencing (NGS), are currently the best tools
with much improved resolution for detecting CNAs precisely. In
terms of detecting CNAs, it is widely accepted that array-CGH is
relatively more matured and cost-effective technology than NGS
(11). However, there has been no
report exploring CNA profiles in HNSCCs with CCRT and their
prognostic implications through high resolution array-CGH.
To explore genetic alterations predicting the
responsiveness to radical surgery plus postoperative CCRT in HNSCC
patients, we analyzed the CNAs across the whole genome using
oligoarray-CGH for the HNSCC patients who had been administered
postoperative CCRT.
Materials and methods
Patients and tumor samples
We retrospectively selected 18 patients who were
diagnosed with HNSCC and underwent primary tumor resection and neck
dissection followed by CCRT at the Department of
Otolaryngology-Head and Neck Surgery from April 2008 to June 2012.
Tumor stage was determined according to the standard
tumor-node-metastasis classification of AJCC guidelines (7th
edition). Frozen tissues (tumor and adjacent normal tissue pairs)
were obtained from the patients with previously untreated HNSCC.
Indications for adjuvant CCRT are as follows: positive or close
margins found on the resection, advanced T stage, lymphovascular
invasion, perineural invasion, multiple nodal metastasis, or
extracapsular spread. All 18 HNSCC patients received postoperative
radiotherapy (RT) that consisted of conventionally fractionated
doses of 1.8–2 Gy each in five weekly sessions. A volume including
the primary tumor site and all the draining lymph nodes at risk
received a dose of 50–55 Gy. Regions that had an inadequate
resection margin, extracapsular nodal spread or the initially
involved lymph node area were boosted up to 65 Gy. The patients
received chemotherapy that consisted of 100 mg
cisplatin/m2 of body-surface area on days 1, 22 and 43
during RT or weekly 30 mg cisplatin/m2 of body surface
area. The Institutional Review Board of Seoul St. Mary's Hospital
approved the retrospective review of the medical records and the
use of archived tumor specimens.
Array comparative genomic
hybridization
For array-CGH analysis, 10-µm-thick frozen
sections of tumor cell-rich areas (>60%) were microdissected.
Genomic DNA was extracted from these sections using a DNeasy Blood
and Tissue kit (Qiagen, Hilden, Germany). The Agilent SurePrint G3
Human CGH Microarray kit, 4×180K (Agilent technologies, Santa
Clara, CA, USA) was used for the array-CGH analysis. This array
contains 180,880 probes with a median probe spacing of 13 kb. The
array hybridization procedure was performed as described elsewhere
(12). In brief, 1 µg of
genomic DNA (gDNA) from tumor tissue was labeled with Cy5-dCTP and
1 µg of gDNA from paired normal tissue with Cy3-dCTP. After
purifying the labeled product by using Amicon Ultra-0.5 centrifugal
Filter Device (Millipore, Billerica, MA, USA), the labeled DNA was
applied on the Agilent 4×180K array with the blocking agent
(Agilent technologies, Santa Clara, CA, USA) containing 50
µg of human Cot-1 DNA (HybMasker, ConnectaGen, Korea). Array
slides were incubated for 24 h at 65°C in the hybridization oven.
After the hybridization, the arrays were washed with wash buffer 1
and 2 according to the manufacturer's protocol and scanned using
High-Resolution Microarray Scanner (Agilent technologies).
Determination of CNAs and recurrently
altered regions
Microarray hybridization images were analyzed with
Feature Extraction Software version 10.7.3.1 using CGH-107-Sep09
protocol for normalization as described previously (12). Data quality was assessed using
CGH_QCTM_Sep09 QC metric set. Only the data that passed the quality
check (in good or excellent range) was used for CNA calling. CNAs
of samples were defined using the ADM-2 algorithm in Agilent
Genomic Workbench Lite Edition 6.0 software (Agilent technologies).
CNAs were defined using the following criteria; a minimum number of
probes in region=3, a maximum number of probes in aberrant
region=100,000, a signal intensity ratio of >0.3 in
log2 scale for gains, and of <−0.3 in log2
scale for losses. High-level amplification was defined as a probe
signal intensity ratio of 1 or higher in log2 scale.
Likewise, a homozygous deletion (HD) was defined as a ratio of -1
or lower in log2 scale. After defining CNAs, recurrently
altered regions (RAR) were defined using the CNVRuler program as
chromosomal segments consisting of multiple overlapping CNAs found
in ≥30% of the samples (13).
Real-time quantitative PCR (qPCR)
analysis
We performed qPCR validation of candidate RARs using
the gDNA extracted from 18 pairs of head and neck cancer/normal
tissues. As a diploid internal control, we used a genomic region on
chromosome 13 (13q32.1) which was observed to have no genomic
alteration in the array-CGH data. Sequence and location of primers
used for the genomic qPCR experiment are as follows. RAR-G2
(7p12.1-p11.2 containing EGFR) forward, TTTGCCAAGGCACGAGTAA
and reverse, CAAGGACCACCTCACAGTTATT; RAR-G8 (18p11.32 containing
TYMS) forward, ATGTGGTGAACAGTGAGCTGTCCT and reverse, AATCA
TGTACGTGAGCAGGGCGTA; RAR-L4 (9p21.3 containing CDKN2A/B)
forward, TTTGGAGGACAAGCTGGTGCA ATG and reverse,
TCCCAGATGAGGACAATGAGGCAA; Diploid control (16p12.2), forward,
GACCTACCCACTTTCAACTCTG and reverse, CTCTCCAAGCGCAAGGTATT. Genomic
qPCR were performed using ViiA 7 Real-Time PCR System and ViiA 7
RUO software (Applied Biosystems, Foster City, CA, USA) as
described previously (14). In
brief, qPCR mixture of 10 µl contains 10 ng of gDNA,
Maxima® SYBR-Green qPCR Master Mix (2X), ROX solution
provided (Fermentas, Foster, CA, USA), and 5 pmol primers. Thermal
cycling conditions consist of one cycle of 10 min at 95°C, followed
by 40 cycles of 5 sec at 95°C, 10 sec at 61°C and 20 sec at 72°C.
The dissociation value of the amplified PCR product was measured as
the temperature inched up at a rate of 0.1°C/sec from 55°C to 95°C.
All qPCR experiments were triplicated and relative quantification
was performed by the 2−ΔΔCT (15). Relative ratios were used to
determine whether the status of a CNA is copy number gain (>2)
or loss (<0.5).
Statistical analysis
To explore associations of chromosomal copy number
changes and clinicopathologic phenotypes, phenotype variables were
treated as categorical variables, that is, age, gender, primary
site, histologic grade, T stage, cervical lymph node metastasis,
presence or absence of lymphovascular invasion, extracapsular
spread and recurrence. Differences in the distributions of RARs for
each categorical variable were tested using two-sided Fisher's
exact test. Disease-specific survival was determined using the
Kaplan-Meier method. The Cox proportional hazards model with
likelihood ratio statistics was used to identify variables
significantly and independently related to survival. P<0.05 was
considered to indicate a statistically significant difference, and
all calculations were performed using SPSS version 16.0 (SPSS,
Chicago, IL, USA).
Results
Clinicopathological characteristics
The median age was 59.2 years (range, 49–77 years)
and all patients were males. The sites of the original primary
tumor were oral cavity (11.1%), oropharynx (44.4%), larynx (16.7%)
and hypopharynx (27.8%). A majority of them (17/18, 94.4%) were
stage T2 and T3, and sixteen patients (89.0%) were cervical lymph
node positives. Thirteen patients (72.2%) were lymphovascular
invasion positives and nine (50.0%) had extracapsular spread of a
metastatic neck node. A total of nine patients (50.0%) had tumor
recurrence after CCRT. There was no clinicopathological factors
significantly associated with the treatment failure defined as
tumor recurrence. Details of clinicopathological characteristics of
the study subjects are summarized in Table I.
 | Table IClinicopathological characteristics
of the 18 HNSCC patients. |
Table I
Clinicopathological characteristics
of the 18 HNSCC patients.
| Parameter | No. of patients
(%) |
|---|
| Age (years) | |
| ≥60 | 7 (38.9) |
| <60 | 11 (61.1) |
| Gender | |
| Male | 18 (100) |
| Female | 0 (0) |
| Primary tumor | |
| Oral cavity | 2 (11.1) |
| Oropharynx | 8 (44.4) |
| Larynx | 3 (16.7) |
| Hypopharynx | 5 (27.8) |
| T stage | |
| T1 | 1 (5.6) |
| T2 | 11 (61.1) |
| T3 | 6 (33.3) |
| Cervical lymph node
stage | |
| N0 | 2 (11.1) |
| N1 | 4 (22.2) |
| N2 | 11 (61.1) |
| N3 | 1 (5.6) |
| Histologic
grade | |
| Well
differentiated | 2 (11.1) |
| Moderate
differentiated | 15 (83.3) |
| Poorly
differentiated | 1 (5.6) |
| Lymphovascular
invasion | |
| Yes | 13 (72.2) |
| No | 5 (27.8) |
| Extracapsular
spread | |
| Yes | 9 (50.0) |
| No | 9 (50.0) |
| Margin
involvement | |
| Yes | 3 (16.7) |
| No | 15 (83.3) |
| Recurrence | |
| Yes | 9 (50.0) |
| No | 9 (50.0) |
Copy number alteration profiles in
HNSCC
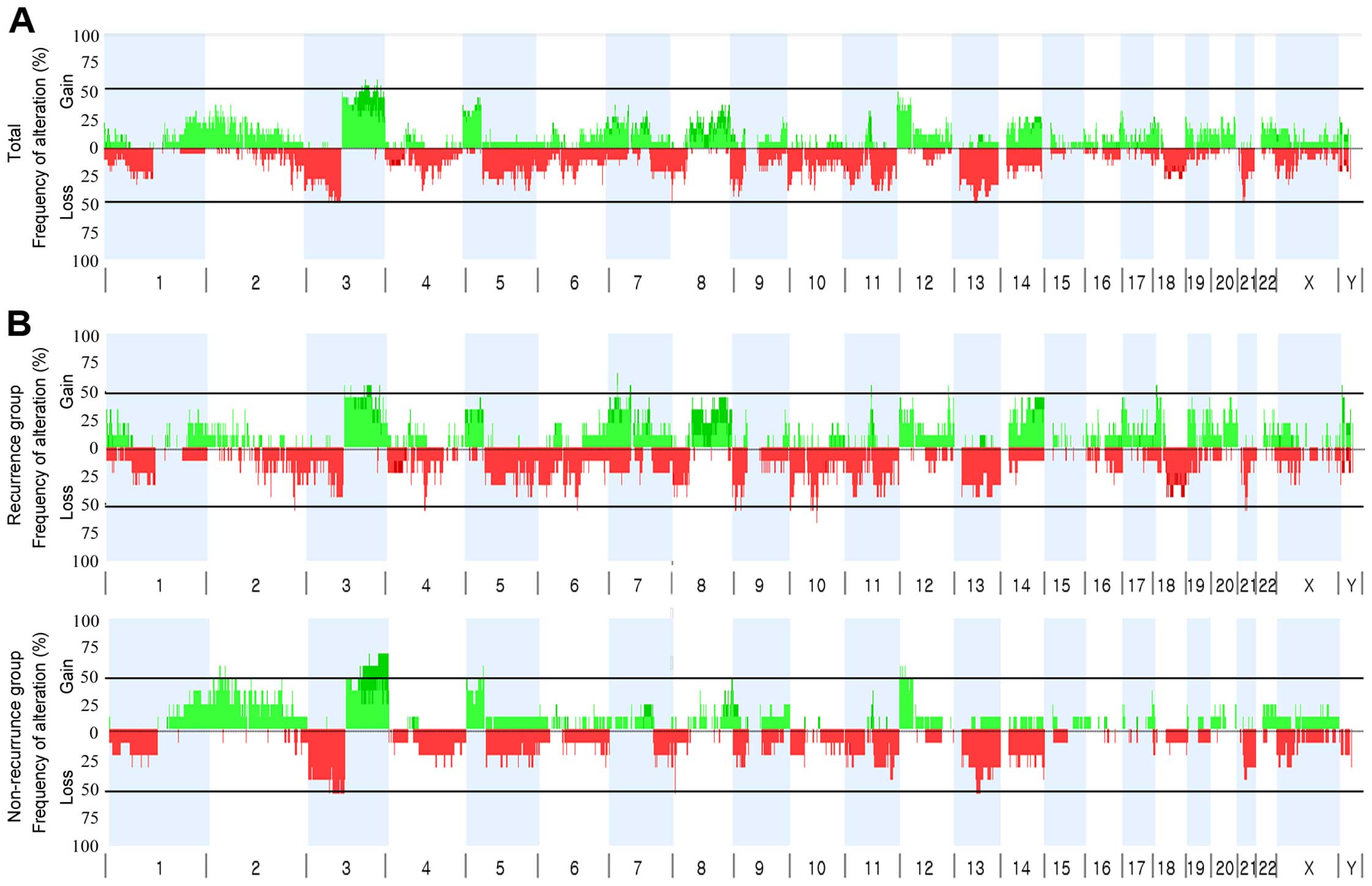
The overall frequency plot of the CNAs detected in
the 18 HNSCCs (9 with and 9 without recurrence) showed that CNAs
were not randomly distributed but rather clustered in certain
chromosomal segments across the genome (Fig. 1A). The mean number of CNAs per each
HNSCC genome was 41.3±18.6. Patients with recurrence had more CNAs
than those without recurrence (mean frequency 57.3±24.9 vs.
25.3±7.2), but the difference did not reach statistical
significance. As a whole, 8 chromosomal arms were recurrently
(appeared in ≥2 cases) gained on: 2p (11%), 3q (28%), 5p (11%), 7p
(11%), 8q (11%), 12p (11%), 18p (11%) and 22q (11%). Also, 8
chromosomal arms were recurrently deleted: 1p (11%), 3p (22%), 4p
(11%), 5q (22%), 10p (17%), 13q (17%), 14q (11%) and 21q (17%). Of
the recurrent chromosomal arm changes, copy number gains on 7p, 8q
and 18p were observed only in the treatment failure group but these
differences were not statistically significant.
Recurrently altered chromosomal
regions
In addition to the entire chromosomal arm changes,
many of the CNAs were regional changes. Some of them were observed
recurrently, suggesting their potential contribution to
tumorigenesis and progression. To identify the commonly implicated
CNAs, we defined RARs. In the present study, chromosomal segments
as the union of overlapping CNAs in ≥20% of the 18 HNSCCs were
defined as RARs (RAR-G for gains and RAR-L for losses,
respectively). A total of 15 RARs (8 RAR-Gs and 7 RAR-Ls) were
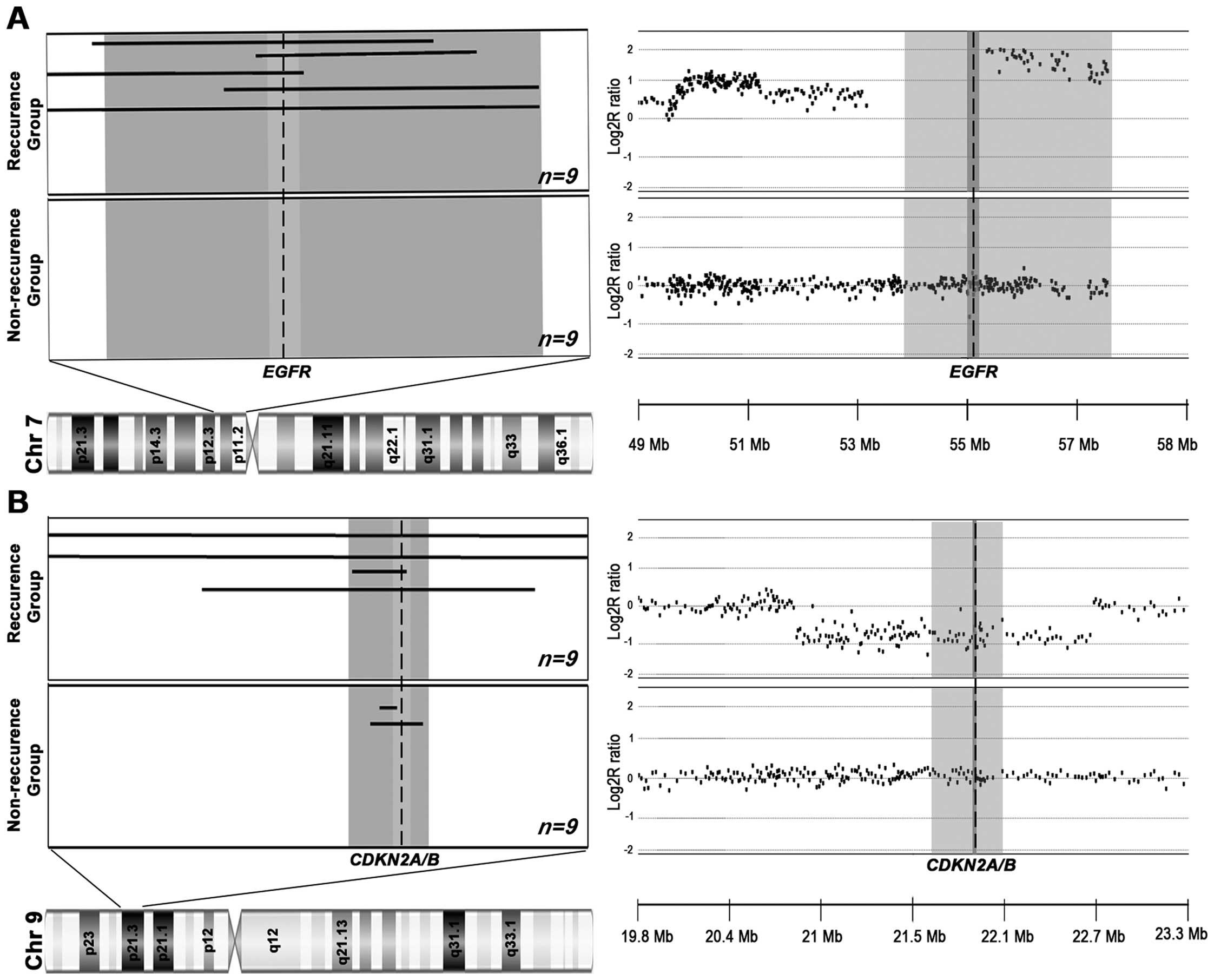
identified in the 18 HNSCC patients (Table II). Fig. 2 illustrates the RAR-G on 7p11.2 and
RAR-L on 9p21.3 as examples. Of the 15 RARs, a RAR-G on 7p11.2
(RAR-G2: 4.22 Mb-sized, 55.6% in recurrence group vs. 0% in
non-recurrence group, P=0.029), and a RAR-G on 18p11.32 (RAR-G8:
4.35 Mb-sized, 44.4% in recurrence group vs. 0% in non-recurrence
group, P=0.029) were significantly more common in the HNSCC with
recurrence group than in the HNSCC without recurrence group. The
well-known oncogene EGFR (7p11.2) and the potential oncogene
TYMS (18p11.32) are located in the recurrence-group dominant
RAR-Gs. A RAR-L on 9p21.3 (0.42 Mb-sized RAR-L4), where CDKN2A/B
are located, more commonly occurred in the HNSCC with recurrence
group (44.4%) than in non-recurrence group (22.2%) but it was not
statistically significant. To validate the three recurrence group
dominant RARs identified by array-CGH, we performed genomic qPCR
validations for those loci (7p11.2, 9p21.3 and 18p11.32). All the
CNAs identified by array-CGH were consistently detected by the qPCR
and as a whole 93.9% (46/49) of the qPCR results were consistent
with the copy number status by array-CGH (data not shown).
 | Table IIRecurrently altered regions in 18
HNSCC. |
Table II
Recurrently altered regions in 18
HNSCC.
| RARs | Chr | Position (Mb) | Size (Mb) | Cytoband | Recur n (%) | Non-re n (%) | Total n (%) | P-value | Cancer-related
genes |
|---|
| RAR-G1 | 3 | 134.3–197.8 | 63.53 | 3q22.2-q29 | 3 (33.3) | 5 (55.5) | 8 (44.4) | 0.347 | PIK3CA,
PIK3CB, BCL6, EIF4A2, ETV5,
FOXL2, LPP, MLF1, PIK3CA, SOX2,
TBL1XR1, TFRC, WWTR1 |
| RAR-G2 | 7 | 53.3–57.6 | 4.22 | 7p12.1-p11.2 | 5 (55.6) | 0 (0) | 5 (27.8) |
0.029* | EGFR |
| RAR-G3 | 8 | 128.2–128.9 | 0.67 | 8q24.21 | 3 (33.3) | 3 (33.3) | 6 (33.3) | 1.000 | MYC |
| RAR-G4 | 8 | 142.7–146.3 | 3.64 | 8q24.3 | 3 (33.3) | 4 (44.4) | 7 (38.9) | 0.637 | RECQL4 |
| RAR-G5 | 11 | 70.5–70.9 | 0.44 | 11q13.4 | 3 (33.3) | 1 (11.1) | 4 (22.2) | 0.576 | - |
| RAR-G6 | 12 | 0.2–7.3 | 7.11 |
12p13.33-p13.31 | 3 (33.3) | 1 (11.1) | 4 (22.2) | 0.576 | FGF23,
FGF6, CCND2, ZNF384 |
| RAR-G7 | 12 | 20.1–20.6 | 0.57 | 12p12.2 | 3 (33.3) | 1 (11.1) | 4 (22.2) | 0.576 | PIK3C2G |
| RAR-G8 | 18 | 0.1–4.5 | 4.35 |
18p11.32-p11.31 | 4 (44.4) | 0 (0) | 4 (22.2) | 0.029 | TYMS,
YES1 |
| RAR-L1 | 3 | 59.4–71.1 | 11.74 | 3p14.2-p13 | 1 (11.1) | 3 (33.3) | 4 (22.2) | 0.294 | FHIT,
MITF, ADAMTS9 |
| RAR-L2 | 5 | 64.1–70.7 | 6.52 | 5q12.3-q13.2 | 3 (33.3) | 1 (11.1) | 4 (22.2) | 0.576 | PIK3R1,
ADAMTS6, CDK7, CENPH, CENPK,
ERBB2IP, MAST4, OCLN, PIK3R1,
PPWD1, RAD17, TRIM23 |
| RAR-L3 | 8 | 4.4–4.8 | 0.46 | 8p23.2 | 3 (33.3) | 1 (11.1) | 4 (22.2) | 0.576 | - |
| RAR-L4 | 9 | 21.7–22.1 | 0.42 | 9p21.3 | 4 (44.4) | 2 (22.2) | 6 (33.3) | 0.620 |
CDKN2A/B |
| RAR-L5 | 10 | 0.1–3.7 | 3.54 | 10p15.3-p15.2 | 3 (33.3) | 1 (11.1) | 4 (22.2) | 0.576 | - |
| RAR-L6 | 11 | 104.6–105.9 | 1.24 | 11q22.3 | 1 (11.1) | 3 (33.3) | 4 (22.2) | 0.294 | - |
| RAR-L7 | 13 | 20.1–21.0 | 0.93 | 13p12.11 | 3 (33.3) | 1 (11.1) | 4 (22.2) | 0.576 | - |
High-level CNAs in HNSCC
Of the CNAs identified in the 18 HNSCCs, 25 loci
were found to be high-level CNAs (defined as ≥1 for amplification
or ≤−1 for HD on the log2 scale), with 15 amplifications
and 10 HDs (Table III). Five
high-level CNAs were detected in >20% of the samples;
amplifications on 7p11.2 and 11q13.3, and HDs on 1p31.1, 4q13.2 and
9p21.3, respectively (Table III).
Well-known oncogenes (EGFR and CCND1) and tumor
suppressor gene (CDKN2A/B) are located in the recurrent
(>20%) high-level CNA regions.
 | Table IIIHigh-level copy number changes in
HNSCC. |
Table III
High-level copy number changes in
HNSCC.
| CNAs | Chr | Position (MP) | Size (Mb) | Cytoband | Freq % (Recur) | Freq %
(Non-re) | Freq % (Total) | Cancer-related
genes |
|---|
| Amp1 | 1 | 59.96–61.93 | 1.97 | p32.1-p31.3 | 11.1 | 0.0 | 11.1 | – |
| Amp2 | 3 | 181.62–183.18 | 1.56 | q26.33-q27.1 | 11.1 | 0.0 | 11.1 | – |
| Amp3 | 7 |
54.87–56.57 | 1.70 | p11.2 | 22.2 | 0.0 | 22.2 |
EGFR |
| Amp4 | 7 | 64.10–66.63 | 2.53 | q11.21 | 11.1 | 0.0 | 11.1 | SBDS |
| Amp5 | 10 | 35.15–36.00 | 0.85 | p11.21 | 0.0 | 11.1 | 11.1 | – |
| Amp6 | 10 | 55.04–55.13 | 0.09 | q21.1 | 11.1 | 0.0 | 11.1 | – |
| Amp7 | 11 |
69.48–69.57 | 0.09 | q13.3 | 22.2 | 11.1 | 33.3 |
CCND1 |
| Amp8 | 11 | 84.35–84.55 | 0.20 | q14.1 | 0.0 | 11.1 | 11.1 | – |
| Amp9 | 11 | 100.89–101.44 | 0.55 | q22.1 | 0.0 | 11.1 | 11.1 | – |
| Amp10 | 13 | 64.76–67.46 | 2.70 | q21.31-q21.32 | 11.1 | 0.0 | 11.1 | – |
| Amp11 | 13 | 73.64–74.37 | 0.73 | q22.1 | 11.1 | 0.0 | 11.1 | – |
| Amp12 | 13 | 77.48–78.10 | 0.62 | q22.3 | 11.1 | 0.0 | 11.1 | – |
| Amp13 | 16 | 51.03–53.37 | 2.34 | q12.1-q12.2 | 0.0 | 11.1 | 11.1 | – |
| Amp14 | 17 | 46.30–46.99 | 0.69 | q21.32 | 11.1 | 0.0 | 11.1 | – |
| Amp15 | 18 | 0.12–4.47 | 4.35 | p11.32 | 11.1 | 0.0 | 11.1 | – |
| HD1 | 1 |
72.77–72.80 | 0.03 | p31.1 | 11.1 | 11.1 | 22.2 | – |
| HD2 | 2 | 141.17–141.27 | 0.10 | q22.1 | 11.1 | 0.0 | 11.1 | – |
| HD3 | 3 | 4.13–4.49 | 0.36 | p26.1 | 11.1 | 0.0 | 11.1 | – |
| HD4 | 3 | 162.51–162.62 | 0.10 | q26.1 | 11.1 | 0.0 | 11.1 | – |
| HD5 | 4 |
69.39–69.46 | 0.07 | q13.2 | 11..1 | 11.1 | 22.2 | – |
| HD6 | 5 | 81.35–81.52 | 0.17 | q14.1-q14.2 | 0.0 | 11.1 | 11.1 | – |
| HD7 | 8 | 2.76–3.18 | 0.42 | p23.2 | 11.1 | 0.0 | 11.1 | – |
| HD8 | 9 |
21.96–22.03 | 0.07 | p21.3 | 22.2 | 11.1 | 33.3 |
CDKN2A |
| HD9 | 10 | 88.47–90.75 | 2.28 | q23.2-q23.31 | 11.1 | 0.0 | 11.1 | BMPR1A,
PTEN |
| HD10 | 11 | 12.32–12.94 | 0.62 | p15.3-p15.2 | 0.0 | 11.1 | 11.1 | – |
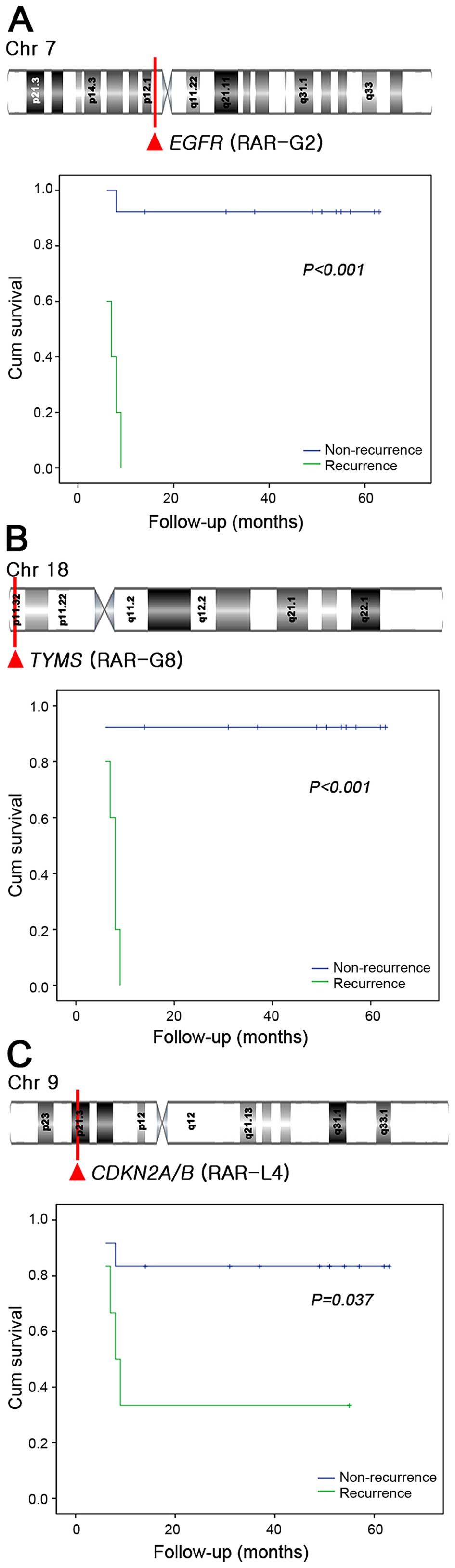
RARs associated with the prognosis in
HNSCC
Univariate survival analysis was performed to assess
the implications of the RARs on patient survival. Univariate
analysis was also performed to identify clinicopathologic features
(age, gender, primary site, histologic grade, T stage, cervical
lymph node metastasis, lymphovascular invasion, extracapsular
spread and recurrence) for inclusion as covariates for Cox
regression. Five-year disease-specific survival rates (DSSR) in our
cohort were 66%. Among the RARs and clinicopathologic factors,
cervical lymph node metastasis (P=0.008), recurrence (P=0.003),
RAR-G2 (P<0.001), RAR-G8 (P<0.001), and RAR-L4 (P=0.037) were
significantly associated with poor DSSR in the univariate analysis
(Fig. 3). Multivariate analysis
with the three significant RARs and two clinical variables
identified on univariate analysis revealed that RAR-G2 [hazard
ratio (HR)=40.68, 95% confidence interval (CI): 3.56–464.69,
P=0.003] and cervical metastasis (HR=16.38, 95% CI: 1.20–223.11,
P=0.036) were significantly associated with poor DSSR (Table IV).
 | Table IVResults of multivariate analysis. |
Table IV
Results of multivariate analysis.
| Variables | HR | 95% CI
| P-value |
|---|
| Lower | Upper |
|---|
| Cervical
metastasis | 16.38 | 1.20 | 223.11 | 0.036 |
| Recurrence | 0.940 | 0.00 | 17.30 | 0.332 |
| RAR-G02 | 40.68 | 3.56 | 464.69 | 0.003 |
| RAR-G08 | 2.16 | 0.16 | 22.46 | 0.142 |
| RAR-L04 | 1.21 | 0.00 | 7.76 | 0.217 |
Discussion
Despite the various treatment modalities, the poor
prognosis of HNSCC has not improved significantly over the last
four decades (16). The lack of
success in HNSCC treatment may be primarily due to lack of
understanding on the etiology and heterogeneous nature of HNSCC. In
the present study, we aimed to explore CNA profiles associated with
the resistance after CCRT in HNSCCs. For this, we analyzed the CNAs
using high-resolution array-CGH (180K oligoarray) and delineated
RARs under the assumption that commonly altered chromosomal
segments in CCRT-resistant subgroup may contain essential genes
contributing to CCRT resistance. Through this approach, we
identified the genetic markers predicting the responsiveness to
CCRT following surgery in HNSCC patients.
The key alterations identified in the present study,
such as amplifications of PIK3CA, EGFR and MYC
and deletions of CDKN2A/B and FHIT, were consistent
with previous observations in HNSCC, suggesting the reliability of
our study (10,12,17,18).
We also identified several novel RARs which have not been reported
to be associated with HNSCC previously; RAR-Gs in 12p13 and 12p12
harboring FGF6, FGF23 and PIK3C2G genes and a
RAR-L in 8p23.2 harboring CSMD1 gene. To validate the
array-CGH results, we performed genomic qPCR validation for the
three interesting RARs (RAR-G2, RAR-G8 and RAR-L4) identified by
array-CGH and all of the CNAs were consistently defined in the
qPCR, suggesting the reliability of our array-CGH analysis and
advantage of higher resolution array analysis.
Two RARs, RAR-G2 (7p11.2 encompassing EGFR)
and RAR-G8 (18p11.32 encompassing TYMS), were significantly
more common in the HNSCC patients with recurrence (CCRT
non-responders) than in those without recurrence (CCRT-responders).
In the observations of Gao et al, EGFR overexpression was
associated with poor survival in HNSCC patients receiving CCRT
(19). Szabó et al examined
expression and genetic alteration of EGFR gene by FISH with
71 HNSCC patients and observed that increased copy number and
overexpression of the EGFR gene was associated with poor
prognosis in HNSCC (20). However,
neither group examined the CNAs involving the responsiveness to
CCRT in terms of recurrence. To our knowledge, this is the first
report which explored the CNA profiles across the whole genome in
HNSCC using high-resolution array-CGH and demonstrated that the
copy number gain of 17p11.2 containing EGFR was
significantly associated with the CCRT failure. When we assessed
the prognostic implications of RARs, the RAR-G2 containing
EGFR was the only significantly associated with poor DSSR in
multivariate analysis. Previously, increased copy number of
EGFR gene was also strongly associated with poorer survival,
both overall and recurrence-free, in HNSCC (21).
In addition to the EGFR, the copy number gain
of TYMS (RAR-G8), which codes thymidylate synthase, was also
found to be significantly more common in the CCRT non-responders.
TYMS catalyzes the conversion of dUMP to dTMP, which is
essential for DNA biosynthesis (22). Consistent with our finding,
overexpression of TYMS, caused by gene amplification, has
been reported to be associated with poor prognosis and resistance
to chemotherapeutic agents that target thymidylate biosynthesis
such as 5-FU (23–25). In the study of Yasumatsu et
al exploring the effect of TYMS knockdown in a HNSCC
cell line (YCU-N), repressed expression of TYMS increased the
cytotoxicity of 5-FU and inhibited cell proliferation (26). These data consistently suggest the
implication of copy number gain of TYMS in acquiring resistance to
5-FU, which may contribute to the resistance to CCRT.
In this study, a copy number loss of 0.46 Mb-sized
locus in 8p23.2, where CSMD1 gene is located, were
recurrently observed in the HNSCC patients. The deletion of 8p23
locus has been frequently reported in HNSCCs and through the
deletion mapping analysis, CSMD1 gene in this locus has been
suggested as a tumor suppressor (27–29).
The deletion of CSMD1 has been also frequently reported in
diverse solid tumors and the underexpression of CSMD1 was
reported to be associated with poor prognosis or higher tumor grade
(30–32). Although copy number loss of
CSMD1 gene did not show significant association with
CCRT-resistance in our study, there was a tendency of non-responder
dominance (3 in non-responders vs. 1 in responders). To our
knowledge, this is the first report to identify the frequent copy
number loss of CSMD1 gene in HNSCC with CCRT resistance.
Larger scale analysis will be required to verify this
possibility.
In spite of the interesting findings, there were
several limitations to the present study. First of all, due to the
limited number of patients treated with primary surgery plus CCRT,
some potential CCRT-associated CNAs could be missed. Second, the
median follow-up period of this study was relatively short.
Prospective, randomized studies using a larger sample will be
required to assess the implications of the CNAs identified in the
present study on clinical results following different treatment
modalities.
In conclusion, we successfully defined novel
recurrent genomic alteration regions and known copy number changes
in 18 HNSCC with a well-designed high-resolution array CGH. Copy
number gain of EGFR (RAR-G2) and TYMS (RAR-G8) were
associated with CCRT failure and RAR-G2 was also found to be an
independent factor of poor DSSR in HNSCC. Our results and the
strategy of analysis will provide useful clues for further studies
to elucidate the molecular pathogenesis of HNSCC and to develop
biomarkers for designing personalized treatment by predicting the
prognosis.
Acknowledgments
The present study was supported by the Cancer
Evolution Research Center (grant no. 2012R1A5A2047939) and the
Institute of Clinical Medicine Research of Bucheon St. Mary's
Hospital Research Fund, 2015.
References
|
1
|
Pignon JP, Bourhis J, Domenge C and
Designé L: Chemotherapy added to locoregional treatment for head
and neck squamous-cell carcinoma: Three meta-analyses of updated
individual data. MACH-NC Collaborative Group Meta-Analysis of
Chemotherapy on Head and Neck Cancer. Lancet. 355:949–955. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Smith RV, Kotz T, Beitler JJ and Wadler S:
Long-term swallowing problems after organ preservation therapy with
concomitant radiation therapy and intravenous hydroxyurea: Initial
results. Arch Otolaryngol Head Neck Surg. 126:384–389. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Cinelli M, Magnelli L and Chiarugi V:
Redundant down-regulation pathways for p53. Pharmacol Res.
37:83–85. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Osman I, Sherman E, Singh B, Venkatraman
E, Zelefsky M, Bosl G, Scher H, Shah J, Shaha A, Kraus D, et al:
Alteration of p53 pathway in squamous cell carcinoma of the head
and neck: Impact on treatment outcome in patients treated with
larynx preservation intent. J Clin Oncol. 20:2980–2987. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Nam TK, Lee JH, Cho SH, Chung IJ, Ahn SJ,
Song JY, Yoon MS, Chung WK and Nah BS: Low hMLH1 expression prior
to definitive chemoradiotherapy predicts poor prognosis in
esophageal squamous cell carcinoma. Cancer Lett. 260:109–117. 2008.
View Article : Google Scholar
|
|
6
|
Li SW, Wang H, Liu ML, Zhang HB, Xiang YQ,
Lv X, Xia WX, Zeng MS, Mai HQ, Hong MH, et al: Positive effect of
high RKIP expression on reduced distant metastasis by chemotherapy
when combined with radiotherapy in locoregionally advanced
nasopharyngeal carcinoma: A prospective study. Med Oncol.
30:3222013. View Article : Google Scholar
|
|
7
|
Cheng SH, Jian JJ, Tsai SY, Chan KY, Yen
LK, Chu NM, Tan TD, Tsou MH and Huang AT: Prognostic features and
treatment outcome in locoregionally advanced nasopharyngeal
carcinoma following concurrent chemotherapy and radiotherapy. Int J
Radiat Oncol Biol Phys. 41:755–762. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Chen C, Chen S, Le QT, Chen J, Chen Z and
Li D, Zhou M and Li D: Prognostic model for distant metastasis in
locally advanced nasopharyngeal carcinoma after concurrent
chemoradiotherapy. Head Neck. 37:209–214. 2015. View Article : Google Scholar
|
|
9
|
Wang TL, Diaz LA Jr, Romans K, Bardelli A,
Saha S, Galizia G, Choti M, Donehower R, Parmigiani G, Shih IeM, et
al: Digital karyotyping identifies thymidylate synthase
amplification as a mechanism of resistance to 5-fluorouracil in
metastatic colorectal cancer patients. Proc Natl Acad Sci USA.
101:3089–3094. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
van den Broek GB, Wreesmann VB, van den
Brekel MW, Rasch CR, Balm AJ and Rao PH: Genetic abnormalities
associated with chemoradiation resistance of head and neck squamous
cell carcinoma. Clin Cancer Res. 13:4386–4391. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Przybytkowski E, Aguilar-Mahecha A, Nabavi
S, Tonellato PJ and Basik M: Ultradense array CGH and discovery of
micro-copy number alterations and gene fusions in the cancer
genome. Methods Mol Biol. 973:15–38. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Joo YH, Park SW, Jung SH, Lee YS, Nam IC,
Cho KJ, Park JO, Chung YJ and Kim MS: Recurrent loss of the FHIT
gene and its impact on lymphatic metastasis in early oral squamous
cell carcinoma. Acta Otolaryngol. 133:992–999. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kim JH, Hu HJ, Yim SH, Bae JS, Kim SY and
Chung YJ: CNVRuler: A copy number variation-based case-control
association analysis tool. Bioinformatics. 28:1790–1792. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Jung SH, Yim SH, Hu HJ, Lee KH, Lee JH,
Sheen DH, Lim MK, Kim SY, Park SW, Kim SH, et al: Genome-wide copy
number variation analysis identifies deletion variants associated
with ankylosing spondylitis. Arthritis Rheumatol. 66:2103–2112.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
16
|
Oddone N, Morgan GJ, Palme CE, Perera L,
Shannon J, Wong E, Gebski V and Veness MJ: Metastatic cutaneous
squamous cell carcinoma of the head and neck: The
Immunosuppression, Treatment, Extranodal spread, and Margin status
(ITEM) prognostic score to predict outcome and the need to improve
survival. Cancer. 115:1883–1891. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Bernaldo de Quirós S, Merlo A, Secades P,
Zambrano I, de Santa María IS, Ugidos N, Jantus-Lewintre E, Sirera
R, Suarez C and Chiara MD: Identification of TRPC6 as a possible
candidate target gene within an amplicon at 11q21-q22.2 for
migratory capacity in head and neck squamous cell carcinomas. BMC
Cancer. 13:1162013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Shi ZZ, Shang L, Jiang YY, Hao JJ, Zhang
Y, Zhang TT, Lin DC, Liu SG, Wang BS, Gong T, et al: Consistent and
differential genetic aberrations between esophageal dysplasia and
squamous cell carcinoma detected by array comparative genomic
hybridization. Clin Cancer Res. 19:5867–5878. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Gao Z, Meng X, Mu D, Sun X and Yu J:
Prognostic significance of epidermal growth factor receptor in
locally advanced esophageal squamous cell carcinoma for patients
receiving chemoradiotherapy. Oncol Lett. 7:1118–1122.
2014.PubMed/NCBI
|
|
20
|
Szabó B, Nelhubel GA, Kárpáti A, Kenessey
I, Jóri B, Székely C, Peták I, Lotz G, Hegedus Z, Hegedus B, et al:
Clinical significance of genetic alterations and expression of
epidermal growth factor receptor (EGFR) in head and neck squamous
cell carcinomas. Oral Oncol. 47:487–496. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chung CH, Ely K, McGavran L,
Varella-Garcia M, Parker J, Parker N, Jarrett C, Carter J, Murphy
BA, Netterville J, et al: Increased epidermal growth factor
receptor gene copy number is associated with poor prognosis in head
and neck squamous cell carcinomas. J Clin Oncol. 24:4170–4176.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wilson PM, Danenberg PV, Johnston PG, Lenz
HJ and Ladner RD: Standing the test of time: Targeting thymidylate
biosynthesis in cancer therapy. Nat Rev Clin Oncol. 11:282–298.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Johnston PG, Fisher ER, Rockette HE,
Fisher B, Wolmark N, Drake JC, Chabner BA and Allegra CJ: The role
of thymidylate synthase expression in prognosis and outcome of
adjuvant chemotherapy in patients with rectal cancer. J Clin Oncol.
12:2640–2647. 1994.PubMed/NCBI
|
|
24
|
Murata S, Adachi M, Kioi M, Torigoe S,
Ijichi K, Hasegawa Y, Ogawa T, Bhayani MK, Lai SY, Mitsudo K, et
al: Etodolac improves 5-FU sensitivity of head and neck cancer
cells through inhibition of thymidylate synthase. Anticancer Res.
31:2893–2898. 2011.PubMed/NCBI
|
|
25
|
Yeh KH, Shun CT, Chen CL, Lin JT, Lee WJ,
Lee PH, Chen YC and Cheng AL: High expression of thymidylate
synthase is associated with the drug resistance of gastric
carcinoma to high dose 5-fluorouracil-based systemic chemotherapy.
Cancer. 82:1626–1631. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yasumatsu R, Nakashima T, Uryu H, Ayada T,
Wakasaki T, Kogo R, Masuda M, Fukushima M and Komune S:
Correlations between thymidylate synthase expression and
chemosensitivity to 5-fluorouracil, cell proliferation and clinical
outcome in head and neck squamous cell carcinoma. Chemotherapy.
55:36–41. 2009. View Article : Google Scholar
|
|
27
|
Sunwoo JB, Sun PC, Gupta VK, Schmidt AP,
El-Mofty S and Scholnick SB: Localization of a putative tumor
suppressor gene in the sub-telomeric region of chromosome 8p.
Oncogene. 18:2651–2655. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ishwad CS, Shuster M, Bockmühl U, Thakker
N, Shah P, Toomes C, Dixon M, Ferrell RE and Gollin SM: Frequent
allelic loss and homozygous deletion in chromosome band 8p23 in
oral cancer. Int J Cancer. 80:25–31. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Toomes C, Jackson A, Maguire K, Wood J,
Gollin S, Ishwad C, Paterson I, Prime S, Parkinson K, Bell S, et
al: The presence of multiple regions of homozygous deletion at the
CSMD1 locus in oral squamous cell carcinoma question the role of
CSMD1 in head and neck carcinogenesis. Genes Chromosomes Cancer.
37:132–140. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ma C, Quesnelle KM, Sparano A, Rao S, Park
MS, Cohen MA, Wang Y, Samanta M, Kumar MS, Aziz MU, et al:
Characterization CSMD1 in a large set of primary lung, head and
neck, breast and skin cancer tissues. Cancer Biol Ther. 8:907–916.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhang R and Song C: Loss of CSMD1 or 2 may
contribute to the poor prognosis of colorectal cancer patients.
Tumour Biol. 35:4419–4423. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Kamal M, Shaaban AM, Zhang L, Walker C,
Gray S, Thakker N, Toomes C, Speirs V and Bell SM: Loss of CSMD1
expression is associated with high tumour grade and poor survival
in invasive ductal breast carcinoma. Breast Cancer Res Treat.
121:555–563. 2010. View Article : Google Scholar
|