Introduction
Glioblastoma multiforme (GBM) is the most aggressive
tumor of the central nervous system and represents over 70% of all
brain malignancies. GBM is incurable by conventional therapeutic
strategies (1). In fact, despite
recent advances in surgery, radiotherapy and chemotherapy with
temozolomide (TMZ) (2), the median
survival of glioma patients remains poor and amounts to ~14 months.
One of the factors underlying tumor recurrence and poor long-term
survival is the presence of a cancer stem-like cell population,
termed glioma stem cells (GSCs), that is particularly resistant to
chemotherapy and radiotherapy and supports tumor self-renewal
(3). Another explanation for drug
resistance can be ascribed to intratumor heterogeneity due to the
development of independent clones derived from a unique progenitor
(4,5).
Increasing evidence suggests the role of epigenetic
events, apart from genetic alterations, in the development and/or
progression of several types of tumors, such as gliomas (6–8). The
potential reversibility of epigenetic alterations represents an
attractive approach to 'reset' the abnormal cancer epigenome by
using epigenetic drugs such as histone deacetylase inhibitors
(HDACi). Most recent data suggest the role of this class of drugs
in tumor growth inhibition, promotion of apoptosis and induction of
differentiation (9,10). Total eradication of the stem
subpopulation may be achieved by using pro-differentiative drugs,
which are able to induce differentiation of GSCs affecting their
self-renewal capabilities (11).
Thus, HDACi can induce de-repression of genes epigenetically
silenced in cancer (12,13) and involved in different cellular
processes, including cell cycle control, differentiation, DNA
repair and apoptosis (14).
Valproic acid (VPA), apart from being an
anticonvulsant and mood stabilizing drug, is also an HDACi that has
shown potent antitumor effects in a variety of in vitro and
in vivo glioma studies (15–18).
In recent years, the discovery of the ability of VPA to affect TMZ
sensitivity in GBM cell lines suggests the use of this drug as a
chemosensitizing agent (19–21).
In the present study, we investigated and compared
for the first time the effects of short-term and long-term VPA
treatments on cell morphology, differentiation behavior and DNA
methylation profile of GSCs. Moreover, we examine whether long-term
VPA treatment induces chemoresistance to VPA. Finally, we utilized
VPA to chemosensitize the GSC subpopulation to TMZ action.
Materials and methods
Cell lines and cell culture
conditions
The seven GSC lines used in this study (GBM2, GBM7,
GBM04, G144, G166, G179 and GliNS2) were isolated from patients
affected by GBM (22,23) and, in 2013, our research group
characterized their cytogenomic and epigenomic profiles (24). Cells were cultured in neural stem
cell-specific medium, and their stem-like properties were
periodically monitored, as previously described (22–24).
Cells were cultured in an adherent culture condition using 10
µg/ml laminin (Invitrogen) in a proliferation permissive
medium composed of Dulbecco's modified Eagle's medium (DMEM)/F-12
and Neurobasal 1:1 (Invitrogen), B-27 supplement without vitamin A
(Invitrogen), 2 mM L-glutamine, 10 ng/ml recombinant human bFGF and
20 ng/ml recombinant human EGF (Miltenyi Biotec), 20 UI/ml
penicillin and 20 µg/ml streptomycin (Euroclone).
Drug treatments
Valproic acid (sodium salt; Sigma) was dissolved in
sterile water to a stock concentration of 0.5 M. Temozolomide
(Sigma) was dissolved in sterile DMSO at the final concentration of
0.05 M. All drugs were stored at −20°C.
In vitro treatments were performed using 2 mM
VPA for 24, 48,72, 96 h and 14 and 30 days; with regard to TMZ, we
administered 50, 100, 200 or 400 µM TMZ for 48 or 72 h.
Cell viability by MTT assay
The methyl-thiazolyl tetrazolium (MTT) assay was
performed to evaluate VPA efficacy in VPA long-term treated (30
days) cells compared with cells that were not previously treated
(naïve). We also performed the cell viability analysis on naïve
cells treated with the combination of 2 mM VPA plus several
concentrations of TMZ (50, 100, 200 and 400 µM) for 96 h.
The combined treatment was divided into a pretreatment phase (only
VPA for 48 h) and a combination phase (VPA plus TMZ for an
additional 48 h).
Cells were seeded at a density of 4×104
cells/well in a 96-well-plate in 100 µl of culture medium
and incubated at 37°C. On the following day, we added VPA or TMZ at
the selected final concentrations. After 24, 48, 72 and 96 h of
treatment, 100 µl of 1 mg/ml MTT solution (Sigma) was added
to each well and incubation was carried out for 3 h at 37°C.
Therefore, formazan granules were solubilized in absolute ethanol.
The dye absorbance was measured spectrophotometrically at a 595-nm
wavelength with the FLUOstar Omega microplate reader (BMG Labtech).
The percentage of metabolic activity inhibition was determined by
comparing the absorbance values of the treated cells with those of
the untreated controls: (Treated cell absorbance/untreated cell
absorbance) × 100. Results are the mean values of two independent
experiments performed in quadruplicate.
Trypan blue dye exclusion assay
GBM2 (0.25×106) or G144 cells
(0.5×106) were seeded in T25 flasks and then treated
with different pharmacological regimens: i) 2 mM VPA for 96 h; ii)
50 µM or iii) 200 µM TMZ for 72 h; iv) 50 µM
or v) 200 µM TMZ for 72 h after pretreatment with 2 mM VPA
for 96 h. Then, cells were harvested and counted to evaluate the
number of live cells and cell mortality, using trypan blue dye to
discriminate live from dead cells. The results reported are the
mean values of three different experiments performed at least in
triplicate.
Cooperative index
In order to evaluate the effects of the combined
VPA-TMZ treatments, the cooperative index (CI) (26) was calculated comparing the sum of
the metabolic activity reduction percentages obtained for each
single agent to the percentages obtained upon combined treatments
(CI = VPA% + TMZ%/combined treament cell%). CI values <1
indicate a synergistic effect, CI values =1 indicate an additive
effect, while CI values >1 indicate an antagonistic effect.
Morphological analysis
To evaluate the cellular morphological changes after
VPA treatment, cells were seeded in serum-free medium at
3×103–104 cells/ml, depending on the cell
growth rate, specific for each GSC line. After 24 h, cells were
treated with 2 mM VPA concentration for 14 and 30 days. Every three
days the culture medium was changed and the drug was administered
again. The morphological changes were evaluated by phase contrast
microscopic observation, comparing VPA-treated and untreated cells.
Representative images were captured for each cell line and for each
treatment.
Immunofluorescence
The immunofluorescence assays were performed on
untreated and 2 mM VPA-treated GSC cultures for 14 and 30 days
using rabbit anti-CD133 (1:50; Santa Cruz Biotechnology, Santa
Cruz, CA, USA) and mouse anti-nestin (1:50; Millipore, Billerica,
MA, USA), rabbit anti-glial fibrillary acidic protein (GFAP, 1:200;
Dako), rabbit anti-βIII-tubulin (1:100; Covance) and goat
anti-myelin basic protein (MBP, 1:50; Santa Cruz Biotechnology) as
primary antibodies. Alexa Fluor 488-conjugated goat anti-mouse or
anti-rabbit (1:200; Molecular Probes, Eugene, OR, USA) was used as
the secondary antibody. Alexa Fluor 647-conjugated phalloidin
(1:200; Molecular Probes) was used to visualize the actin
filaments. Propidium iodide (PI) was used to counterstain the
nuclei.
Briefly, untreated cells were placed onto slides by
means of Cytospin, while VPA-treated cells spotaneously adhered to
the slides. Cells were washed with Dulbecco's modified
phosphate-buffered saline (PBS), fixed with 4% paraformaldehyde for
15 min and treated for 10 min with 0.1 M glycine (in PBS). The
slides were incubated for 30 min at room temperature (RT). in
blocking solution [5% bovine serum albumin (BSA), 0.6% Triton X-100
in PBS] and treated for 30 min with 70 U/mg RNAse (1:30;
Sigma-Aldrich, Milan, Italy) in blocking solution. Cells were
incubated with the primary antibodies at 4°C overnight. Then, the
slides were rinsed with washing buffer (0.3% Triton X-100 in PBS)
and incubated with secondary fluorescent antibodies, phalloidin and
2.5 mg/ml PI for 1 h at RT. The cells were then washed with PBS and
coverslips were mounted using polyvinyl alcohol mounting medium
(Fluka Analytical, Milan, Italy). Fluorescent cell preparations
were examined using a Radiance 2100 confocal microscope (Bio-Rad,
Hercules, CA, USA).
In order to perform a quantitative-like analysis,
the number of immunereactive cells for each marker was counted,
evaluating at least 100 cells/sample over different areas of the
slide.
MeDIP-Chip
GBM2 and G144 cell lines were treated with 2 mM VPA
for 96 h and 30 days and then DNA extraction from the control and
treated cultures was performed using the Wizard Genomic DNA
Purification kit (Promega, Milan, Italy), according to the
manufacturer's protocol.
Methylated DNA immunoprecipitation and chip
hybridization were performed as previously described (24) following the guidelines of the
Agilent Microarray Analysis of Methylated DNA Immunoprecipitation
protocol (version 1.0; Agilent Technologies, Santa Clara, CA,
USA).
The raw data, expressed as combined z-score values,
were analyzed according to the methodological approach conceived by
Dr Ravid Straussman (27). This
method allowed us to classify the methylation status of each CpG
island (CGI) as methylated, unmethylated, or undetermined (CGIs
with uncertain methylation status). Then, we decided to consider
only data referred to CGIs located in gene promoters for the
subsequent analysis due to the well-known biological effects of
methylation in these regions. When a promoter contained two or more
CGIs, we averaged the related combined z-score values in order to
establish the correct methylation status. Finally, we compared data
from the untreated and VPA-treated samples to highlight the
methylation changing promoters.
Bioinformatic analysis
The Gene Ontology (GO) analysis was performed, as
previously described (24), using
GOstat software (http://gostat.wehi.edu.au/) (28).
Statistical analysis
Statistical analysis was carried out performing
Fisher's exact and Student's t-tests. The critical level of
significance was set at P<0.05.
Results
Effect of short- and long-term VPA
treatments on GSC morphology
In a previous study, we evaluated the effect on GSC
morphology induced by VPA after 24, 48 and 72 h (25). We revealed that short-term VPA
treatment severely modified cell morphology. In this study, we
extended the evaluation to 14 and 30 days of treatment.
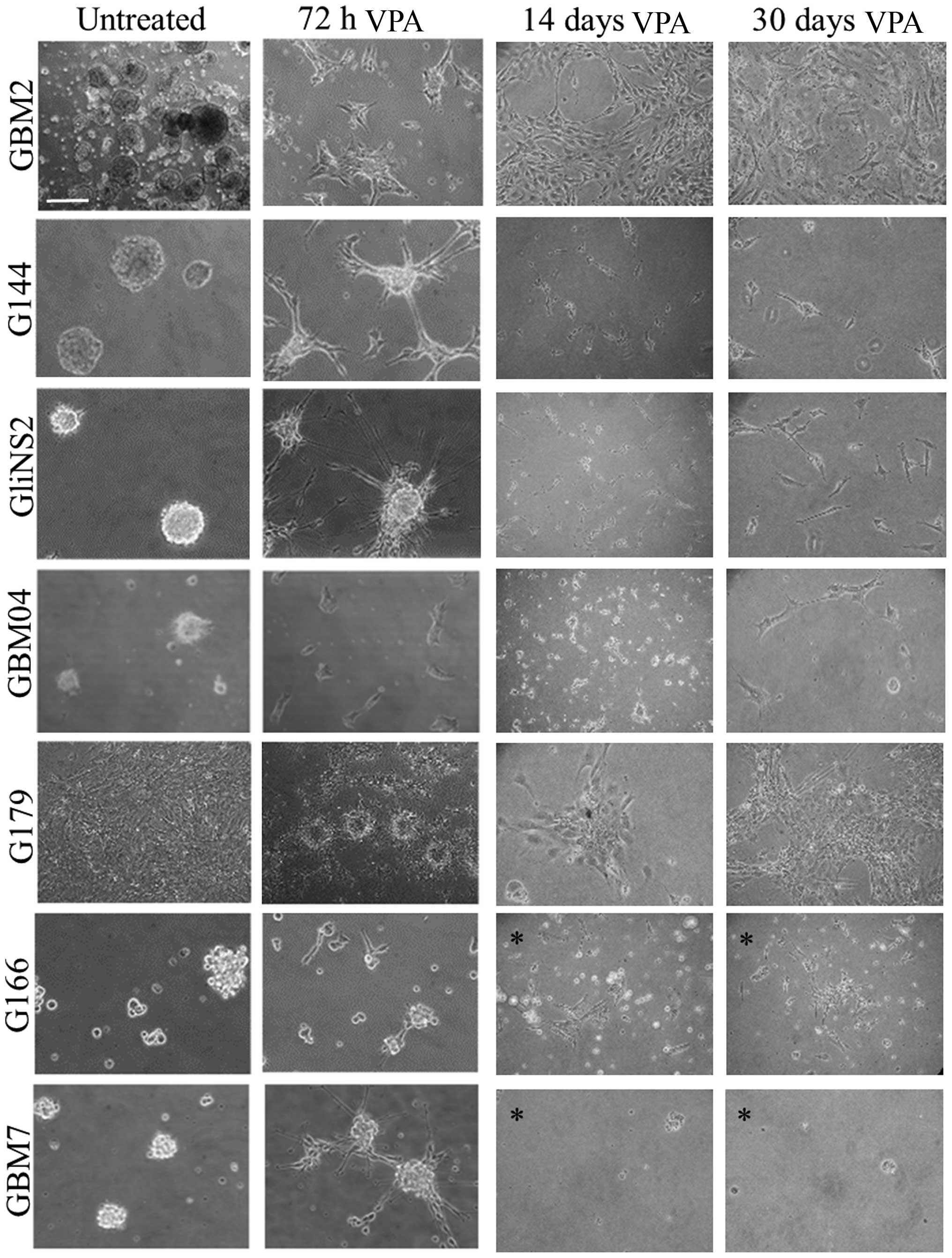
After 14 and, particularly, 30 days of exposure, the
pro-differentiating effect of VPA was evident: most of the cells
were star-shaped and displayed neurite-like processes (Fig. 1). G166 and GBM7 cells showed a high
number of dead cells, demonstrating the predominant cytotoxic
effect of VPA or the induction of apoptosis following terminal
differentiation. Therefore, these two cell lines were treated with
a lower drug concentration (1 mM) for 14 and 30 days. While G166
cells showed important morphological modifications at this lower
concentration treatment, the GBM7 cell line was still extremely
sensitive, showing a prevalence of cell death.
Differentiation behavior of GSCs in
response to short- and long-term VPA treatment
The pro-differentiation ability of VPA was evaluated
analyzing the expression of markers of stemness (CD133 and Nestin)
and differentiation (GFAP, βIII-tubulin and MBP) by
immunofluorescence in the untreated and 2 mM VPA-treated cells for
14 and 30 days. We compared these data with those we had obtained
after VPA short-term treatment (25).
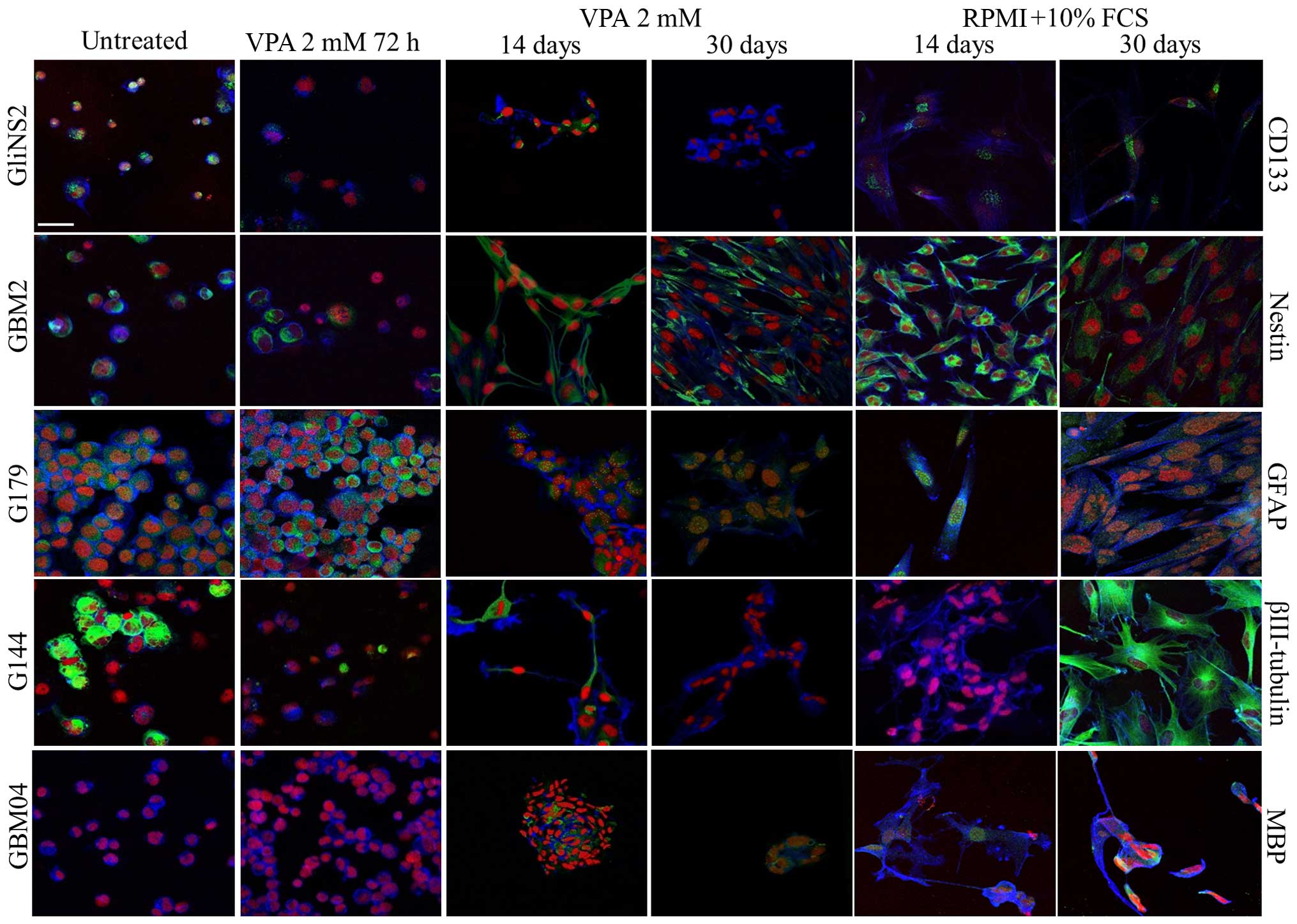
The summary of the immunofluorescence results is
reported in Table I, and
representative images are shown in Fig.
2. After 14 and 30 days of VPA exposure, 100% of the GBM2 cells
maintained nestin, βIII-tubulin and GFAP expression. Regarding
CD133 and MBP markers, we observed a significant increase in cells
expressing these two proteins with the prolongation of the
treatment time. The G144 cell line manifested a reduction in cells
expressing both stemness and differentiation markers after 30 days
compared to the control cells. An unusual behavior was observed for
GFAP, for which there was a decrease in cells expressing GFAP after
14 days, while this number increased after 30 days, but remained
significantly below the untreated control. The G166 cell line
showed a very high number of cells expressing differentiation
markers after 14 days of treatment; after 30 days these values were
drastically decreased. In contrast, few cells were positive for
both Nestin and CD133 after 14 days of treatment. The GliNS2 cell
line showed a significant reduction in CD133, GFAP and MBP
immunoreactive cells after 14 and 30 days of treatment.
Contrariwise, the percentage of cells positive for βIII-tubulin was
increased after both treatment times. An unusual behavior was
observed for Nestin as after 14 days almost all cells were
negative, while after 30 days nearly all cells were positive. After
30 days, the GBM04 cell line exhibited an enrichment of cells
expressing MBP, probably indicating the capability to induce
oligodendrocyte differentiation, while all other markers were
negative. Almost 100% of G179 cells retained the expression of
CD133, βIII-tubulin and GFAP after 14 and 30 days. The longest
treatment caused a significant decrease in cells expressing Nestin;
in contrast, a strong increase in cells expressing MBP was observed
after this specific time.
 | Table IAnalysis of stemness and
differentiation marker expression in GSC lines. |
Table I
Analysis of stemness and
differentiation marker expression in GSC lines.
| Cell line | Treatment | CD133 (%) | Nestin (%) | βIII Tub (%) | GFAP (%) | MBP (%) |
|---|
| GBM2 | Untreated | 62.96 | 75.44 | 19.27 | 63.81 | 100.00 |
| 2 mM VPA 72 h | 56.48 | 53.77b | 36.45a | 64.81 | 94.96a |
| 2 mM VPA 14
days | 43.39a | 100.00c | 100.00c | 100.00c | 0.00c |
| 2 mM VPA 30
days | 100.00c | 100.00c | 100.00c | 100.00c | 93.00a |
| RPMI 10% FCS 14
days | 97.09c | 100.00c | 100.00c | 8.26c | 100.00 |
| RPMI 10% FCS 30
days | 78.38a | 68.90 | 100.00c | 5.93c | 100.00 |
| G144 | Untreated | 78.38 | 91.35 | 45.79 | 82.46 | 100.00 |
| 2 mM VPA 72 h | 67.86 | 87.85 | 13.79c | 80.90 | 96.82 |
| 2 mM VPA 14
days | 32.00c | 0.00c | 100.00c | 0.00c | 0.00c |
| 2 mM VPA 30
days | 0.00c | 0.00c | 18.00c | 32.71c | 0.00c |
| RPMI 10% FCS 14
days | 100.00c | 100.00a | 0.00c | 100.00c | 100.00 |
| RPMI 10% FCS 30
days | 100.00c | 100.00a | 100.00c | 100.00c | 100.00 |
| G166 | Untreated | 7.34 | 78.30 | 25.92 | 83.90 | 100.00 |
| 2 mM VPA 72 h | 1.92 | 53.70b | 43.52a | 81.31 | 100.00 |
| 1 mM VPA 14
days | 0.00a | 0.00c | 99.21c | 100.00c | 100.00 |
| 1 mM VPA 30
days | 0.00a | 0.00c | 0.00c | 0.00c | 0.00c |
| RPMI 10% FCS 14
days | 100.00c | 54.37b | 100.00c | 100.00c | 100.00 |
| RPMI 10% FCS 30
days | 98.30c | 79.17 | 100.00c | 100.00c | 100.00 |
| G179 | Untreated | 100.00 | 99.17 | 100.00 | 98.21 | 0.00 |
| 2 mM VPA 72 h | 96.32 | 100.00 | 100.00 | 100.00 | 0.00 |
| 2 mM VPA 14
days | 100.00 | 73.56c | 100.00 | 100.00 | 0.00 |
| 2 mM VPA 30
days | 100.00 | 0.00c | 100.00 | 100.00 | 100.00c |
| RPMI 10% FCS 14
days | 100.00 | 96.48 | 98.20 | 100.00 | 95.28c |
| RPMI 10% FCS 30
days | 100.00 | 94.07 | 100.00 | 100.00 | 100.00c |
| GliNS2 | Untreated | 100.00 | 79.63 | 71.43 | 93.64 | 100.00 |
| 2 mM VPA 72 h | 55.50c | 88.46 | 62.96 | 89.32 | 90.74b |
| 2 mM VPA 14
days | 28.15c | 0.00c | 95.93 | 0.00 | 0.00c |
| 2 mM VPA 30
days | 0.00c | 100.00c | 100.00 | 0.00 | 0.00c |
| RPMI 10% FCS 14
days | 90.48a | 100.00c | 100.00 | 100.00 | 100.00 |
| RPMI 10% FCS 30
days | 93.14a | 97.17b | 100.00 | 100.00 | 100.00 |
| GBM04 | Untreated | 86.29 | 96.58 | 76.78 | 86.08 | 0.00 |
| 2 mM VPA 72 h | 100.00c | 97.33 | 88.28c | 44.17c | 0.00 |
| 2 mM VPA 14
days | 11.29c | 0.00c | 54.20c | 100.00c | 76.03c |
| 2 mM VPA 30
days | 0.00c | 0.00c | 0.00c | 0.00c | 100.00c |
| RPMI 10% FCS 14
days | 100.00c | 100.00 | 100.00c | 100.00c | 96.36c |
| RPMI 10% FCS 30
days | 100.00c | 100.00 | 100.00c | 88.24 | 100.00c |
In our previous study (25), we showed that GSCs expressed both
stemness and differentiation markers at variable levels in
untreated and 72 h VPA-treated samples. This behavior persisted
also after VPA long-term treatments in 3 out of 6 cell lines (GBM2,
G179 and GliNS2). Notably, the others did not show immunoreactive
cells for stemness markers after 30 days of treatment.
Methylation profiles of GSCs after short-
and long-term VPA treatments
We investigated the effect of 2 mM VPA
administration for 96 h and 30 days on the CGI methylation status
of two GSC lines characterized by a different susceptibility to
this drug. In particular, the GBM2 cell line showed a reduced
viability after a 72 h VPA treatment and co-expressed stemness and
differentiation markers also after 30 days of VPA exposure, while
the G144 cell line did not display any reduction in cell viability,
but it seemed able to differentiate terminally after long-term VPA
exposure.
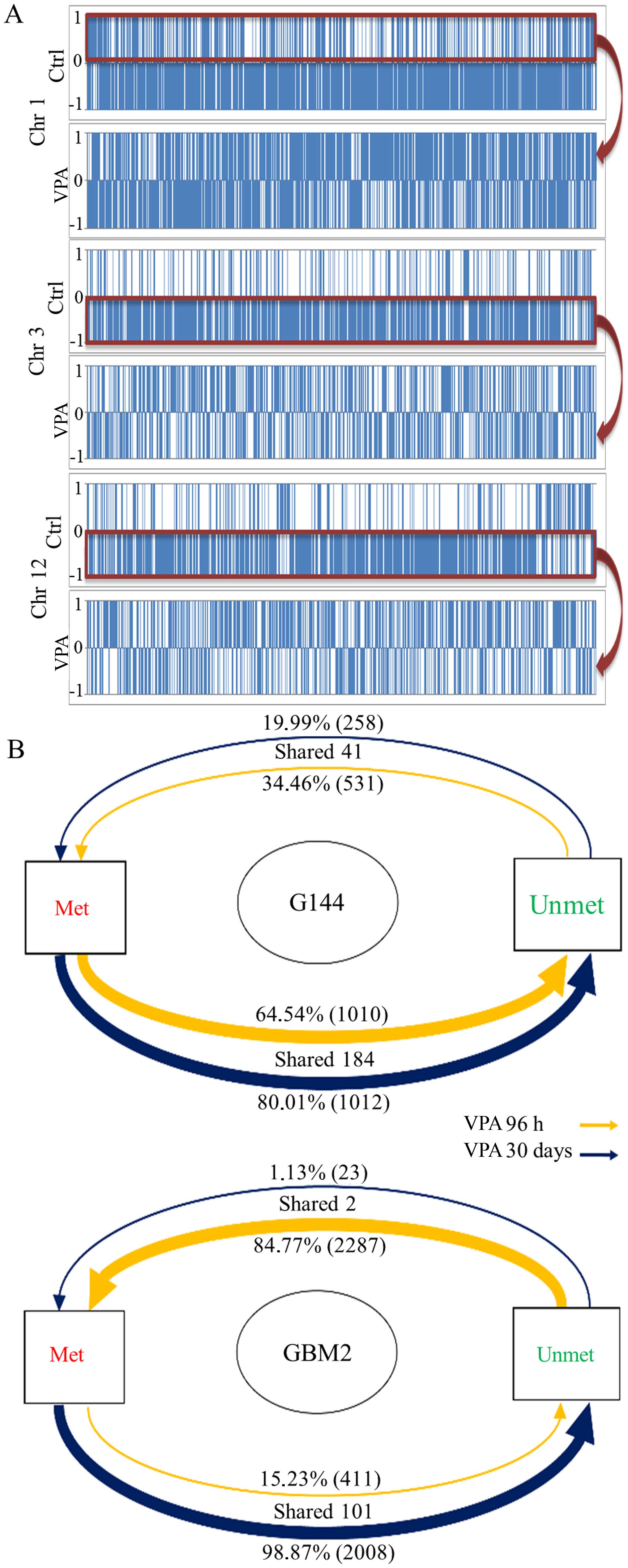
Following observation of the raw data of the three
chromosomes taken as examples (Fig.
3A), it is clear that VPA severely affected CGIs within the
gene promoters. The results of the two treatments were matched with
the respective untreated control cells at 96 h and 30 days of
culture in order to normalize the spontaneous fluctuations of the
methylation status due to in vitro expansion, and finally
the two regimens were compared to each other.
We calculated the percentage of genes that did not
change their methylation status after treatment (Table II, 'unchanged' columns), and the
percentage of genes that, instead, had a modified methylation
status after the regimen (Table
II, 'modified' columns).
 | Table IIPercentages of gene promoters with
unchanged or modified methylation status in CTRL vs. VPA 96 h, CTRL
vs. VPA 30 days and VPA 96 h vs. VPA 30 day comparisons. |
Table II
Percentages of gene promoters with
unchanged or modified methylation status in CTRL vs. VPA 96 h, CTRL
vs. VPA 30 days and VPA 96 h vs. VPA 30 day comparisons.
| Cell line | Comparisons | Unchanged
methylation status promoters (%)
| Modified
methylation status promoters (%)
|
|---|
| Methylated | Unmethylated | Total
unchanged | Met→Unmet | Unmet→Met | Total modified |
|---|
| G144 | CTRL vs. VPA 96
h | 17.96 | 64.56 | 82.52 | 11.82 | 5.66 | 17.48 |
| CTRL vs. VPA 30
days | 24.76 | 60.48 | 85.24 | 11.81 | 2.95 | 14.76 |
| VPA 96 h vs. VPA 30
days | 13.91 | 63.88 | 77.79 | 9.59 | 12.62 | 22.21 |
| GBM2 | CTRL vs. VPA 96
h | 16.93 | 54.60 | 71.53 | 5.20 | 23.27 | 28.47 |
| CTRL vs. VPA 30
days | 14.94 | 63.07 | 78.01 | 21.73 | 0.26 | 21.99 |
| VPA 96 h vs. VPA 30
days | 10.38 | 54.99 | 65.37 | 29.95 | 4.68 | 34.63 |
The two cell lines did not show substantial
differences between each other in the 'unchanged' category, in
which the class of unmethylated genes was the most highly
represented. Instead, in the 'modified' category they behaved
somewhat differently: in the GBM2 cell line a high percentage of
genes switched from an unmethylated to a methylated status after 96
h (23.27%), while a similar percentage of genes moved in the
opposite direction after 30 days (21.73%). Conversely, in the G144
cell line VPA produced a comparable change in regards to both
short- and long-term treatments, since an analogous percentage of
genes was found in both directions (11.82 and 5.66% after 96 h,
11.81 and 2.95% after 30 days). These data can also be appreciated
by comparing the two regimens with each other directly. Indeed, the
percentages of the 'modified' category were similar in the G144
cell line (9.59 and 12.62%), reflecting the homogeneity of both
short- and long-term VPA treatments (Table II, row 3). On the contrary, in the
GBM2 cell line the percentage of the 'modified' category 96 h
versus 30 days was very different (29.95 vs. 4.68%) (Table II, row 6).
In order to evaluate the methylation changes
specifically caused by VPA treatment and not by fluctuations due to
in vitro expansion, we chose the genes that showed a change
in the same direction following both treatments, thus assuming that
the changes occurred during the short-term treatment and were
maintained until the second time-point, i.e., the change was
stable. Judging from the fraction of the genes with modified
methylation status that were shared at the two treatment times, the
changes made by VPA in the G144 cell line, although less relevant,
appeared more stable and maintained than those recorded in the GBM2
cell line, where few genes retained the same methylation profile in
the two treatments (Fig. 3B).
Indeed, a large fraction of genes changed from unmethylated to
methylated status after 96 h of VPA, but after 30 days of VPA a
similar number of genes returned back from a methylated to an
unmethylated status (Fig. 3B). With
regard to the G144 cell line, we observed that the majority of
'modified' genes underwent a variation of the methylation pattern
from methylated to unmethylated status in both treatments. These
behaviors could be assessed to the different initial VPA
sensitivities of the two cell lines and to a sort of resistance
mechanism acquired during long-term VPA treatment by the GBM2 cell
line.
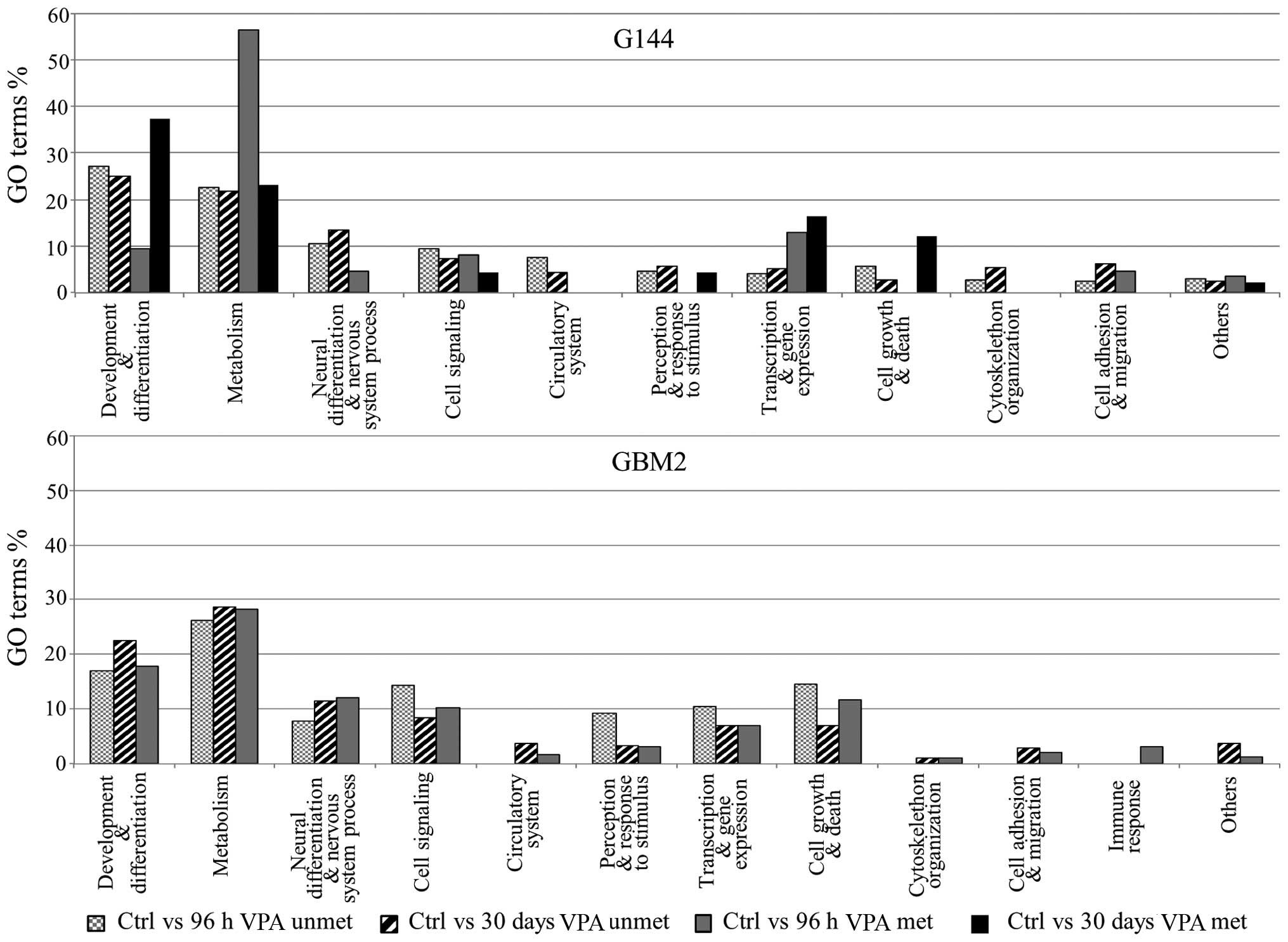
Gene Ontology analysis revealed that two functional
categories were more represented after short- and long-term VPA
treatments in both cell lines: 'development and
differentiation' and 'metabolism' (Fig. 4). Interestingly, VPA treatments
induced in both cell lines a general trend to the unmethylated
status for the 'neural differentiation and nervous system
process' category. Once again, the G144 and GBM2 cell lines
showed differences related to the effect of long-term treatment.
Indeed, in the GBM2 cell line the gene list of the 'switched from
unmethylated to methylated status' did not show any statistically
significant GO terms after long-term treatment (Fig. 4).
Finally, we investigated whether the short- and
long-term treatments influence the methylation status of specific
genes involved in critical signaling pathways responsible for GBM
initiation, migration and invasion, such as WNT, SHH, PDGFR and
EGFR pathways (29,30). First of all, following analysis of
the methylation status of several critical genes in the two
untreated cell lines, we observed, for example in the 96 h
controls, that they showed several homologies such as the
unmethylated status of many tumor-suppressor genes (TSC1, NF2,
SMAD4 and MEN1, EXT1, GPC3 and RB1), but for other genes the
results were discrepant. For example, PTEN and E-cadherin (CDH1)
were methylated in the GBM2 cell line and unmethylated in the G144
cell line, while CTNNB1, several WNT ligands and RUNX1 were
methylated in G144 and unmethylated in GBM2 (Table III).
 | Table IIIChanges in the methylation status of
genes involved in several glioma-related pathways, in neural
differentiation and in stemness maintenance after short- and
long-term VPA treatment. |
Table III
Changes in the methylation status of
genes involved in several glioma-related pathways, in neural
differentiation and in stemness maintenance after short- and
long-term VPA treatment.
| Pathway/genes | G144
| GBM2
|
|---|
| 96 h Ctrl→VPA | 30 days
Ctrl→VPA | 96 h Ctrl→VPA | 30 days
Ctrl→VPA |
|---|
| Receptor tyrosine
kinase pathway |
| ALK | U | U | U | U |
| EGFR | U | M→U | ND→U | U |
| PDGFRL | U | U | U | U |
| PDGFC | U→ND | M→U | U | U |
| PDGFD | M→U | U | U →M | M→U |
| PDGFB | U | M→U | U | M→U |
| SHC1 | U | U | U | U |
| SHC2 | M→U | M | U →ND | M→U |
| SHC3 | U | U | U | U |
| SHC4 | U | U | U | U |
| GRB2 | U | U | U | U |
| SOS1 | U | U | U | U |
| SOS2 | U | U | U | U |
| RAS | U | M→U | U | U |
| RAF | U | U | U | M→U |
| MAP2K1 | U | U→M | U | U |
| MAP2K2 | U | M→ND | U | U |
| MAP2K3 | U | U | U | U |
| MAP2K4 | U | U | U | U |
| MAP2K5 | U | U | U | U |
| MAP2K7 | U | U | U | M→U |
| MAPK1 | M→U | U | U | U |
| NF1 | U | M→U | U | U |
| ABL1 | U | U | U | U |
| ABL2 | U | U→M | U | U |
| FES | U | M | U→M | M |
| JAK1 | U | U | U | U |
| JAK2 | U | U | ND→U | M→U |
| JAK3 | M→U | M | M | M |
| STAT1 | U | U | U | U |
| STAT2 | U→M U | | U | U |
| STAT3 | U | U | U | U |
| RB pathway |
| CDKN2A | U | U | ND→M | M |
| CDK6 | U | U | U | U |
| CCND1 | M→ND | M | ND→M | M→ND |
| RB1 | U | U | U | U |
| E2F1 | U | U | U | U |
| E2F2 | U | U | U | U |
| E2F3 | U | U | U | U |
| E2F5 | U→ND | U | U | U |
| E2F6 | U→M | U | U | U |
| E2F7 | U | U | U | U |
| E2F8 | U | M | U→M | M |
| TFE3 | U | U | U | U |
| p53 pathway |
| TP53BP2 | U | U | U | U |
| p53 pathway |
| TP53I3 | U | U→M | U | M→U |
| TP53INP1 | M | M | M | M→ND |
| TP53I11 | U | M | U→M | M→U |
| TP53INP2 | U | U | M→U | M→U |
| MDM2 | M→ND | M | M→U | U |
| NOTCH1 | U | U | U | U |
| PUMA | M→U | U →M | U | M→U |
| NOXA | M | M | U | U |
| BCL2 | U | M→U | ND | M→U |
| BAX | U | M→U U | | U |
| P21 | U | U | U | U |
| CDK6 | U | U | U | U |
| CDK7 | U | U | U | U |
| CDK8 | M→U | U | U | U |
| CDK10 | U | U | U | U |
| CCND1 | M→ND | M | ND→M | M→ND |
| MSH2 | U | M→U | M→U | M→U |
| BRCA1 | U | U | M | M |
| PI3K/AKT
pathway |
| PDK2 | U→ND | U | U | U |
| PTEN | U | U | M | M |
| PIK3R1 | U | U | U→M | M |
| PIK3R2 | U→M | U | U | U |
| PIK3R3 | U | M→U | U | U |
| PIK3R4 | U | U | ND→U U | |
| PIK3CA | M→U | U | U | U |
| PIK3C2B | U | U | U | U |
| PIK3CD | U | U | U | U |
| AKT1 | U | U | U→M | M→U |
| AKT3 | U | U | U | U |
| BCL2 | U | M→U | ND | M→U |
| FOXO1 | U | U | U | U |
| FOXO3 | U | U→M U | | U |
| FOXO4 | U | U | U | M→U |
| TSC1 | U | M→U U | | U |
| RHEB | U | M→U U | | U |
| MTOR | U | U | U | U |
| 4EBP1 | U→M | U | U →ND | M→U |
| RAC3 | U | M→U | ND→U | U |
| RAC1 | M→U | M→U | U | U |
| NF2 | U | U | U | U |
| SHH pathway |
| SHH | U | U | U | U |
| EXT1 | U | U | U | U |
| PTCH1 | U | U | U | U |
| PTCH2 | M | M | M→U | M→U |
| SMO | M→U | M→U | U | M→U |
| GLI1 | U | U | U | U |
| GLI3 | U | U | U | U |
| GLI4 | U→M | U | U | U |
| WNT pathway |
| GPC3 | U→ND | M→U | U | U |
| WNT2B | U→ND | M | U | ND→U |
| WNT9A | M | M | U | U |
| WNT3A | M | M | U→M | M |
| WNT6 | M→U | M→U | U | M→U |
| WNT10A | M→U | M U | →M | M |
| WNT7A | U | U | U | U |
| WNT5A | U | U | U | U |
| WNT2 | U | M→U | U | M→U |
| WNT11 | M→U | M→U | U | M→U |
| WNT3 | M | M | M | M→ND |
| WNT9B | M | M→U | U | U |
| FZD7 | U | M→U | U | U |
| FZD5 | U | U | U | U |
| FZD9 | U | M→U | M | M |
| FZD1 | U | U | U | U |
| FZD6 | U | M→U | ND→M | M |
| FZD8 | U | U | U | U |
| FZD4 | U | ND→M | U→M | M→ND |
| FZD10 | M | M | M | M |
| FZD2 | U | U | M→U | M→U |
| LRP6 | U | U | U | U |
| GSK3β | U | U | U | U |
| β-catenin | M→U | ND→U | U | U |
| APC2 | M→U | M→U | ND | M→U |
| AXIN1 | M | M | M | M |
| AXIN2 | U | U | U | U |
| CDH1 | U | M | M | M→ND |
| α-catenin | U | ND→M | U | M→U |
| ACTB | U | U | U | U |
| ACTG1 | M | M | ND→U | U |
| ACTA1 | M→U | M | U | M→U |
| TCF7 | M | M | M | M |
| TCF12 | M | M | M | M |
| TCF25 | M→U | M→U | U | M→U |
| TCF4 | U | U | U | U |
| TCF15 | U | M | U | M→U |
| C-MYC | U | U | U | U |
| BMP4 | U | M | U→M | M→U |
| BMP/TGFβ
pathway |
| BMP2 | U | U | U | U |
| BMP3 | U | M | U | M→U |
| BMP4 | U | M | U→M | M→U |
| BMP6 | U→ND | U | U | U |
| BMP7 | M→U | M→U | ND→M | M→ND |
| BMP8A | M | M | M | M→ND |
| BMP8B | M→U | M | ND→U | M→U |
| TGFβ1 | M | M | U →M | M→ND |
| TGFβ2 | M | M→U | U | U |
| TGFβR1 | U | M→U | ND→U | M→U |
| BMP/TGFβ
pathway |
| TGFβR2 | U | U | M→U | U |
| SMAD2 | U | U | U | M→U |
| SMAD3 | U | U | U | U |
| SMAD4 | U | U | U | U |
| RUNX1 | M→U | U | U→M | M→U |
| MEN1 | U →M | M | U→M | U |
| Neural
differentiation |
| BMPR1B | U | U | U→M | U |
| TUBB3 | U | U | U | U |
| MAP2 | M | U | U→M | U |
| OLIG1 | U | M→U | U | U |
| OLIG2 | M→U | M | M | M |
| OLIG3 | M→ND | M | M | M→U |
| Stemness
maintenance |
| OCT4 | M | M | M→U | M |
| NOTCH1 | U | U | U→M | U |
| CD44 | U | ND→U | U→M | ND→U |
| PROM1 | U | U | U→M | U |
| NES | U | U | U | M→U |
| MGMT | M | M | U→M | U |
In the second instance, we considered the
methylation status of the same genes after the drug treatments. The
general effect attributable to VPA in both cell lines was towards
an unmethylated status of the genes in all pathways considered.
Furthermore, a greater number of genes showed this switch (from
methylated to unmethylated status) after 30 days of VPA treatment,
compared to 96 h (Table III).
VPA resistance in VPA long-term treated
cells
To confirm the hypothesis of the acquisition of a
resistant phenotype after VPA long-term treatment by the GBM2 cell
line, we performed analysis of the cell viability by MTT assay on
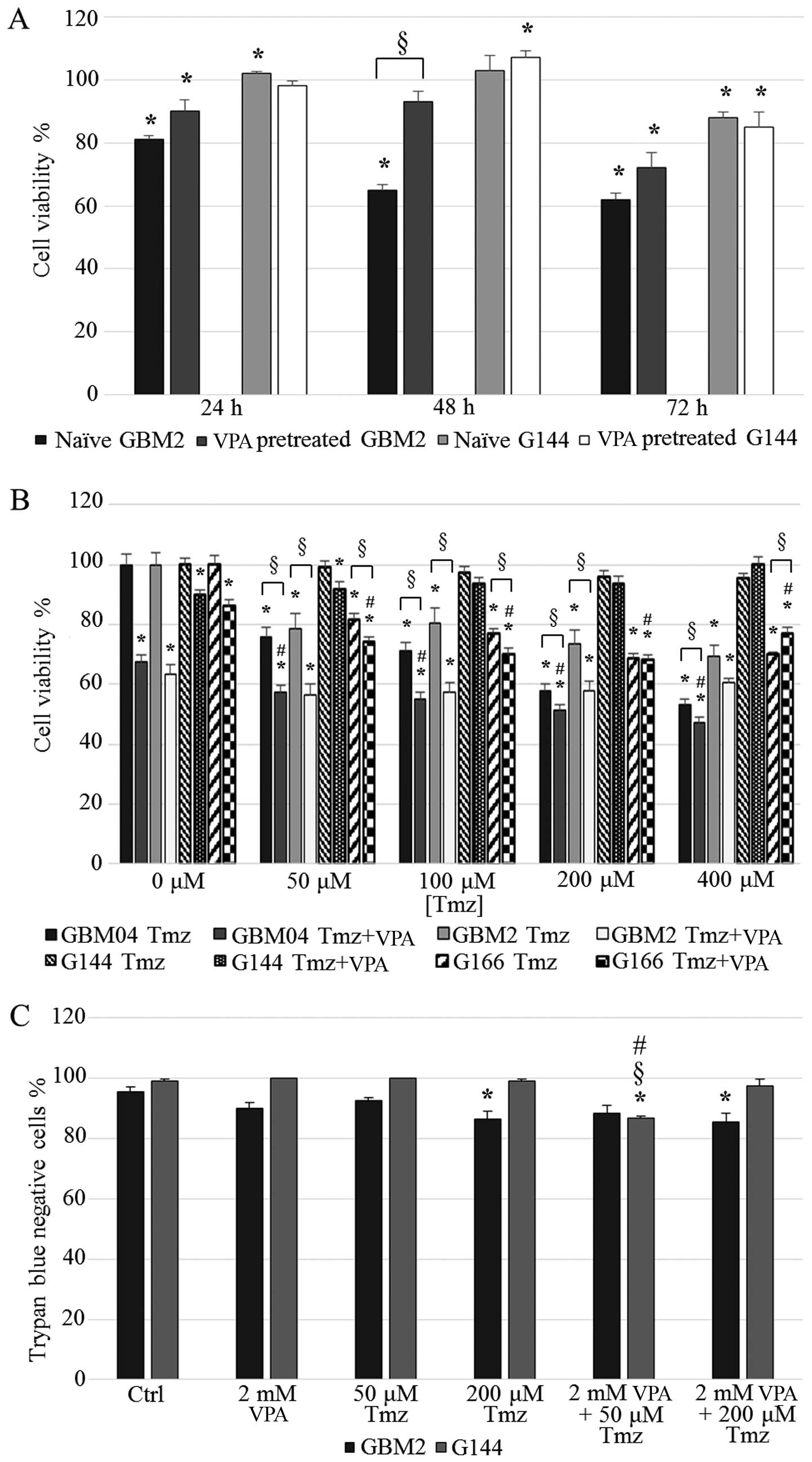
naïve and VPA-pretreated G144 and GBM2 cells.
The results showed that G144 cell viability after
VPA treatment was generally not modified by the pretreatment for 30
days, as expected. In fact, this cell line was already resistant,
as confirmed by the low VPA sensitivity of naïve cells (Fig. 5A).
With regard to the GBM2 cell line, we already
observed a consistent cell viability reduction in the naïve cells,
especially at 48 and 72 h of treatment (65 and 62%, respectively).
VPA long-term pretreated GBM2 cells showed a reduced sensitivity to
the drug when compared to the naïve GBM2 cells at all the
time-points tested; the induction of a resistant phenotype in the
GBM2 cell line, probably due to the long-term in vitro
exposure to 2 mM VPA, was particularly evident at 48 h of treatment
in which the cell viability of the 30 day-exposed cells reached 92%
(P<0.0001) (Fig. 5A).
VPA treatment failed to sensitize GSCs to
TMZ
In GBM, first-line treatment includes radical
surgical resection followed by concurrent radiotherapy and
chemotherapy, typically temozolomide (TMZ) (31). TMZ exerts cytotoxicity against GBM
cells by creating O6-methylguanine lesions, which leads
to DNA fragmentation (32). The
response to TMZ is favorably affected by the promoter methylation
of the methylguanine-DNA methyltransferase (MGMT) gene. This enzyme
removes, if expressed, the methyl groups added by TMZ, thereby
preventing GBM cell death (33). In
this study we evidenced that, in GBM2 cells, the MGMT promoter
switched from an unmethylated to a methylated status after 96 h of
VPA treatment (Table III). To
verify whether VPA treatment was able to increase TMZ efficacy by
virtue of this change in MGMT promoter methylation, we analyzed
cell viability after 2 mM VPA (96 h) plus several doses of TMZ (48
h). Unexpectedly, the results showed that VPA exposure did not
produced any relevant increase in TMZ efficacy in the GBM2 cell
line. In fact, cell viability was reduced at all drug combinations
in a statistically significant manner when compared to TMZ only
treatments (except for VPA plus 400 µM TMZ), but no
statistically significant difference was observed with the VPA
single treatment (P>0.05) (Fig.
5B). Furthermore, CI values for the combined treatments clearly
indicated the presence of antagonism between the two compounds
(CI>1; Table IV).
 | Table IVCooperative index (CI) of VPA + TMZ
combined treatments (MTT assay). |
Table IV
Cooperative index (CI) of VPA + TMZ
combined treatments (MTT assay).
| Cooperative
index | VPA 2 mM
|
|---|
| GBM2 | G144 | G166 | GBM04 |
|---|
| TMZ 50
µM | 1.33 | 1.35 | 1.25 | 1.33 |
| TMZ 100
µM | 1.31 | 2.01 | 1.22 | 1.37 |
| TMZ 200
µM | 1.50 | 2.21 | 1.43 | 1.54 |
| TMZ 400
µM | 1.71 | 445.80 | 1.90 | 1.51 |
The MGMT promoter in the G144 cell line was
methylated and this status was not altered by VPA; this cell line
could be virtually sensitive to TMZ. Surprisingly, the G144 cells
were resistant to TMZ and this issue was not modified by VPA
treatment, suggesting the presence of a complex pattern of drug
resistance mechanisms (CI>1; Table
IV) (Fig. 5B).
Considering the heterogeneity of GBM, we extended
this analysis to two additional cell lines (GBM04 and G166). In
both lines, all drug combinations displayed a statistically
significant reduction in cell viability compared to the single
treatments (except for VPA plus 200 µM TMZ in the G166 cell
line), but the CI values clearly indicated that there was no
synergism between the two drugs (CI>1; Table IV).
Moreover, we analyzed the percentages of live and
dead cells by trypan blue dye-exclusion assay in the GBM2 and G144
cell lines treated with only VPA or TMZ, and VPA plus TMZ. The
percentage of live GBM2 cells after the combined treatments was
slightly reduced with respect to the single treatments and the
differences were not statistically significant. With regard to the
percentage of live G144 cells after the combined treatments, we
observed a weak decrease only in the 2 mM VPA plus 50 µM TMZ
combination compared to the control culture and single treatments
(13.4%) (Fig. 5C).
Discussion
VPA is an approved drug for the treatment of
epileptic seizures, and is effective for bipolar disorders and
migraine. VPA is also an HDACi (34,35)
that has shown potent antitumor effects in vitro and in
vivo in a variety of cancers, including GBM (36,37).
This drug is a chromatin remodeling agent, consequently able to
strongly modify gene expression (38–40),
inducing cancer cell differentiation, apoptosis and growth arrest.
We previously demonstrated that short-term VPA treatment is able to
modulate the cancer stem-like properties of GSCs by its
pro-differentiating ability, overcoming their perpetual stem cell
state. In particular, VPA induced a sort of initial differentiation
process and a sharp growth arrest (25).
The risk of GBM patients to experience epileptic
seizures is in the range of 30–50% (41) and the majority are treated with VPA
(42–44). For this reason, it is interesting to
evaluate in vitro the potential effects of long-term VPA
administration on GSCs, in order to also consider the application
of this drug for anticancer purposes.
We firstly investigated the effects of long-term VPA
treatment on cell morphology. As expected, the VPA
pro-differentiating power was more marked after long-term
treatment, with respect to short-term administration. Five out of
seven GSC lines showed consistent morphological changes after 14
and 30 days (from rolling spheres to attached star-shaped cells
with neurite-like structures), while in the G166 and GBM7 cell
lines a cytotoxic effect or apoptosis prevailed following terminal
differentiation. Overall, these results demonstrated that VPA
induced several morphological variations, as previously described
in other tumors (45): we
speculated that the HDAC inhibitory action of VPA could trigger
reactivation of genes involved in differentiation, usually silenced
in cancer stem cells (12,13). In fact, for example, after 30 days
of treatment, we identified a potentially reactivating methylation
change of OLIG1 and OLIG3 promoters, which are involved in
oligodendrocyte and neuronal differentiation (46,47),
from the methylated to the unmethylated status in G144 and GBM2
cells, respectively (Table
III).
The expression analysis of stemness and
differentiation markers highlighted the variability of VPA
pro-differentiating ability among the GSC lines; this issue is
representative of GBM heterogeneity. In our previous study, the
immunofluorescence analysis, performed after 2 mM VPA for 72 h,
revealed the co-expression of stemness and differentiation markers
(25). Therefore, in the present
study, we investigated the expression of such markers after 14 and
30 days of treatment to verify the real capability of GSCs to
terminally differentiate in the consequence of a long
pro-differentiating stimulus. In particular, the G144 and GBM04
cell lines after 30 days expressed only one differentiation marker,
GFAP or MBP respectively, suggesting peculiar differentiation
behaviors towards the astrocytic and oligodendrocytic lineages.
Conversely, the G166 cell line had a particular response; after 14
days it showed a high percentage of cells positive for the
differentiation markers, while after 30 days, these cells did not
express any one marker. We thus speculated that the G166 cell line
is subjected to a transdifferentiation process (48). The analysis also revealed that 3 out
of 6 cell lines maintained the co-expression of stemness and
differentiation markers (GBM2, G179 and GliNS2), suggesting an
impairment of any gene involved in the regulation of
differentiation. Alternatively, resistance mechanisms to VPA may
arise in these cell lines and VPA-resistant cells selectively grow
during drug treatment (49,50).
In the second instance, we analyzed the DNA
methylation profiles of the GBM2 and G144 cell lines after 96 h and
30 days of VPA treatment to study and compare the epigenetic
impacts of the two pharmacological regimens. Specifically, we
selected two cell lines showing deep differences in VPA
responsiveness in order to verify whether these features were
consequent to a drug-specific activity on chromatin remodeling.
Overall, VPA strongly modified the DNA methylation pattern of these
cell lines. Our data confirmed the described VPA property to modify
the methyloma (51,52); it determines not only alterations in
histone acetylation, but also, indirectly, changes in DNA
methylation.
After 96 h of VPA treatment, the percentages of
genes that modified their methylation status in the two cell lines
could directly reflect their initial VPA sensitivity: we observed
that 28.47% of genes had an altered methylation status in the GBM2
cell line, which is VPA sensitive, while few genes (17.48%) were
altered in the G144 cells, a resistant cell line. Furthermore, the
different VPA susceptibilities of the two cell lines were also
evidenced by the different trends in the methylation change after
short-term treatment. In fact, in GBM2 cells, a high percentage of
genes switched from an unmethylated to a methylated status
(23.27%), while the majority of 'modified' genes moved in the
opposite direction compared with the G144 cells (11.82%). The
latter also maintained this issue after the long-term regimen. In
contrast, the GBM2 modifying trend was completely revolutionized
and became similar to the G144 behavior. The long-term exposure
probably promotes in the GBM2 cell line the acquisition of
resistance and the clonal expansion of resistant cells. This
hypothesis was confirmed by the MTT assay in the GBMS cells
folloiwng a 30-day VPA pretreatment, which showed a reduced
sensitivity to 2 mM VPA compared to the naïve cells.
Moreover, we observed by means of GO analysis that
the 'development and differentiation' and 'neural
differentiation and nervous system process' functional
categories were the most represented ones, together with
'metabolism', after VPA treatments in both cell lines. These
data further confirmed the VPA pro-differentiating ability on the
GSC lines.
Nevertheless, it is important to explore the
function and the role of genes influenced by these changes in
regards to the methylation status after VPA treatment. Thus, we
decided to investigate the methylation status of genes involved in
critical signaling pathways important in GBM, such as MGMT, before
and after short- and long-term treatments. MGMT promoter
methylation is currently considered a predictive biomarker for the
responsiveness to chemotherapy with alkylating agents such as TMZ;
it is associated with an increased survival of GBM patients treated
with TMZ (33,53). In the present study, we found that
VPA treatment for 96 h induced a methylation shift in the MGMT
promoter only in the GBM2 cell line, from an unmethylated to a
methylated status, while the long-term treatment did not modify the
status. Conversely, G144 cells did not show any modification of
MGMT promoter methylation after both regimens (methylated).
In consequence of these data, we finally verified
whether a short-term VPA pretreatment is able to sensitize GSCs to
TMZ. Here, we used VPA pretreatment, as a chemo-sensitizer, to
augment TMZ sensitivity of the GBM2, G144, GBM04 and G166 cell
lines. Unexpectedly, VPA did not produce any relevant increase in
the TMZ efficacy in all cell lines in terms of metabolic activity
and percentage of live cells. These data clearly indicate that the
DNA methylation status of the MGMT promoter and its epigenetic
switch induced by VPA are not the only determinant factors to
predict and induce TMZ chemosensitivity of GSCs. A complex and
extensive chemo-resistance landscape characterizes these cancer
cells and other molecular mechanisms, such as the overexpression of
p-glycoprotein (54), are involved
in TMZ resistance (55–58).
The development of new therapeutic approaches that
selectively target GSCs is mandatory to defeat GBM. Specifically,
differentiation-inducing therapies are promising treatments to
affect GSC self-renewal ability. Furthermore, restoring the
physiological epigenetic pattern could be a useful strategy to
fight GBM chemoresistance, yet it is unable to be the unique and
definitive resolution to overcome this devastating feature.
Short-term VPA treatment could combine in a single pharmacological
regimen both of these two therapeutic strategies. Unfortunately,
long-term VPA treatment induced the development of a resistant
phenotype and, although the therapeutic properties and side effects
of VPA are known since the 1970s, the question of whether a
long-term antineoplastic treatment with this drug may be harmful
for patients has never been exhaustively answered (59).
However, the overall data obtained warrant further
investigation into the VPA effects on this fatal tumor in order to
benefit patient prognosis. Additional in vitro and in
vivo studies are necessary to clarify various key aspects of
this pharmacological approach.
Acknowledgments
The authors gratefully acknowledge Professor Austin
Smith (University of Cambridge, Cambridge, UK), ISE (Integrated
Systems Engineering) and Dr Antonio Daga (IRCCS-AOU San
Martino-IST, Genova, Italy) for kindly providing us with the cell
lines used in this study. This study was supported by FAR 2011
n.12-1-62 and n.12-155 from the University of Milano-Bicocca (to
L.D. and A.B., respectively) and by FIRB project from the Ministry
of University and Scientific Research (to M.L.).
Abbreviations:
|
GBM
|
glioblastoma multiforme
|
|
GSCs
|
glioma stem cells
|
|
VPA
|
valproic acid
|
|
HDACi
|
histone deacetylase inhibitor
|
|
TMZ
|
temozolomide
|
References
|
1
|
Louis DN, Ohgaki H, Wiestler OD, Cavenee
WK, Burger PC, Jouvet A, Scheithauer BW and Kleihues P: The 2007
WHO classification of tumours of the central nervous system. Acta
Neuropathol. 114:97–109. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Brandes AA, Franceschi E, Ermani M, Tosoni
A, Albani F, Depenni R, Faedi M, Pisanello A, Crisi G, Urbini B, et
al: Pattern of care and effectiveness of treatment for glioblastoma
patients in the real world: Results from a prospective
population-based registry. Could survival differ in a high-volume
center? Neurooncol Pract. 1:166–171. 2014.
|
|
3
|
Nduom EK, Hadjipanayis CG and Van Meir EG:
Glioblastoma cancer stem-like cells: Implications for pathogenesis
and treatment. Cancer J. 18:100–106. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Sottoriva A, Spiteri I, Piccirillo SG,
Touloumis A, Collins VP, Marioni JC, Curtis C, Watts C and Tavaré
S: Intratumor heterogeneity in human glioblastoma reflects cancer
evolutionary dynamics. Proc Natl Acad Sci USA. 110:4009–4014. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Piccirillo SG, Combi R, Cajola L, Patrizi
A, Redaelli S, Bentivegna A, Baronchelli S, Maira G, Pollo B,
Mangiola A, et al: Distinct pools of cancer stem-like cells coexist
within human glioblastomas and display different tumorigenicity and
independent genomic evolution. Oncogene. 28:1807–1811. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Esteller M: Epigenetics in cancer. N Engl
J Med. 358:1148–1159. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hatziapostolou M and Iliopoulos D:
Epigenetic aberrations during oncogenesis. Cell Mol Life Sci.
68:1681–1702. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Nagarajan RP and Costello JF: Molecular
epigenetics and genetics in neuro-oncology. Neurotherapeutics.
6:436–446. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Khan O and La Thangue NB: HDAC inhibitors
in cancer biology: Emerging mechanisms and clinical applications.
Immunol Cell Biol. 90:85–94. 2012. View Article : Google Scholar
|
|
10
|
Song SH, Han SW and Bang YJ:
Epigenetic-based therapies in cancer: Progress to date. Drugs.
71:2391–2403. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Cinatl J Jr, Cinatl J, Scholz M, Driever
PH, Henrich D, Kabickova H, Vogel JU, Doerr HW and Kornhuber B:
Antitumor activity of sodium valproate in cultures of human
neuroblastoma cells. Anticancer Drugs. 7:766–773. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Blaheta RA, Michaelis M, Driever PH and
Cinatl J Jr: Evolving anticancer drug valproic acid: Insights into
the mechanism and clinical studies. Med Res Rev. 25:383–397. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Van Lint C, Emiliani S and Verdin E: The
expression of a small fraction of cellular genes is changed in
response to histone hyperacetylation. Gene Expr. 5:245–253.
1996.PubMed/NCBI
|
|
14
|
Chateauvieux S, Morceau F, Dicato M and
Diederich M: Molecular and therapeutic potential and toxicity of
valproic acid. J Biomed Biotechnol. 2010:4793642010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Bacon CL, O'Driscoll E and Regan CM:
Valproic acid suppresses G1 phase-dependent sialylation of a 65 kDa
glycoprotein in the C6 glioma cell cycle. Int J Dev Neurosci.
15:777–784. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Knüpfer MM, Hernáiz-Driever P, Poppenborg
H, Wolff JE and Cinatl J: Valproic acid inhibits proliferation and
changes expression of CD44 and CD56 of malignant glioma cells in
vitro. Anticancer Res. 18:3585–3589. 1998.PubMed/NCBI
|
|
17
|
Chavez-Blanco A, Perez-Plasencia C,
Perez-Cardenas E, Carrasco-Legleu C, Rangel-Lopez E, Segura-Pacheco
B, Taja-Chayeb L, Trejo-Becerril C, Gonzalez-Fierro A, Candelaria
M, et al: Antineoplastic effects of the DNA methylation inhibitor
hydralazine and the histone deacetylase inhibitor valproic acid in
cancer cell lines. Cancer Cell Int. 6:22006. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Das CM, Aguilera D, Vasquez H, Prasad P,
Zhang M, Wolff JE and Gopalakrishnan V: Valproic acid induces p21
and topoisomerase-II (alpha/beta) expression and synergistically
enhances etoposide cytotoxicity in human glioblastoma cell lines. J
Neurooncol. 85:159–170. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Van Nifterik KA, Van den Berg J, Slotman
BJ, Lafleur MV, Sminia P and Stalpers LJ: Valproic acid sensitizes
human glioma cells for temozolomide and γ-radiation. J Neurooncol.
107:61–67. 2012. View Article : Google Scholar
|
|
20
|
Ryu CH, Yoon WS, Park KY, Kim SM, Lim JY,
Woo JS, Jeong CH, Hou Y and Jeun SS: Valproic acid downregulates
the expression of MGMT and sensitizes temozolomide-resistant glioma
cells. J Biomed Biotechnol. 2012:9874952012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chen CH, Chang YJ, Ku MS, Chung KT and
Yang JT: Enhancement of temozolomide-induced apoptosis by valproic
acid in human glioma cell lines through redox regulation. J Mol Med
Berl. 89:303–315. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Pollard SM, Yoshikawa K, Clarke ID, Danovi
D, Stricker S, Russell R, Bayani J, Head R, Lee M, Bernstein M, et
al: Glioma stem cell lines expanded in adherent culture have
tumor-specific phenotypes and are suitable for chemical and genetic
screens. Cell Stem Cell. 4:568–580. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Griffero F, Daga A, Marubbi D, Capra MC,
Melotti A, Pattarozzi A, Gatti M, Bajetto A, Porcile C, Barbieri F,
et al: Different response of human glioma tumor-initiating cells to
epidermal growth factor receptor kinase inhibitors. J Biol Chem.
284:7138–7148. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Baronchelli S, Bentivegna A, Redaelli S,
Riva G, Butta V, Paoletta L, Isimbaldi G, Miozzo M, Tabano S, Daga
A, et al: Delineating the cytogenomic and epigenomic landscapes of
glioma stem cell lines. PLoS One. 8:e574622013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Riva G, Baronchelli S, Paoletta L, Butta
V, Biunno I, Lavitrano M, Dalprà L and Bentivegna A: In vitro
anticancer drug test: A new method emerges from the model of glioma
stem cells. Toxicol Rep. 1:188–199. 2014. View Article : Google Scholar
|
|
26
|
Aouali N, Palissot V, El-Khoury V, Moussay
E, Janji B, Pierson S, Brons NH, Kellner L, Bosseler M, Van Moer K,
et al: Peroxisome proliferator-activated receptor gamma agonists
potentiate the cytotoxic effect of valproic acid in multiple
myeloma cells. Br J Haematol. 147:662–671. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Straussman R, Nejman D, Roberts D,
Steinfeld I, Blum B, Benvenisty N, Simon I, Yakhini Z and Cedar H:
Developmental programming of CpG island methylation profiles in the
human genome. Nat Struct Mol Biol. 16:564–571. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Beissbarth T and Speed TP: GOstat: Find
statistically overrepresented Gene Ontologies within a group of
genes. Bioinformatics. 20:1464–1465. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kanu OO, Hughes B, Di C, Lin N, Fu J,
Bigner DD, Yan H and Adamson C: Glioblastoma multiforme
oncogenomics and signaling pathways. Clin Med Oncol. 3:39–52.
2009.PubMed/NCBI
|
|
30
|
Wardak Z and Choe KS: Molecular pathways
and potential therapeutic targets in glioblastoma multiforme.
Expert Rev Anticancer Ther. 13:1307–1318. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Stupp R, Mason WP, van den Bent MJ, Weller
M, Fisher B, Taphoorn MJ, Belanger K, Brandes AA, Marosi C, Bogdahn
U, et al European Organisation for Research and Treatment of Cancer
Brain Tumor and Radiotherapy Groups; National Cancer Institute of
Canada Clinical Trials Group: Radiotherapy plus concomitant and
adjuvant temozolomide for glioblastoma. N Engl J Med. 352:987–996.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhang J, Stevens MF and Bradshaw TD:
Temozolomide: Mechanisms of action, repair and resistance. Curr Mol
Pharmacol. 5:102–114. 2012. View Article : Google Scholar
|
|
33
|
Hegi ME, Diserens AC, Gorlia T, Hamou MF,
de Tribolet N, Weller M, Kros JM, Hainfellner JA, Mason W, Mariani
L, et al: MGMT gene silencing and benefit from temozolomide in
glioblastoma. N Engl J Med. 352:997–1003. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Göttlicher M, Minucci S, Zhu P, Krämer OH,
Schimpf A, Giavara S, Sleeman JP, Lo Coco F, Nervi C, Pelicci PG,
et al: Valproic acid defines a novel class of HDAC inhibitors
inducing differentiation of transformed cells. EMBO J.
20:6969–6978. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Phiel CJ, Zhang F, Huang EY, Guenther MG,
Lazar MA and Klein PS: Histone deacetylase is a direct target of
valproic acid, a potent anticonvulsant, mood stabilizer, and
teratogen. J Biol Chem. 276:36734–36741. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Osuka S, Takano S, Watanabe S, Ishikawa E,
Yamamoto T and Matsumura A: Valproic acid inhibits angiogenesis in
vitro and glioma angiogenesis in vivo in the brain. Neurol Med Chir
(Tokyo). 52:186–193. 2012. View Article : Google Scholar
|
|
37
|
Knüpfer MM, Pulzer F, Schindler I, Hernaíz
Driever P, Knüpfer H and Keller E: Different effects of valproic
acid on proliferation and migration of malignant glioma cells in
vitro. Anticancer Res. 21:347–351. 2001.PubMed/NCBI
|
|
38
|
Cameron EE, Bachman KE, Myöhänen S, Herman
JG and Baylin SB: Synergy of demethylation and histone deacetylase
inhibition in the re-expression of genes silenced in cancer. Nat
Genet. 21:103–107. 1999. View
Article : Google Scholar : PubMed/NCBI
|
|
39
|
De la Cruz-Hernández E, Perez-Plasencia C,
Pérez-Cardenas E, Gonzalez-Fierro A, Trejo-Becerril C,
Chávez-Blanco A, Taja-Chayeb L, Vidal S, Gutiérrez O, Dominguez GI,
et al: Transcriptional changes induced by epigenetic therapy with
hydralazine and magnesium valproate in cervical carcinoma. Oncol
Rep. 25:399–407. 2011.
|
|
40
|
Suzuki H, Gabrielson E, Chen W, Anbazhagan
R, van Engeland M, Weijenberg MP, Herman JG and Baylin SB: A
genomic screen for genes upregulated by demethylation and histone
deacetylase inhibition in human colorectal cancer. Nat Genet.
31:141–149. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
41
|
van Breemen MS, Wilms EB and Vecht CJ:
Epilepsy in patients with brain tumours: Epidemiology, mechanisms,
and management. Lancet Neurol. 6:421–430. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Sizoo EM, Koekkoek JA, Postma TJ, Heimans
JJ, Pasman HR, Deliens L, Taphoorn MJ and Reijneveld JC: Seizures
in patients with high-grade glioma: A serious challenge in the
end-of-life phase. BMJ Support Palliat Care. 4:77–80. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Guthrie GD and Eljamel S: Impact of
particular antiepileptic drugs on the survival of patients with
glioblastoma multiforme. J Neurosurg. 118:859–865. 2013. View Article : Google Scholar
|
|
44
|
Vecht CJ, Kerkhof M and Duran-Pena A:
Seizure prognosis in brain tumors: New insights and evidence-based
management. Oncologist. 19:751–759. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Duenas-Gonzalez A, Candelaria M,
Perez-Plascencia C, Perez-Cardenas E, de la Cruz-Hernandez E and
Herrera LA: Valproic acid as epigenetic cancer drug: Preclinical,
clinical and transcriptional effects on solid tumors. Cancer Treat
Rev. 34:206–222. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Dai J, Bercury KK, Ahrendsen JT and
Macklin WB: Olig1 function is required for oligodendrocyte
differentiation in the mouse brain. J Neurosci. 35:4386–4402. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Liu Z, Li H, Hu X, Yu L, Liu H, Han R,
Colella R, Mower GD, Chen Y and Qiu M: Control of precerebellar
neuron development by Olig3 bHLH transcription factor. J Neurosci.
28:10124–10133. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Soda Y, Marumoto T, Friedmann-Morvinski D,
Soda M, Liu F, Michiue H, Pastorino S, Yang M, Hoffman RM, Kesari
S, et al: Transdifferentiation of glioblastoma cells into vascular
endothelial cells. Proc Natl Acad Sci USA. 108:4274–4280. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Liu T, Liu PY, Tee AE, Haber M, Norris MD,
Gleave ME and Marshall GM: Over-expression of clusterin is a
resistance factor to the anti-cancer effect of histone deacetylase
inhibitors. Eur J Cancer. 45:1846–1854. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Juengel E, Makarević J, Tsaur I, Bartsch
G, Nelson K, Haferkamp A and Blaheta RA: Resistance after chronic
application of the HDAC-inhibitor valproic acid is associated with
elevated Akt activation in renal cell carcinoma in vivo. PLoS One.
8:e531002013. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Milutinovic S, D'Alessio AC, Detich N and
Szyf M: Valproate induces widespread epigenetic reprogramming which
involves demethylation of specific genes. Carcinogenesis.
28:560–571. 2007. View Article : Google Scholar
|
|
52
|
Detich N, Bovenzi V and Szyf M: Valproate
induces replication-independent active DNA demethylation. J Biol
Chem. 278:27586–27592. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Brandes AA, Franceschi E, Tosoni A, Blatt
V, Pession A, Tallini G, Bertorelle R, Bartolini S, Calbucci F,
Andreoli A, et al: MGMT promoter methylation status can predict the
incidence and outcome of pseudoprogression after concomitant
radiochemotherapy in newly diagnosed glioblastoma patients. J Clin
Oncol. 26:2192–2197. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Munoz JL, Rodriguez-Cruz V, Greco SJ,
Nagula V, Scotto KW and Rameshwar P: Temozolomide induces the
production of epidermal growth factor to regulate MDR1 expression
in glioblastoma cells. Mol Cancer Ther. 13:2399–2411. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Munoz JL, Rodriguez-Cruz V, Greco SJ,
Ramkissoon SH, Ligon KL and Rameshwar P: Temozolomide resistance in
glioblastoma cells occurs partly through epidermal growth factor
receptor-mediated induction of connexin 43. Cell Death Dis.
5:e11452014. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Munoz JL, Walker ND, Scotto KW and
Rameshwar P: Temozolomide competes for P-glycoprotein and
contributes to chemoresistance in glioblastoma cells. Cancer Lett.
367:69–75. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Tivnan A, Zakaria Z, O'Leary C, Kögel D,
Pokorny JL, Sarkaria JN and Prehn JH: Inhibition of multidrug
resistance protein 1 (MRP1) improves chemotherapy drug response in
primary and recurrent glioblastoma multiforme. Front Neurosci.
9:2182015. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Kitange GJ, Mladek AC, Carlson BL,
Schroeder MA, Pokorny JL, Cen L, Decker PA, Wu W, Lomberk GA, Gupta
SK, et al: Inhibition of histone deacetylation potentiates the
evolution of acquired temozolomide resistance linked to MGMT
upregulation in glioblastoma xenografts. Clin Cancer Res.
18:4070–4079. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Zighetti ML, Fontana G, Lussana F, Chiesa
V, Vignoli A, Canevini MP and Cattaneo M: Effects of chronic
administration of valproic acid to epileptic patients on
coagulation tests and primary hemostasis. Epilepsia. 56:e49–e52.
2015. View Article : Google Scholar : PubMed/NCBI
|