Introduction
EGFR molecular profiling predicts non-small
cell lung cancer (NSCLC) patients' responsiveness to tyrosine
kinase inhibitors (TKIs), such as erlotinib and gefitinib
(reversible TKI EGFR) and afatinib (irreversible TKI
EGFR) (1,2). EGFR mutational analyses are
performed exclusively in patients with adenocarcinoma (ADC) or
adenosquamous carcinoma (ADCSCC) because of their higher rates of
EGFR gene mutations relative to other NSCLC types (3).
Caucasian NSCLC patients experience EGFR
mutational rates ranging from 10 to 15.7%. The most representative
EGFR mutations are the exon 19 deletions and the exon 21
L858R point mutations, which account for ~90% of cases (1,4).
Differences in the reported incidence of EGFR mutations
could be related to patient selection and the use of different
methodologies to test for EGFR. Direct Sanger sequencing
requires a proportion of >40–50% of tumor cells, whereas
pyrosequencing and matrix-assisted laser desorption ionization-time
of flight (MALDI-TOF) (5) using
Myriapod Lung Status CE-IVD kits (Diatech Pharmacogenetics, Jesi,
Italy) require as few as 20% tumor cells. Moreover, the DNA
quantity and quality affect EGFR mutational analyses and may
cause a proportion of cases to be missed (6,7).
The majority of recent lung cancer diagnoses have
been based on small biopsies or cytological smears of patients who
cannot undergo surgery because of advanced disease. Thus, small
biopsy and cytosmear samples often represent the only source of
NSCLC tumor cell DNA for molecular characterization. However, the
validity of EGFR testing on small biopsies and cytological
smears remains debatable (4,8,9).
KRAS mutations that are located mainly in
codons 12 and 13 of exon 2 have been reported in up to 30.0% of
NSCLC patients (10,11). Although the role of KRAS
mutations as predictive markers of treatment response in NSCLCs is
still under debate, TKI administration has recently been shown to
have potentially detrimental effects on patients with KRAS
mutations (12). Moreover, the
clinical value of KRAS mutation testing may increase if the
development of a MEK inhibitor in NSCLCs with KRAS mutations
leads to drug approval (5).
The first aim of this study was to analyze a large
consecutive and homogeneous series of Italian NSCLCs to determine
the incidence of EGFR and KRAS mutations over the
last 5 years. This study also aimed to compare two different
routinely used testing methods and investigate the viability of a
multi-target methodology for use in clinical practice by
considering its feasibility for different specimen types (e.g.,
cytological, small biopsy and surgical samples). We also evaluated
the effect of analytical turnaround time (TAT) on the success rate
and clinical utility of a multi-target analysis in routine clinical
care.
Materials and methods
Patients
Samples from 2,387 NSCLC patients between January
2010 and September 2015 were analyzed at the Molecular Pathology
Laboratory of the Unit of Anatomic Pathology 3 of the Azienda
Ospedaliero-Universitaria Pisana to determine their EGFR
mutational status. The majority of patients were diagnosed and
treated in northern Tuscany's Oncology Departments (Italy). The
tumor samples were obtained from the respective Pathological
Anatomy Departments. The oncologists required EGFR
mutational testing based on the individual clinical situation of
each patient, and the majority of patients were also recommended to
undergo KRAS mutational testing.
Each sample was accompanied by a histological
diagnosis performed by hematoxylin and eosin (HE) staining for FFPE
sections and Papanicolaou staining for cytological smears. Clinical
pathological information concerning gender and age were available
for all patients. Informed consent was collected by the oncologist
upon each patient's first visit.
DNA purification and mutation
detection
DNA was extracted from the FFPE and cytological
smears using a commercial kit (Qiagen, Milan, Italy) following the
manufacturer's instructions.
The status of EGFR exons 18–21 from January
2010 to February 2013 was analyzed by the SSCP-Sanger sequencing
method. The MALDI-TOF method was used on a Sequenom (Agena
Bioscience, San Diego, CA, USA) platform. The SSCP-Sanger analysis
[performed as previously described by Rotella et al
(12)] revealed that all of the
mutations were detected with a sensitivity of ~10% of the mutated
allele. The MALDI-TOF dedicated Myriapod Lung Status CE-IVD kit
(Diatech Pharmacogenetics) detects more rep resentative EGFR
mutations and has a sensitivity ranging from 2.5 to 10%. This kit
can also simultaneously analyze EGFR, KRAS, BRAF,
NRAS, PIK3CA, ALK, ERBB2, DDR2, RET and MAP2K1 mutations.
When required, the mutational status of KRAS codons
12 and 13 from 2010 to February 2013 was analyzed with a
pyrosequencing Anti-EGFR MoAb response® (KRAS
status) CE-IVD marked kit (Diatech Pharmacogenetics) following the
manufacturer's instructions. This step showed a sensitivity of
5–10% of mutated alleles. The pyrosequencing method was replaced by
the method using MALDI-TOF dedicated Myriapod Lung Status CE-IVD
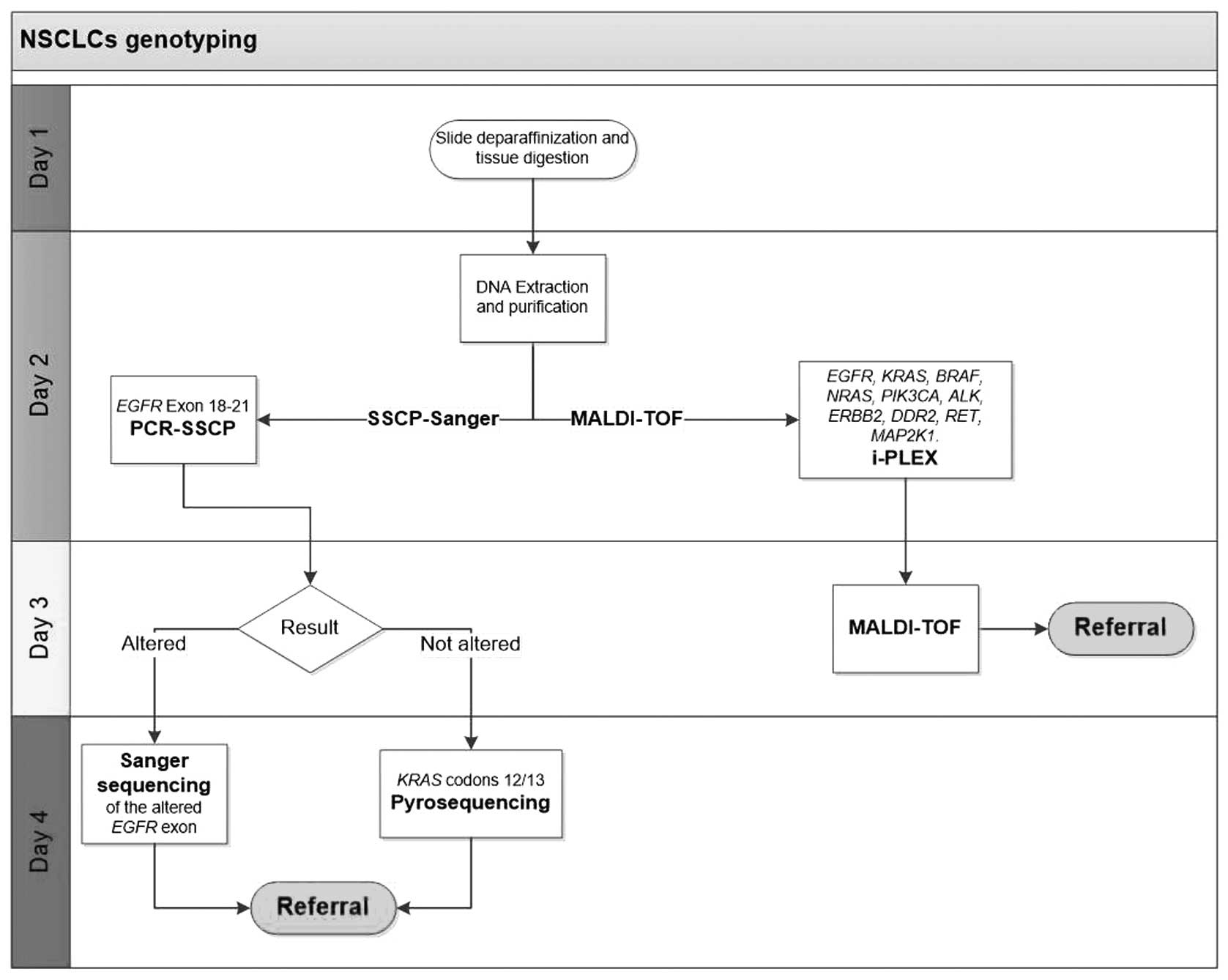
kits (Diatech Pharmacogenetics) in March 2013. Fig. 1 shows the operative flow chart for
the tests described above. Cell/tissue sample enrichment was
performed to ensure the highest tumor content. The SSCP-Sanger
method required a tumor cell representativeness of >20%, whereas
pyrosequencing and the Myriapod Lung Status CE-IVD kits each
required tumor cell representativeness of >10%.
Evaluation of the analytical TAT
The analytical TAT is the mean time from sample
receipt to results interpretation. This value was recorded for each
sample according to the different methods of analysis and then
compared.
Statistical analyses
Patients were classified as mutated or wild-type
based on the presence of EGFR and KRAS mutations. The
quality and quantity of the material were measured by the total
number or percentage of neoplastic cells to determine whether the
samples could be analyzed.
Differences in the variables between the two groups
and their association with the clinical data were tested using
Fisher's exact test or a two-sided Chi-square test. A p-value of
0.05 or less was considered significant. All of the statistical
analyses were performed with STATISTICA software (Dell Software,
Tulsa, OK, USA).
Results
Clinicopathological characteristics
Our analysis revealed that 1993 (83.5%) patients
presented lung adenocarcinoma, 190 (8.0%) presented squamous cell
carcinoma (SCC) and 204 (8.5%) presented not otherwise specified
NSCLC (NSCLC-NOS). In addition, 1,539 patients were males (64.5%)
and 848 (35.5%) were females, and the mean age of the entire series
was 68.1 years (range, 25–91).
Molecular testing adequacy
Table I shows the
overall efficiency of the EGFR molecular tests according to
the method of analysis and type of material. The SSCP-Sanger method
showed analyzable cytological, bioptic and surgical specimen
percentages of 90.3, 90.9 and 98.1%, respectively, whereas the
MALDI-TOF platform showed analyzable cytological, bioptic and
surgical specimen percentages of 94.6, 95.7 and 96.9%,
respectively. The MALDI-TOF platform showed a reduction in the
percentage of non-analyzable cases because of the low amount of
input DNA needed. A significant increase (p=0.03) of analyzable
samples was observed.
 | Table IOverall efficiency of molecular
testing. |
Table I
Overall efficiency of molecular
testing.
| Type of
material | Method | Total | Not analyzable
cases
| Analyzable cases
|
|---|
NE
n (%) | NA
n (%) | n (%) | P-value |
|---|
| Cytology | S-S | 425 | 37 (8.7) | 4 (0.9) | 384 (90.3) | 0.012 |
| M-T | 554 | 29 (5.2) | 1 (−) | 524 (94.6) | |
| Biopsy | S-S | 265 | 18 (6.8) | 6 (2.2) | 241 (90.9) | 0.026 |
| M-T | 304 | 10 (3.3) | 3 (0.9) | 291 (95.7) | |
| Surgical
specimen | S-S | 421 | 1 (−) | 7 (1.6) | 413 (98.1) | 0.6 |
| M-T | 357 | 3 (0.8) | 8 (2.2) | 346 (96.9) | |
| Total | S-S | 1111 | 56 (5.0) | 17 (1.5) | 1038 (93.5) | 0.030 |
| M-T | 1215 | 42 (3.4) | 12 (1.0) | 1161 (95.6) | |
EGFR mutational status
EGFR exon 18–21 mutations were found in 311
of 2,199 (14.1%) cases, and a histological analysis showed that 290
(15.8%) were ADCs, 15 (8.2%) were NSCLCs-NOS and 6 (3.3%) were
SCCs.
Table II summarizes
the EGFR mutational analysis results with regard to the
method and type of analyzed material. The EGFR mutational
ratios were 14.5 and 13.8% for the SSCP-Sanger and MALDI-TOF
platforms, respectively. The MALDI-TOF platform and SSCP-Sanger
method showed ratios of mutated samples for surgical, small biopsy
and cytological samples of 12.0, 16.2 and 17.1%, respectively, and
15.3, 11.4 and20.8%, respectively.
 | Table IIComparison of the EGFR
mutational rate according to the analytical method and sampling
type. |
Table II
Comparison of the EGFR
mutational rate according to the analytical method and sampling
type.
| Type of
material | Method | ADCs
| SCCs
| NSCLCs-NOS
| Total
|
|---|
| Total | MUT
n (%) | Total | MUT
n (%) | Total | MUT
n (%) | Total | MUT
n (%) |
|---|
| Cytology | S-S | 298 | 62 (20.8) | 32 | 1 (3.1) | 54 | 8 (14.8) | 384 | 71 (18.5) |
| M-T | 457 | 78 (17.1) | 20 | 2 (10.0) | 47 | 3 (6.4) | 524 | 83 (15.8) |
| Biopsy | S-S | 175 | 20 (11.4) | 29 | 2 (6.9) | 37 | 3 (8.1) | 241 | 25 (10.4) |
| M-T | 229 | 37 (16.2) | 28 | 0 | 34 | 1 (2.9) | 291 | 38 (13.1) |
| Surgical
specimen | S-S | 352 | 54 (15.3) | 54 | 1 (1.9) | 7 | 0 | 413 | 55 (13.3) |
| M-T | 324 | 39 (12.0) | 19 | 0 | 3 | 0 | 346 | 39 (11.3) |
| Total | S-S | 825 | 136 (16.5) | 115 | 4 (3.5) | 98 | 11 (11.2) | 1038 | 151 (14.5) |
| M-T | 1010 | 154 (15.2) | 67 | 2 (3.0) | 84 | 4 (4.8) | 1161 | 160 (13.8) |
Table III shows
the different types of EGFR mutations according to the
method of analysis. For the ADCs, the number of EGFR exon
18, 19, 20 and 21 mutations were 6, 71, 21 and 70 with the
MALDI-TOF platform, respectively, and 11, 82, 13 and 38 with the
SSCP-Sanger method, respectively. Table IV shows the EGFR exon 19
deletions and insertions according to the SSCP-Sanger and MALDI-TOF
methods. Two simultaneous mutations of the EGFR gene were
found in 23 patients, and the majority of double mutations were
represented by T790M exon 20 mutations concomitant to exon 19
deletion (11 samples) and T790M mutation concomitant to L858R exon
21 mutation (7 cases).
 | Table IIIOverall EGFR mutations
revealed by SSCP-Sanger sequencing and MALDI-TOF. |
Table III
Overall EGFR mutations
revealed by SSCP-Sanger sequencing and MALDI-TOF.
| Exon/mutation | ADCs
| NSCLCs NOS
| SCCs
| Total
|
|---|
| S-S | M-T | S-S | M-T | S-S | M-T | S-S | M-T |
|---|
| Exon 18 |
| E709A | 1 | | | | | | 1 | |
| G719A | 4 | 2 | | | | | 4 | 2 |
| G719C | 2 | | | | | | 2 | |
| G719S | | 1 | | | | | | 1 |
| DEL/INS | 1 | | | | | | 1 | |
| E709A+G719A | | | 1 | | | | 1 | |
| E709A+G719S | 1 | | | | | | 1 | |
| E709K+G719S | 1 | | | | | | 1 | |
| Exon 19 |
| L747P | 1 | | | | | | 1 | |
| DEL/INS | 81 | 71 | 7 | 2 | 3 | 2 | 91 | 75 |
| Exon 20 |
| S768I | | 2 | | | | | | 2 |
| T790Ma | 7 | 11 | | | | 1 | 7 | 12 |
| Insertion | 6 | 6 | | | | | 6 | 6 |
| Exon 21 |
| V834L | | | 1 | | | | 1 | |
| H835L | | | | | 1 | | 1 | |
| P848L | | 1 | | | | | | 1 |
| L858R | 35 | 64 | 2 | 2 | | | 37 | 66 |
| L861Q | 2 | 4 | | | | | 2 | 4 |
| Other |
| E709K+L858R | | 1 | | | | | | 1 |
| G719A+L858R | 1 | | | | | | 1 | |
| G719C+S768I | | 1 | | | | | | 1 |
| G719S+S768I | | 1 | | | | | | 1 |
 | Table IVEGFR exon 19
deletions/insertions revealed by SSCP-Sanger sequencing and
MALDI-TOF. |
Table IV
EGFR exon 19
deletions/insertions revealed by SSCP-Sanger sequencing and
MALDI-TOF.
| S-S | M-T |
|---|
| EGFR exon 19
DELETIONS/INSERTIONS | 77 | 63 |
| p.E746_E749>T
(c.2236_2246>TAC) | 1 | 1 |
|
p.E746_A750delELREA
(c.2235-2249del15) | 33 | 30 |
|
p.E746_A750delELREA
(c.2236-2250del15) | 21 | 17 |
| p.E746_A750>NP
(c.2235_2248>TC) | 1 | |
| p.E746_T751>A
(c.2237_2251del15) | 1 | 1 |
| p.E746_T751>Q
(c.2236_2253>CAA) | 1 | |
| p.E746_T751>S
(c.2235_2252>ATT) | 1 | |
| p.E746_S752>I
(c.2236_2256>ATC | 1 | |
| p.E746_S752>V
(c.2237_2255>T) | 9 | 9 |
| p.E746_P753>VS
(c.2237_2257>TCT) | 1 | |
| p.E746_A750>VP
(c.2237_2248>TAC) | 1 | |
| p.L747_E749delLRE
(c.2239_2247del9) | 3 | |
| p.L747_A750>P
(c.2239_2248>C) | 1 | |
|
p.L747_T751delLREAT
(c.2240_2254del15) | 5 | |
| p.L747_T751>P
(c.2239_2251>C) | 1 | |
|
p.L747_S752delLREATS
(c.2239_2256del18) | 1 | |
| p.L747_P753>S
(c.2240_2257del18) | 7 | |
| p.L747_A755>AN
(c.2239_2264>GCCAA) | 1 | |
|
p.S752_I759delSPKANKEI
(c.2254_2277del24) | 1 | |
|
p.L747_P753>S/p.L747_T751delLREAT | | 8 |
|
p.L747_A750>P/p.L747_S752 | | 4 |
| p.E746_E749
delELRE/p.K745_E746insIPVAIK | | 1 |
|
p.E746_A750>QP/p.E746_S752 | | 1 |
|
p.E746_T751>VA/E746_T751>V | | 1 |
|
p.K745_E746insIPVAIK | | 2 |
Among the 6 SCCs with EGFR alterations, we
found 4 exon 19 deletions, 1 H835L exon 21 mutation and 1 T790M
mutation concomitant with exon 19 deletion.
Of the 15 NSCLCs-NOS EGFR mutated cases, we
found 9 exon 19 deletions, 5 exon 21 mutations (4 L858R and 1
V834L) and 1 exon 18 mutation in two different codons (E709A and
G719A).
EGFR mutations versus gender and age
EGFR mutations were found in 193 (24.5%) of
786 female patients and 118 (8.3%) of 1,412 male patients. A
significant correlation (p<0.0001) was found between EGFR
mutations and female patients. The mean age of the patients with
mutation was 67.9 years, which was identical to the mean age of the
patients without mutation. However, the EGFR mutational rate
was significantly higher (p=0.02) in female patients 65 years and
over (28.6%; 132 of 462) compared with younger women (18.9%; 60 of
316). Age-dependent differences were not observed for the male
EGFR mutation rates.
KRAS status
Within the KRAS codons, 12/13 mutations were
found in 584 (30.5%) lung carcinomas, including 528 of 1597 (33.0%)
ADCs, 44 of 163 (26.9%) NSCLCs-NOS and 12 of 155 (7.7%) SCCs.
Table V summarizes
the results of the KRAS mutational analysis in ADCs
according to the method and type of analyzed material. The KRAS
mutational ratios by pyrosequencing and the MALDI-TOF platform were
28.7 and 31.7%, respectively. The MALDI-TOF platform demonstrated
that in the ADCs, mutated samples occurred in 122 of 322 (37.9%)
surgical samples, 69 of 218 (31.7%) small biopsies and 145 of 444
(32.7%) cytological specimens, whereas pyrosequencing demonstrated
that mutated samples occurred in 96 of 281 (34.2%) surgical
samples, 40 of 139 (28.8%) small biopsies and 56 of 193 (29.0%)
cytological specimens. MALDI-TOF indicated that there were 27 exon
3 codon 61 mutations, including 19 p.Q61H, 7 p.Q61L and 1
p.Q61R.
 | Table VComparison of KRAS mutational
rates according to the analytical methods and sampling type. |
Table V
Comparison of KRAS mutational
rates according to the analytical methods and sampling type.
| Type of
material | Method | ADCs
| SCCs
| NSCLC-NOS
| Total
|
|---|
| Total | MUT | Total | MUT n (%) | Total | MUT n (%) | Total | MUT n (%) |
|---|
| Cytology | PYRO | 193 | 56 (29.0) | 26 | 3 (11.5) | 42 | 11 (26.2) | 261 | 70 (26.8) |
| M-T | 444 | 145 (32.7) | 20 | 1 (5.0) | 47 | 13 (27.7) | 511 | 159 (31.1) |
| Biopsy | PYRO | 139 | 40 (28.8) | 22 | 4 (18.2) | 30 | 11 (36.7) | 191 | 55 (28.8) |
| M-T | 218 | 69 (31.7) | 28 | 2 (7.1) | 34 | 7 (20.6) | 280 | 78 (27.9) |
| Surgical
specimen | PYRO | 281 | 96 (34.2) | 40 | 2 (5.0) | 7 | 1 (14.3) | 328 | 99 (30.2) |
| M-T | 322 | 122 (37.9) | 19 | 0 | 3 | 1 (33.3) | 344 | 123 (35.8) |
| Total | PYRO | 613 | 192 (31.3) | 88 | 9 (10.2) | 79 | 23 (29.1) | 780 | 224 (28.7) |
| M-T | 984 | 336 (34.1) | 67 | 3 (4.5) | 84 | 21 (25.0) | 1135 | 360 (31.7) |
KRAS mutations versus gender and age
KRAS mutations were found in 173 (25.2%) of
686 female patients and 412 (33.5%) of 1,229 male patients. A
significant correlation (p=0.0002) was observed between the
KRAS mutations and male patients. Significant correlations
were not observed between the mutated and not-mutated samples
according to patient age (p=0.21).
Other gene mutations
The Myriapod Lung Status CE-IVD kit (Diatech
Pharmacogenetics) analysis revealed 4 NRAS gene mutations (1
p.G12D, 2 p.Q61L and 1 p.Q61K), 26 BRAF gene mutations (2 p.G466A,
1 p.G466E, 2 p.G466V, 1 p.D594G and 20 p.V600E), 5 ERBB2 gene
mutations (p.A775 G776insYVMA), 16 PIK3CA gene mutations (2
p.E542K, 6 p.E545K and 8 p.H1047R), 1 ALK gene mutation (p.C1156K)
and 1 MAPK2K1 gene mutation (p.Q56P).
Analytical TAT
The mean TAT for the SSCP-Sanger/pyrosequencing
analyses was four working days (Fig.
1). Overall, the throughput of this protocol was limited by the
number of analyzable samples from the SSCP and Sanger sequencing,
and only 5 patients could be simultaneously tested because of
limitations of the SSCP precasting gel. The MALDI-TOF platform
produced a mean TAT for the simultaneous EGFR and
KRAS analyses of three working days (Fig. 1), and 10 patients could be
simultaneously analyzed. At least one additional day was required
for cytological smear demounting.
Discussion
To the best of our knowledge, this is one of the
larger studies to have performed an EGFR mutational analysis
in a homogeneous series of patients with metastatic NSCLCs from a
single center. These results directly reflect the daily EGFR
testing routine.
Our study presents an appropriate assessment of the
epidemiologic and methodological information related to EGFR
mutational testing of metastatic NSCLC patients in a clinical
setting.
A total of 2,387 patients from Northern Tuscany
(Italy) with metastatic lung cancer were analyzed, and they yielded
an overall EGFR mutational rate of 14.1%. These data are
consistent with that of previous reports for Caucasian patients
(1,2,10,11,13–16).
The predominant EGFR alteration was the exon 19 deletion,
which was followed by the exon 21 L858R point mutation (13,17).
The EGFR mutation rate was significantly higher in ADC
patients (15.8%) than in NSCLC-NOS (8.2%) and SCC (3.3%) patients
(3,18–20).
The strong association between EGFR mutations and female patients
was confirmed in this study. Moreover, a significant association
between EGFR mutations and older age (≥65 years) in female
patients was observed. This finding supports that of Gahr et
al (13).
In our 5 years of experience with daily EGFR
analyses, we have changed our method of analysis for EGFR
and KRAS from the Sanger sequencing and pyrosequencing methods to
the Sequenom multi-marker MALDI-TOF platform.
Recent advances in multiplex genotyping and high
throughput genomic profiling by multi-marker sequencing offer the
possibility of rapidly and comprehensively interrogating individual
patient cancer genomes from small tumor biopsies and cytological
samples. Particular emphasis can be placed on daily molecular
diagnoses of lung cancers. Our experience with the two different
methodologies in a large series of NSCLCs has helped emphasize
certain important factors in genotyping and genomic profiling.
The overall rates of EGFR and KRAS
mutations were not significantly changed after the adoption of a
multi-target technique, although the MALDI-TOF platform nearly
doubled the rate of detection of the L858R mutation.
The number of failed analyses because of low
quantity or damaged DNA and reaction inhibition significantly
decreased within cytological samples and small biopsies. Assessing
the status of multiple genes requires a small amount (as low as 40
ng) of DNA template; however, this amount is crucial for performing
reliable EGFR mutation analyses of cytological samples and
small biopsies, and these samples are often the only material
available to establish a diagnosis and perform a molecular
analysis.
The adoption of the MALDI-TOF platform reduced the
mean analytical TAT, which has important implications for the
management and treatment of patients.
MALDI-TOF testing revealed an insignificant increase
in the KRAS mutational rate. Furthermore, the strong
association between KRAS mutations and male patients and the
mutual exclusivity of EGFR and KRAS gene mutations
were confirmed (21).
The Myriapod Lung Status CE-IVD kit revealed several
mutations in KRAS at exon 3 as well as in NRAS,
BRAF, PIK3CA, ERBB2, ALK and
MAPK2K1. This finding suggests that a more comprehensive
approach to predictive biomarker analysis is needed. The ability to
simultaneously test several relevant genes may be beneficial to
patients because of the potential to identify alternative treatment
options.
In conclusion, although, this study brings nothing
new to the field of NSCLC mutational screening, our results
underline important concepts from a methodological point of view,
first of all it confirms that small biopsy or cytological samples
are adequate for multiple mutational testing of NSCLCs in a large
series of cases. EGFR mutations are detectable with a
similar frequency in the surgical, small biopsy and cytological
samples by using the SSCP-Sanger or MALDI-TOF platforms, even if
the MALDI-TOF method reduces the rate of missed samples when the
DNA quality and quantity is low in the small biopsy and cytological
samples. Moreover, this method is also able to detect a wider range
of mutations using a small amount of DNA. Furthermore, the
MALDI-TOF platform allows for the rapid implementation and
application of newly identified biomarkers for target therapies and
does not negatively affect the time and cost effectiveness of the
analytical procedure.
References
|
1
|
Rosell R, Moran T, Queralt C, Porta R,
Cardenal F, Camps C, Majem M, Lopez-Vivanco G, Isla D, Provencio M,
et al Spanish Lung Cancer Group: Screening for epidermal growth
factor receptor mutations in lung cancer. N Engl J Med.
361:958–967. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Weber B, Hager H, Sorensen BS, McCulloch
T, Mellemgaard A, Khalil AA, Nexo E and Meldgaard P: EGFR mutation
frequency and effectiveness of erlotinib: A prospective
observational study in Danish patients with non-small cell lung
cancer. Lung Cancer. 83:224–230. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Krawczyk P, Ramlau R, Chorostowska-Wynimko
J, Powrózek T, Lewandowska MA, Limon J, Wasąg B, Pankowski J,
Kozielski J, Kalinka-Warzocha E, et al: The efficacy of EGFR gene
mutation testing in various samples from non-small cell lung cancer
patients: A multicenter retrospective study. J Cancer Res Clin
Oncol. 141:61–68. 2015. View Article : Google Scholar :
|
|
4
|
Scarpino S, Pulcini F, Di Napoli A,
Giubettini M and Ruco L: EGFR mutation testing in pulmonary
adenocarcinoma: Evaluation of tumor cell number and tumor percent
in paraffin sections of 120 small biopsies. Lung Cancer. 87:8–13.
2015. View Article : Google Scholar
|
|
5
|
Sherwood JL, Müller S, Orr MC, Ratcliffe
MJ and Walker J: Panel based MALDI-TOF tumour profiling is a
sensitive method for detecting mutations in clinical non small cell
lung cancer tumour. PLoS One. 9:e1005662014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
da Cunha Santos G, Saieg MA, Geddie W and
Leighl N: EGFR gene status in cytological samples of nonsmall cell
lung carcinoma: Controversies and opportunities. Cancer Cytopathol.
119:80–91. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Eberhard DA, Giaccone G and Johnson BE;
Non-Small-Cell Lung Cancer Working Group: Biomarkers of response to
epidermal growth factor receptor inhibitors in Non-Small-Cell Lung
Cancer Working Group: Standardization for use in the clinical trial
setting. J Clin Oncol. 26:983–994. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Aisner DL, Deshpande C, Baloch Z, Watt CD,
Litzky LA, Malhotra B, Sepulveda AR, Langer C, Evans T and Van
Deerlin VM: Evaluation of EGFR mutation status in cytology
specimens: An institutional experience. Diagn Cytopathol.
41:316–323. 2013. View
Article : Google Scholar
|
|
9
|
Aisner DL and Marshall CB: Molecular
pathology of non-small cell lung cancer: A practical guide. Am J
Clin Pathol. 138:332–346. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Helland Å, Skaug HM, Kleinberg L, Iversen
ML, Rud AK, Fleischer T, Sagerup C, Solberg S, Jørgensen L,
Ariansen S, et al: EGFR gene alterations in a Norwegian cohort of
lung cancer patients selected for surgery. J Thorac Oncol.
6:947–950. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ludovini V, Bianconi F, Pistola L, Pistola
V, Chiari R, Colella R, Bellezza G, Tofanetti FR, Siggillino A,
Baldelli E, et al: Optimization of patient selection for EGFR-TKIs
in advanced non-small cell lung cancer by combined analysis of
KRAS, PIK3CA, MET, and non-sensitizing EGFR mutations. Cancer
Chemother Pharmacol. 69:1289–1299. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Rotella V, Fornaro L, Vasile E, Tibaldi C,
Boldrini L, Chella A, D'Incecco A, Cirigliano G, Chioni A, Lupi C,
et al: EGFR and K-Ras mutations in women with lung adenocarcinoma:
Implications for treatment strategy definition. J Exp Clin Cancer
Res. 33:772014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Gahr S, Stoehr R, Geissinger E, Ficker JH,
Brueckl WM, Gschwendtner A, Gattenloehner S, Fuchs FS, Schulz C,
Rieker RJ, et al: EGFR mutational status in a large series of
Caucasian European NSCLC patients: Data from daily practice. Br J
Cancer. 109:1821–1828. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Marchetti A, Martella C, Felicioni L,
Barassi F, Salvatore S, Chella A, Camplese PP, Iarussi T, Mucilli
F, Mezzetti A, et al: EGFR mutations in non-small-cell lung cancer:
Analysis of a large series of cases and development of a rapid and
sensitive method for diagnostic screening with potential
implications on pharmacologic treatment. J Clin Oncol. 23:857–865.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Smits AJ, Kummer JA, Hinrichs JW, Herder
GJ, Scheidel-Jacobse KC, Jiwa NM, Ruijter TE, Nooijen PT,
Looijen-Salamon MG, Ligtenberg MJ, et al: EGFR and KRAS mutations
in lung carcinomas in the Dutch population: Increased EGFR mutation
frequency in malignant pleural effusion of lung adenocarcinoma.
Cell Oncol (Dordr). 35:189–196. 2012. View Article : Google Scholar
|
|
16
|
Locatelli-Sanchez M, Couraud S, Arpin D,
Riou R, Bringuier PP and Souquet PJ: Routine EGFR molecular
analysis in non-small-cell lung cancer patients is feasible: Exons
18–21 sequencing results of 753 patients and subsequent clinical
outcomes. Lung. 191:491–499. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Penzel R, Sers C, Chen Y,
Lehmann-Mühlenhoff U, Merkelbach-Bruse S, Jung A, Kirchner T,
Büttner R, Kreipe HH, Petersen I, et al: EGFR mutation detection in
NSCLC - assessment of diagnostic application and recommendations of
the German Panel for Mutation Testing in NSCLC. Virchows Arch.
458:95–98. 2011. View Article : Google Scholar
|
|
18
|
Miyamae Y, Shimizu K, Hirato J, Araki T,
Tanaka K, Ogawa H, Kakegawa S, Sugano M, Nakano T, Mitani Y, et al:
Significance of epidermal growth factor receptor gene mutations in
squamous cell lung carcinoma. Oncol Rep. 25:921–928.
2011.PubMed/NCBI
|
|
19
|
Perez-Moreno P, Brambilla E, Thomas R and
Soria JC: Squamous cell carcinoma of the lung: Molecular subtypes
and therapeutic opportunities. Clin Cancer Res. 18:2443–2451. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Boch C, Kollmeier J, Roth A,
Stephan-Falkenau S, Misch D, Grüning W, Bauer TT and Mairinger T:
The frequency of EGFR and KRAS mutations in non-small cell lung
cancer (NSCLC): Routine screening data for central Europe from a
cohort study. BMJ Open. 3:42013. View Article : Google Scholar
|
|
21
|
Unni AM, Lockwood WW, Zejnullahu K,
Lee-Lin SQ and Varmus H: Evidence that synthetic lethality
underlies the mutual exclusivity of oncogenic KRAS and EGFR
mutations in lung adenocarcinoma. eLife. 4:e069072015. View Article : Google Scholar : PubMed/NCBI
|