Introduction
The AML1-ETO (AE) fusion protein, which originates
from the t(8;21) chromosomal rearrangement, is one of the most
frequent translocation products found in de novo acute
myeloid leukemia (AML) (1). Murine
experiment data have demonstrated that AE alone is not sufficient
to induce leukemia (2–4), but requires altered signal
transduction pathways for leukemia progression (5), suggesting a model of AML pathogenesis
in which the two groups of genetic alterations are required for the
induction of the full-blown disease. c-KIT mutations are considered
as one of the most important subsequent events (up to 48%) in
leukemia cases harboring AE, and have adverse effects on the
disease outcome (5–7). Furthermore, synergism between c-KIT
mutation and AE in the induction of AML has been demonstrated
(8–9). A recent study (10) further revealed that activated c-KIT
upregulates the PI3K/AKT signaling pathway and reverts AE-induced
DNA damage and apoptosis, which accounts for the increased
chemo-resistance observed in t(8;21)-positive AML patients with
activated c-KIT mutations. However, the reason for the high
incidence of c-KIT mutations and the exact mechanisms involved in
the synergism of AE with mutated-c-KIT in AE leukemia remain
unclear.
Amyloid precursor protein (APP), a type I integral
membrane protein, generated by the APP gene which is located on
21q21.3, is implicated in synapse formation and plasticity. One of
the processed APP products, β-amyloid, is directly related to the
pathogenesis of neurodegenerative disorders such as Alzheimer's
disease (11). APP is also
ubiquitously expressed in nonneuronal tissues, and may also be
involved in the growth of various cell types in both physiological
and abnormal states. It has been shown that APP promotes cancer
cell proliferation and metastasis and its overexpression in oral
squamous cell carcinoma, pancreatic and colorectal cancer has an
adverse effect on prognosis (12–15).
Moreover, APP is also highly expressed in AML harboring complex
karyotypes or t(8;21) (16,17). Notably, APP plays an important role
in AE leukemia and its overexpression enhances migration of
Kasumi-1 cells by MMP-2 (18). In
the present study, we showed that, from the results obtained from
the clinical observation, APP was positively correlated with c-KIT
mutations/overexpression and had prognostic predictive value; from
the data obatined from the cell model experiment, APP regulated
cell apoptosis but not proliferation and was involved in the
regulation of c-Kit expression and the PI3K/AKT signaling pathway,
in AE leukemia. Our study indicates that APP may cooperate with
c-KIT mutations/overexpression in the regulation of cell apoptosis
via the PI3K/AKT signaling pathway in AE leukemia, suggesting that
APP overexpression in AE leukemia may be the reason for the high
incidence of c-KIT mutations and APP may be involved in the
synergism of AE and c-KIT mutations to induce leukemia.
Materials and methods
Patient samples
Sixty-five bone marrow samples were obtained from AE
leukemia patients admitted between February 2006 and June 2013 at
Nanfang Hospital and used for the analysis of APP and c-KIT mRNA
expression and c-KIT mutations. Characteristics of the patients are
documented in Table I. All samples
were obtained upon approval of the Nanfang Hospital, Southern
Medical College of Medicine Institutional Review Boards.
 | Table IClinical characteristics of the
AML1-ETO-positive leukemia patients. |
Table I
Clinical characteristics of the
AML1-ETO-positive leukemia patients.
| Characteristics | APP-H (n=33) | APP-L (n=32) | P-value |
|---|
| Median age (range),
in years | 30 (5–69) | 28.5 (4–58) | 0.890 |
| Gender, male/female
(ratio) | 22/11 (2.0) | 18/14 (1.3) | 0.388 |
| WBC
(×109/l) (range) | 22.3 (3.1–97.6) | 12.4 (1.7–70.3) | 0.008 |
| Marrow blasts, %
(range) | 38.0 (12.0–94.0) | 33.5 (12.0–93.0) | 0.423 |
| Bone marrow
cellularity, % (range) | 91.0 (47.0–99.0) | 83.0 (52.0–98.0) | 0.031 |
| c-KIT mutations/total
cases | 15/33 | 2/32 | <0.001 |
| c-KIT expression
level (range) | 0.00424
(0.00001–0.08479) | 0.00057
(0.00004–0.01418) | 0.001 |
Quantitative real-time polymerase chain
reaction (qPCR) analysis
For each patient, a bone marrow sample was collected
at diagnosis and mononuclear cells were enriched by density
gradient centrifugation with Ficoll solution. Total RNA extraction
and cDNA synthesization were carried out, and PCR primer sequences
for APP, c-KIT and β-actin are presented in Table II. APP and c-KIT mRNA expression as
assessment by qPCR was previously described (18).
 | Table IISequences of the PCR primers for each
gene. |
Table II
Sequences of the PCR primers for each
gene.
| Gene | Primers | Sequences |
|---|
| APP | F |
5′-TGGCCCTGGAGAACTACATC-3′ |
| R |
5′-AATCACACGGAGGTGTGTCA-3′ |
| c-KIT | F |
5′-CACCGAAGGAGGCACTTACAC-3′ |
| R |
5′-GGAATCCTGCTGCCACACA-3′ |
| β-actin | F |
5′-CGTCTTCCCCTCCATCG-3′ |
| R |
5′-CTCGTTAATGTCACGCAC-3′ |
Molecular analysis
DNA from mononuclear cells, isolated from the 65
patient bone marrow samples obtained at diagnosis, was extracted
using a DNA extraction kit (Qiagen) according to the manufacturer's
instructions. The screening of the c-KIT (exon 8 and 17) mutations
was performed by PCR and direct Sanger sequencing. PCR primers are
presented in Table III. The total
reaction volume of 25 µl contained 2 µl DNA (100 ng),
1 µl of each primer (50 pmol), 12.5 µl PCR mix
(Takara) and 8.5 µl ddH2O. Purified PCR products
were sequenced with Sanger sequencing. The results were analyzed
with Chromas software (Technelysium Pty Ltd.).
 | Table IIISequence of PCR primers for c-KIT
mutation detection. |
Table III
Sequence of PCR primers for c-KIT
mutation detection.
| Gene | Primers | Sequences |
|---|
| KIT |
| Exon 8 | F |
5′-CTCCCTGAAAGCAGAAAC-3′ |
| R |
5′-CAGAAAGATAACACCAAAATAG-3′ |
| Exon 17 | F |
5′-GCAAAGGCATATTAGGAACTC-3′ |
| R |
5′-GTTGTAGTAATGTTCAGCATACC-3′ |
Cell cycle distribution, apoptosis and
c-KIT mutation/expression assays in Kasumi-1 cells
To further analyze the correlation of cell
proliferation and apoptosis with APP expression, the Kasumi-1 cell
line, which harbors the AML1-ETO fusion gene, a c-KIT mutation
(19), and has high expression of
APP (17), was chosen as a cell
model. According to our previous study (18), APP was silenced in the Kasumi-1
cells by lentivirus transduction, and the cells were defined as
siRNA-APP-treated (siAPP) Kasumi-1 cells; in the same manner,
another group of Kasumi-1 cells was transfected with scramble siRNA
(TTCTCCGAACGTGTCACGT) and served as a negative control (NC). Using
flow cytometric analysis, we assessed the expression of CD117, the
cell cycle distribution and apoptosis in the siAPP, NC and
wild-type Kasumi-1 cells. Using the PCR method, the screening of
mutations and expression levels of c-KIT were assessed.
Western blot analysis
The proteins, resolved from the siAPP, NC and
wild-type Kasumi-1 cells, respectively, underwent centrifugation
and quantification, and were used for western blot analysis as
previously described (18). The
analysis of Bcl-2 (Abcam, Cambridge, MA, USA), c-Kit and p-AKT
(both from Cell Signal Technology; CST), p53 and NF-κB (both from
Abcam), caspase-3 and caspase-9 (both from CST), with their
relative primary monoclonal antibodies was performed following the
manufacturer's instructions, and GAPDH (Abcam) was chosen as the
standard control.
Statistical analysis
SPSS 17.0 software (SPSS Inc., Chicago, IL, USA) was
used for the statistical analysis. Data are expressed as the means
± standard deviation. Statistical analysis was performed by one-way
ANOVA followed by Fisher's post hoc test procedure for assessment
of significance (P<0.05).
Results
APP is correlated with c-KIT
mutations/overexpression and is indicative of poor disease outcome
in patients with AE leukemia
The 65 patients were divided into an APP high
expression (APP-H) group (n=33, with the level of APP greater than
the median level) and an APP low expression (APP-L) group (n=32,
with the level of APP less than the median level) according to the
median value of APP relative expression levels. The results from
analysis of the clinical characteristics of the patients in the
APP-H and the APP-L group showed that a significantly higher
peripheral white blood cell (WBC) count (29.2±3.9×109/l
vs. 17.9±2.9×109/l, P=0.008) and bone marrow cellularity
(86.7±1.7 vs. 80.3±2.3%, P=0.031) were observed in the APP-H
patients when compared with these values in the APP-L patients,
while there was no statistical difference in the bone marrow
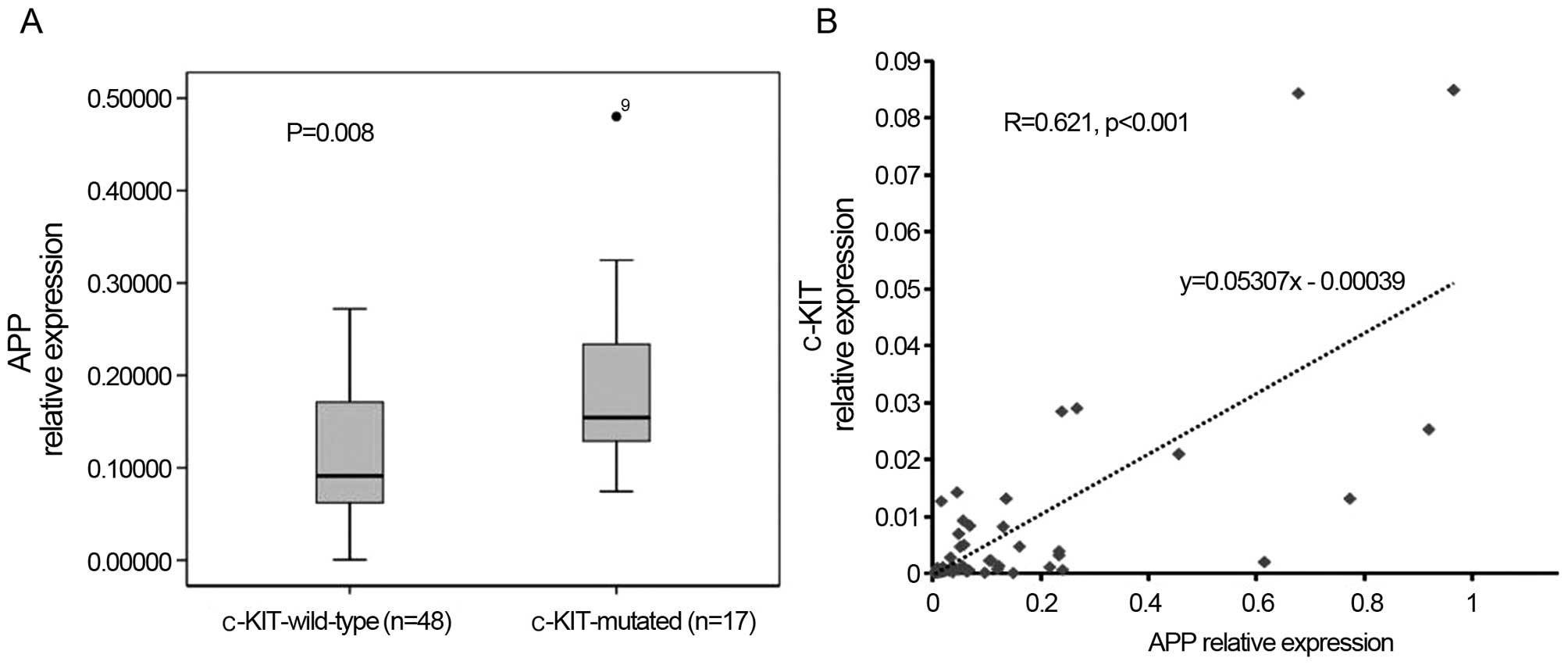
blasts. Moreover, APP was correlated with c-KIT
mutations/overexpression, in that 15 out of 17 c-KIT-mutated cases
belonged to the APP-H group (15/33), whereas only two mutations
were observed in the APP-L group (2/32, P<0.001, Table I). Meanwhile, APP was expressed
apparently higher in the c-KIT-mutated patients (P=0.008, Fig. 1A) and its expression levels were
positively correlated with c-KIT mRNA expression (r=0.621,
P<0.001, Fig. 1B). In addition,
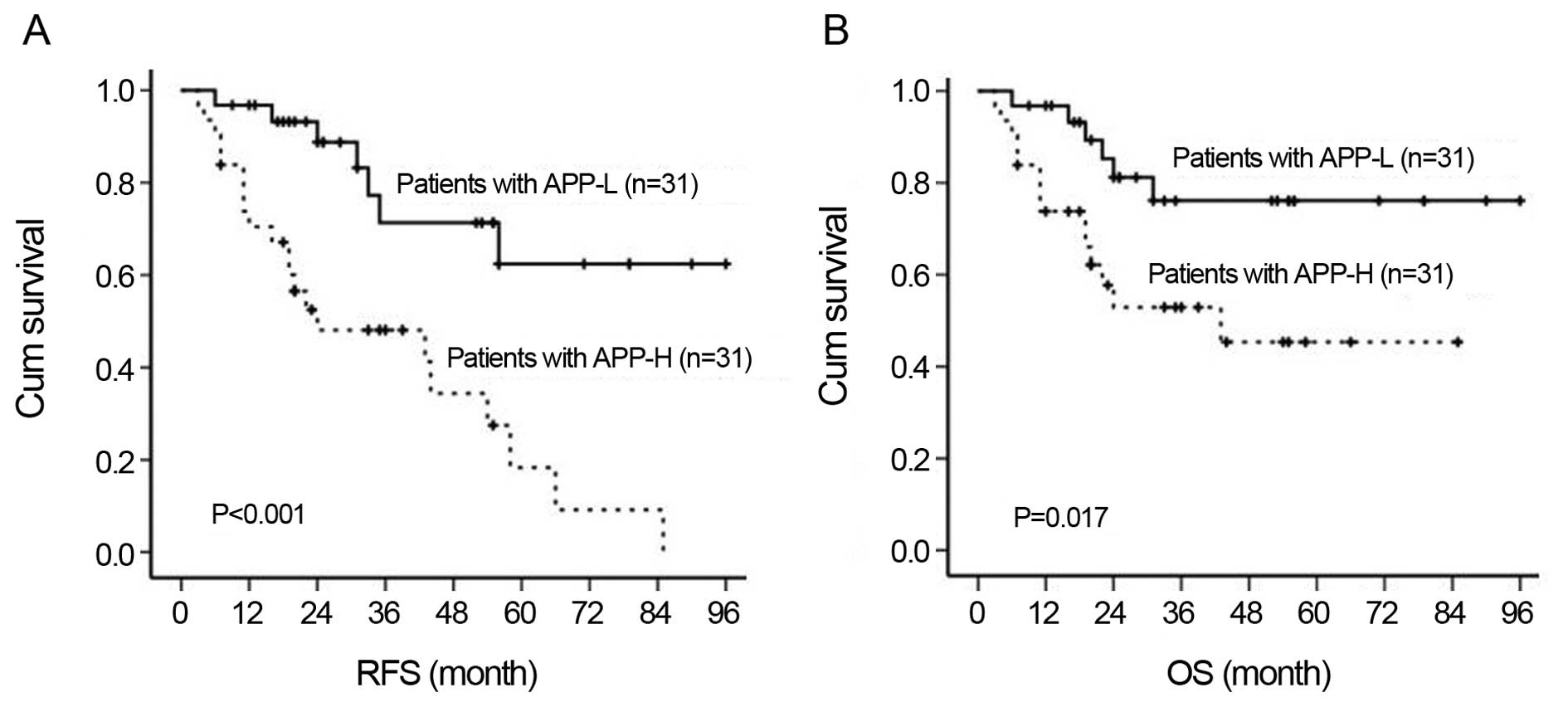
APP overexpression indicated poor disease outcome, in that both the
relapse-free survival (RFS) rate (36.8±5.4 vs. 72.5±7.3%,
P<0.001) and the overall survival (OS) rate (49.7±7.1 vs.
78.0±6.5%, P=0.027) were significantly lower in the APP-H group
than that in the APP-L group (Fig.
2).
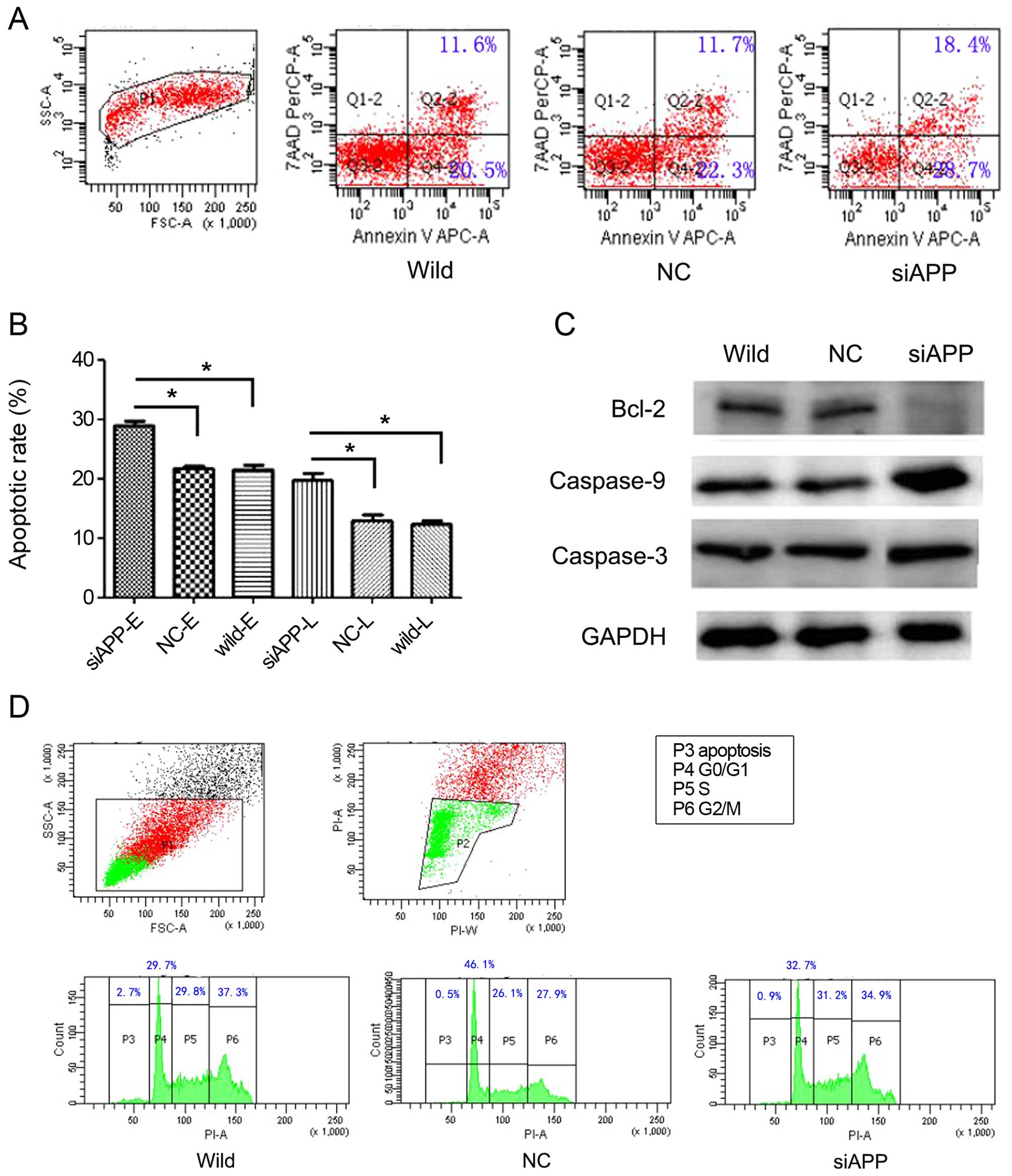
APP is involved in the regulation of cell
apoptosis but not proliferation in Kasumi-1 cells
As APP promotes cell proliferation in some solid
cancers (12,14,15),
we compared the cell cycle distribution of single vs. double
oncogene-expressing cells during the course of ex vivo
expansion. Notably, no obvious differences were observed among the
siAPP, NC and wild-type cells (Fig.
3D), indicating that proliferation was not correlated with APP
expression in AE leukemia. However, cell apoptosis, both early and
late, as estimated by analyzing Annexin-V, increased significantly
when APP was knocked down in the Kasumi-1 cells, in that the early
and late apoptosis rates in the siAPP cells were 29.00±0.98 and
19.80±1.51%, respectively; when compared with 21.43±0.86 and
12.33±0.75% in the wild-type cells and 21.67±0.78 and 12.90±1.25%
in the NC cells, respectively, there were statistical differences
(early apoptosis: F=71.927, P<0.001; late apoptosis rate:
F=35.239, P<0.001, respectively, Fig. 3A and B). In parallel, western
blotting analysis (Fig. 3C)
revealed reduced levels of the anti-apoptotic protein Bcl-2 and
increased levels of activated caspase-3 and caspase-9 in the siAPP
cells, which also implied an increase in the basal apoptosis
rate.
Knockdown of APP downregulates c-KIT
expression
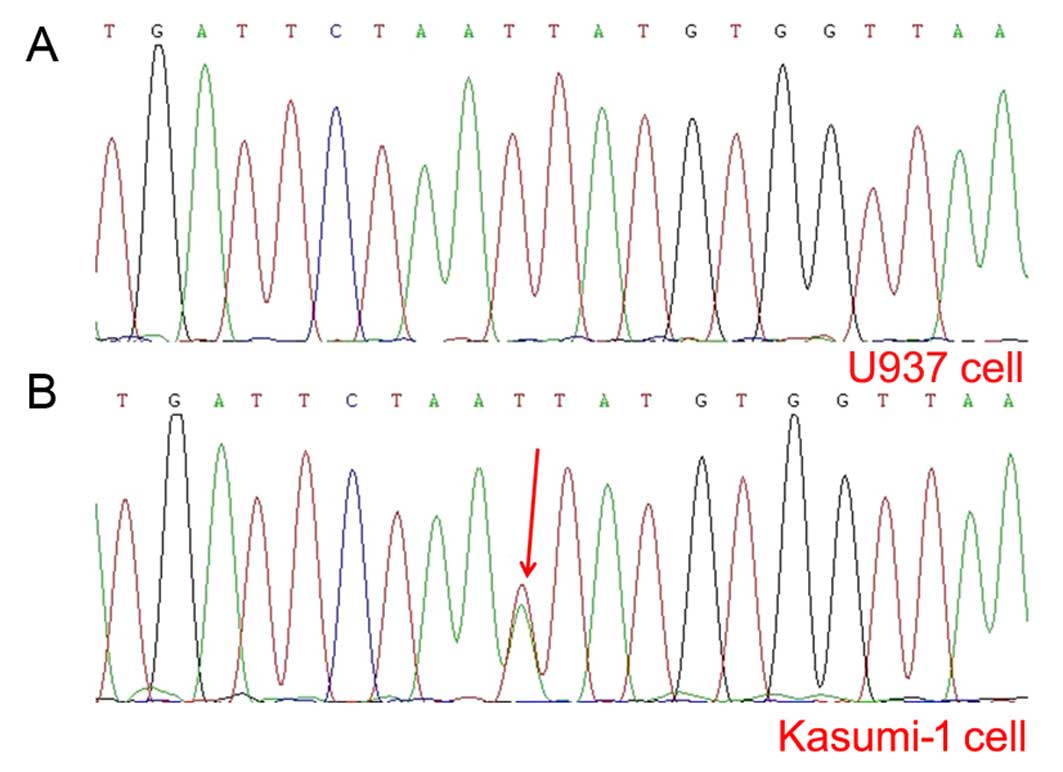
It is known that Kasumi-1 cells harbor c-KIT
mutation (19) and mutated c-KIT
upregulates c-KIT expression and decreases AE leukemia cell
apoptosis (10). Our PCR assay
verified the fact that a c-KIT mutation (N822K) (Fig. 4B) and c-KIT overexpression existed
in the Kasumi-1 cell line. In accordance with the correlation of
APP with c-KIT mutations/overexpression in our clinical
observations, we further analyzed the difference in CD117 or c-KIT
mRNA expression levels among the siAPP, NC and wild-type cells and
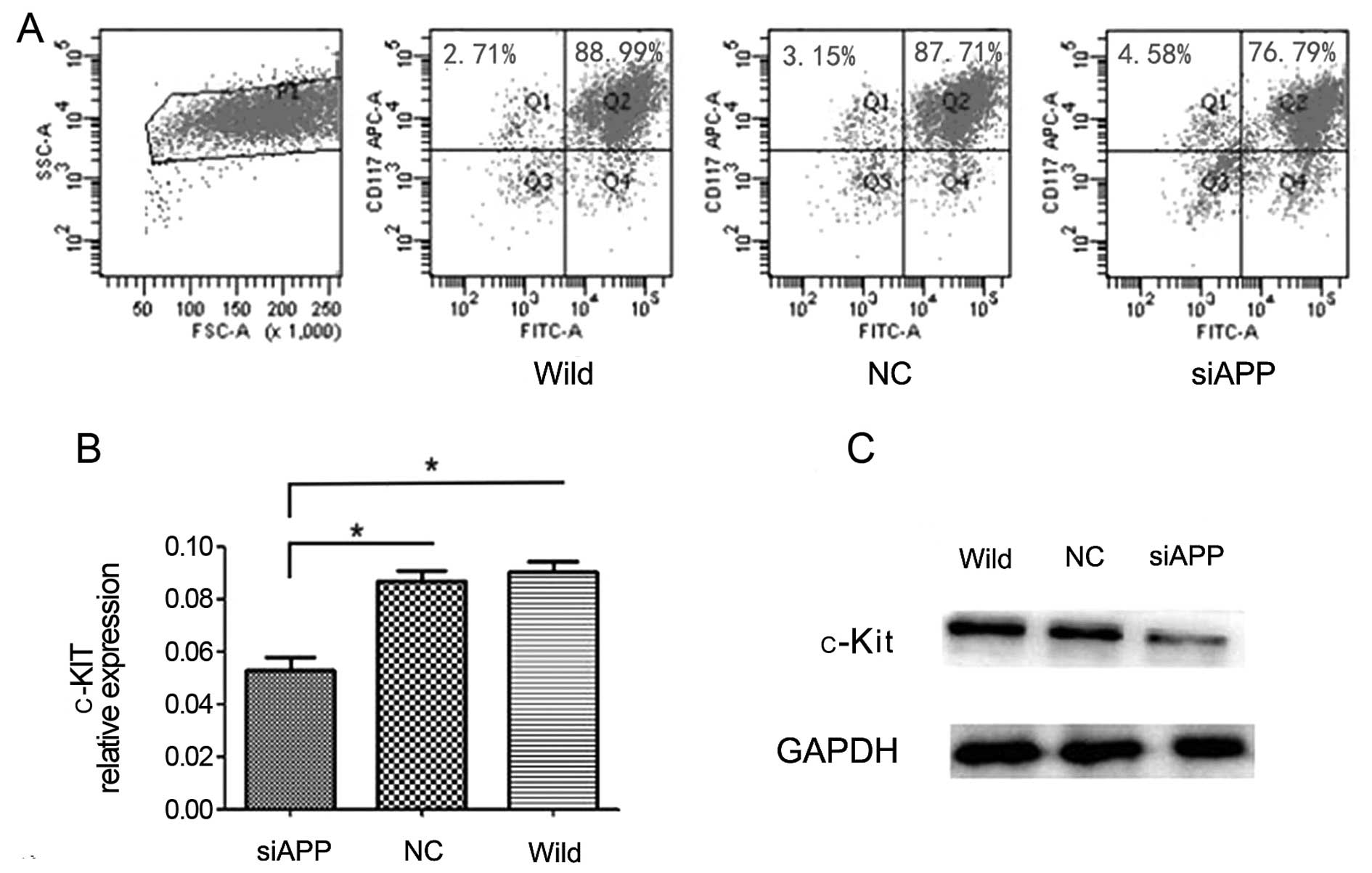
found that both CD117 (by flow cytometric analysis, siAPP:
80.66±0.69%, wild-type: 93.30±0.89%, NC: 91.28±0.42%, respectively;
F=78.71, P=0.006, Fig. 5A) and
c-KIT mRNA expression levels (by qPCR analysis, siAPP:
0.05273±0.00873, wild-type: 0.09008±0.00712, NC: 0.08707±0.00676,
respectively; F=22.46, P=0.002, Fig.
5B) in the siAPP Kasumi-1 cells decreased significantly, as
compared with those in the wild-type and NC Kasumi-1 cells. The
result of western blotting analysis revealed (Fig. 5C) that c-Kit protein decreased when
APP was knocked down further confirming the involvement of APP in
the regulation of c-KIT expression.
siRNA-APP downregulates the PI3K/AKT
signaling pathway
As the PI3K/AKT pathway, enhanced by activated c-KIT
(10), is an intracellular
signaling pathway which is important in the regulation of cell
apoptosis in AE leukemia (20,21),
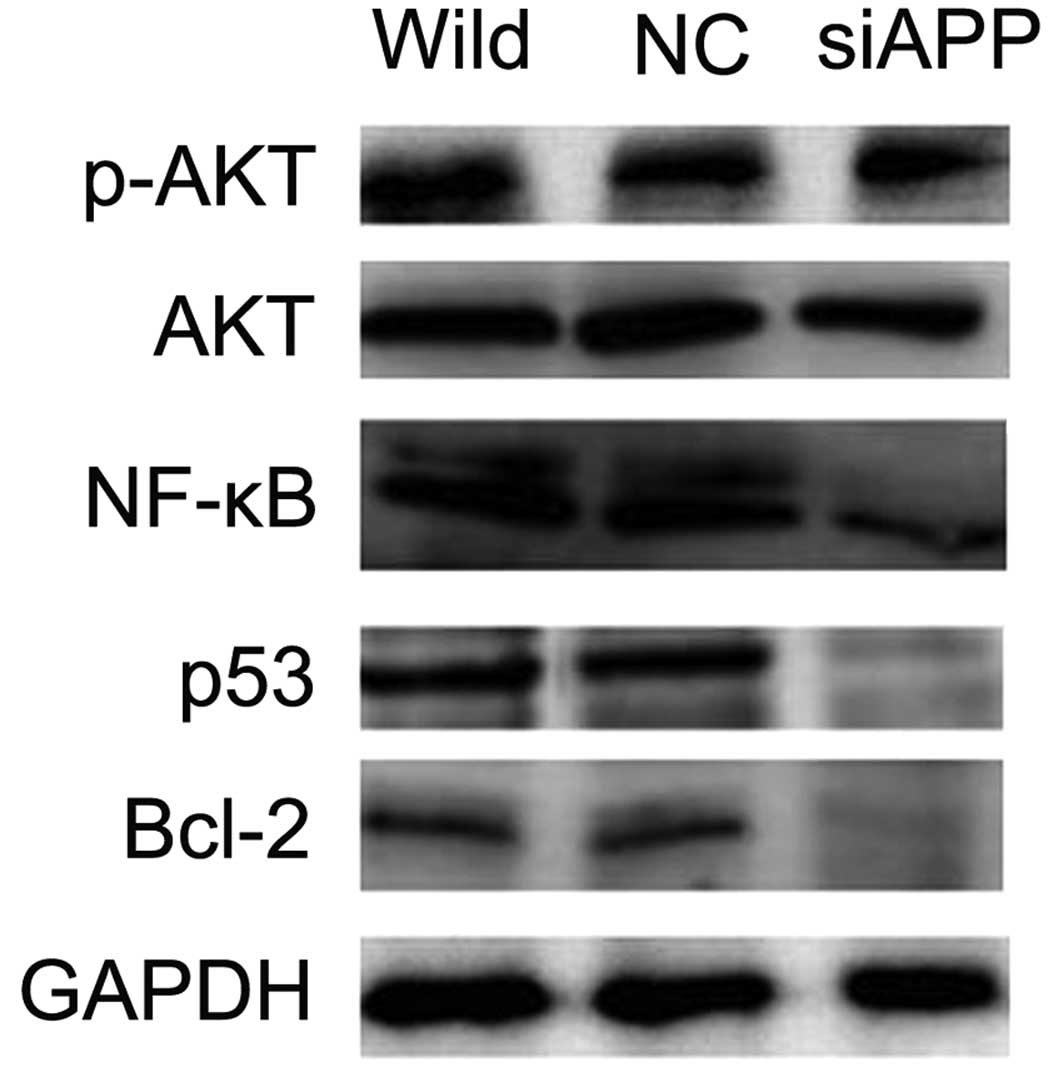
we evaluated the correlation of APP expression levels with the
PI3K/AKT pathway using western blot analysis. The data revealed
decreased levels of AKT phosphorylation concomitant with reduced
transcription factors p53 and NF-κB, both of which are important
downstream receptors in the PI3K/AKT pathway and play a vital role
in the regulation of cell apoptosis (22–24),
in the siAPP Kasumi-1 cells, as compared with the levels in the NC
and wild-type Kasumi-1 cells (Fig.
6), suggesting that APP mediates Kasumi-1 cell apoptosis via
the PI3K/AKT pathway.
Discussion
It is known that APP promotes cancer cell
proliferation and metastasis and has an adverse effect on disease
outcome in various solid cancers (12–15).
The effect of APP on migration and prognosis in AE leukemia was
reported in our previous study (18). In this study, we showed that APP
decreased cell apoptosis but did not promote cell proliferation, in
accordance with the finding that APP may lead to leukemia
progression due to abnormal apoptosis (16); knockdown of APP increased cell
apoptosis but did not significantly affect cell proliferation in AE
leukemia. In parallel, from the clinical data, we observed
significantly a higher peripheral WBC count and bone marrow
cellularity in the APP-H patients than these parameters in the
APP-L cases, while there was no apparent difference in the bone
marrow blasts between the two groups. We also determined that APP
is correlated with c-KIT mutations/overexpression in AE leukemia
patients. In an in vitro study, we further demonstrated the
involvement of APP in the regulation of c-KIT expression and the
PI3K/AKT signaling pathway, which play important roles in cell
apoptosis regulation.
Notably, our clinical data demonstrated that c-KIT
mutations were frequent in the APP-H patients and the expression
levels of APP and c-KIT were positively correlated at the
transcription levels (as evidenced by qPCR analysis). This suggests
that APP is closely correlated with c-KIT mutation/overexpression.
The correlation of APP with c-KIT expression was further confirmed
at both the transcription (as evidenced by qPCR analysis) and
translation (as confirmed by CD117 assay and western blot analysis)
levels in the Kasumi-1 cell experiment. c-KIT mutations which are
considered as one of the most important subsequent events and that
are highly expressed (up to 48%), cooperate with full length
AML1-ETO to induce leukemia, and adversely affect the disease
outcome, in AE leukemia. Since the APP gene is located on 21q21.3
and expresses highly in t(8;21), the correlation of APP with c-KIT
mutations in our data may explain the high incidence of c-KIT
mutations in this subtype of leukemia and indicate the involvement
of APP in the progression of AE leukemia. Furthermore, activating
c-KIT mutations upregulate c-KIT expression and reverse
AML1-ETO-induced DNA damage and apoptosis. Based on these data, we
report that APP may cooperate with c-KIT mutation/overexpression in
the regulation of cell apoptosis in AE leukemia cells.
The PI3K/AKT signaling pathway is of central
importance in AE leukemia and plays an important role in processes
critical for leukemia progression (20,21).
AKT is a major downstream effector molecule of the PI3K/AKT pathway
(25) and increased AKT
phosphorylation implies activation of the PI3K pathway (26). AKT plays a central role in apoptosis
inhibition through its regulatory effects on various downstream
targets such as anti-apoptotic Bcl-2 (27) and activated caspase-3 and -9 and
transcription factors NF-κB and p53 (22-24).
In the present study, we silenced the APP gene and found that p-AKT
and its downstream targets Bcl-2, NF-κB and p53 were significantly
reduced while caspase-3 and caspase-9 were increased, suggesting
that APP regulates cell apoptosis via the PI3K/AKT signaling
pathway. Moreover, activating c-KIT mutations upregulatd the
PI3K/AKT pathway and the PI3K inhibitor increased cell apoptosis in
AE leukemia. These results further support the cooperation of APP
with c-KIT mutation/overexpression in the regulation of cell
apoptosis via the PI3K/AKT pathway in AE leukemia.
In conclusion, APP regulates cell apoptosis but not
cell proliferation in AE leukemia. The regulatory mechanism may
involve the synergism of APP and c-KIT mutation/overexpression to
decrease cell apoptosis via the PI3K/AKT pathway. Its exact
mechanisms require further in-depth study. Moreover, our findings
suggest that APP may be a new biomarker for targeted therapy in AE
leukemia.
Acknowledgments
This study was funded by the National Natural
Science Foundation of China (no. 81500138), the Natural Science
Foundation of Guangdong Province, China (no. 2014A030313270), the
Medical Research Foundation of Guangdong Province, China (no.
B2014250), the Science and Technology Program of Guangzhou, China
(no. 2013J4100109), the Ph.D. Programs Foundation of the Ministry
of Education of China (no. 20124433110001) and the National
High-Tech R&D Program (863 Program) China (no.
2012AA02A505).
References
|
1
|
Rowley JD: Identificaton of a
translocation with quinacrine fluorescence in a patient with acute
leukemia. Ann Genet. 16:109–112. 1973.PubMed/NCBI
|
|
2
|
Yergeau DA, Hetherington CJ, Wang Q, Zhang
P, Sharpe AH, Binder M, Marín-Padilla M, Tenen DG, Speck NA and
Zhang DE: Embryonic lethality and impairment of haematopoiesis in
mice heterozygous for an AML1-ETO fusion gene. Nat Genet.
15:303–306. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Higuchi M, O'Brien D, Kumaravelu P, Lenny
N, Yeoh EJ and Downing JR: Expression of a conditional AML1-ETO
oncogene bypasses embryonic lethality and establishes a murine
model of human t(8;21) acute myeloid leukemia. Cancer Cell.
1:63–74. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Rhoades KL, Hetherington CJ, Harakawa N,
Yergeau DA, Zhou L, Liu LQ, Little MT, Tenen DG and Zhang DE and
Zhang DE: Analysis of the role of AML1-ETO in leukemogenesis, using
an inducible transgenic mouse model. Blood. 96:2108–2115.
2000.PubMed/NCBI
|
|
5
|
Jiao B, Wu CF, Liang Y, Chen HM, Xiong SM,
Chen B, Shi JY, Wang YY, Wang JH, Chen Y, et al: AML1-ETO9a is
correlated with C-KIT overexpression/mutations and indicates poor
disease outcome in t(8;21) acute myeloid leukemia-M2. Leukemia.
23:1598–1604. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Paschka P, Marcucci G, Ruppert AS, Mrózek
K, Chen H, Kittles RA, Vukosavljevic T, Perrotti D, Vardiman JW,
Carroll AJ, et al Cancer and Leukemia Group B: Adverse prognostic
significance of KIT mutations in adult acute myeloid leukemia with
inv(16) and t(8;21): A Cancer and Leukemia Group B Study. J Clin
Oncol. 24:3904–3911. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Schnittger S, Kohl TM, Haferlach T, Kern
W, Hiddemann W, Spiekermann K and Schoch C: KIT-D816 mutations in
AML1-ETO-positive AML are associated with impaired event-free and
overall survival. Blood. 107:1791–1799. 2006. View Article : Google Scholar
|
|
8
|
Wang YY, Zhao LJ, Wu CF, Liu P, Shi L,
Liang Y, Xiong SM, Mi JQ, Chen Z, Ren R, et al: C-KIT mutation
cooperates with full-length AML1-ETO to induce acute myeloid
leukemia in mice. Proc Natl Acad Sci USA. 108:2450–2455. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Nick HJ, Kim HG, Chang CW, Harris KW,
Reddy V and Klug CA: Distinct classes of c-Kit-activating mutations
differ in their ability to promote RUNX1-ETO-associated acute
myeloid leukemia. Blood. 119:1522–1531. 2012. View Article : Google Scholar :
|
|
10
|
Wichmann C, Quagliano-Lo Coco I, Yildiz Ö,
Chen-Wichmann L, Weber H, Syzonenko T, Döring C, Brendel C,
Ponnusamy K, Kinner A, et al: Activating c-KIT mutations confer
oncogenic cooperativity and rescue RUNX1/ETO-induced DNA damage and
apoptosis in human primary CD34+ hematopoietic
progenitors. Leukemia. 29:279–289. 2015. View Article : Google Scholar
|
|
11
|
Zhang MY, Zheng CY, Zou MM, Zhu JW, Zhang
Y, Wang J, Liu CF, Li QF, Xiao ZC, Li S, et al: Lamotrigine
attenuates deficits in synaptic plasticity and accumulation of
amyloid plaques in APP/PS1 transgenic mice. Neurobiol Aging.
35:2713–2725. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Hansel DE, Rahman A, Wehner S, Herzog V,
Yeo CJ and Maitra A: Increased expression and processing of the
Alzheimer amyloid precursor protein in pancreatic cancer may
influence cellular proliferation. Cancer Res. 63:7032–7037.
2003.PubMed/NCBI
|
|
13
|
Ko SY, Lin SC, Chang KW, Wong YK, Liu CJ,
Chi CW and Liu TY: Increased expression of amyloid precursor
protein in oral squamous cell carcinoma. Int J Cancer. 111:727–732.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Krause K, Karger S, Sheu SY, Aigner T,
Kursawe R, Gimm O, Schmid KW, Dralle H and Fuhrer D: Evidence for a
role of the amyloid precursor protein in thyroid carcinogenesis. J
Endocrinol. 198:291–299. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Takayama K, Tsutsumi S, Suzuki T,
Horie-Inoue K, Ikeda K, Kaneshiro K, Fujimura T, Kumagai J, Urano
T, Sakaki Y, et al: Amyloid precursor protein is a primary androgen
target gene that promotes prostate cancer growth. Cancer Res.
69:137–142. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Baldus CD, Liyanarachchi S, Mrózek K, Auer
H, Tanner SM, Guimond M, Ruppert AS, Mohamed N, Davuluri RV,
Caligiuri MA, et al: Acute myeloid leukemia with complex karyotypes
and abnormal chromosome 21: Amplification discloses overexpression
of APP, ETS2, and ERG genes. Proc Natl Acad Sci USA. 101:3915–3920.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang W, Meng FY, Huang ZF, Huang M and Liu
LX: Expression and role of amyloid precrusor protein gene in acute
myeloid leukemia. Chin J Hematol. 31:309–314. 2010.In Chinese.
|
|
18
|
Jiang L, Yu G, Meng W, Wang Z, Meng F and
Ma W: Overexpression of amyloid precursor protein in acute myeloid
leukemia enhances extramedullary infiltration by MMP-2. Tumour
Biol. 34:629–636. 2013. View Article : Google Scholar
|
|
19
|
Larizza L, Magnani I and Beghini A: The
Kasumi-1 cell line: A t(8;21)-kit mutant model for acute myeloid
leukemia. Leuk Lymphoma. 46:247–255. 2005. View Article : Google Scholar
|
|
20
|
Pulikkan JA, Madera D, Xue L, Bradley P,
Landrette SF, Kuo YH, Abbas S, Zhu LJ, Valk P and Castilla LH:
Thrombopoietin/MPL participates in initiating and maintaining
RUNX1-ETO acute myeloid leukemia via PI3K/AKT signaling. Blood.
120:868–879. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kumar A, Fernandez-Capetillo O and Carrera
AC: Nuclear phosphoinositide 3-kinase β controls double-strand
break DNA repair. Proc Natl Acad Sci USA. 107:7491–7496. 2010.
View Article : Google Scholar
|
|
22
|
Sheng S, Qiao M and Pardee AB: Metastasis
and AKT activation. J Cell Physiol. 218:451–454. 2009. View Article : Google Scholar
|
|
23
|
Wang L, Zhao WL, Yan JS, Liu P, Sun HP,
Zhou GB, Weng ZY, Wu WL, Weng XQ, Sun XJ, et al: Eriocalyxin B
induces apoptosis of t(8;21) leukemia cells through NF-κB and MAPK
signaling pathways and triggers degradation of AML1-ETO oncoprotein
in a caspase-3-dependent manner. Cell Death Differ. 14:306–317.
2007. View Article : Google Scholar
|
|
24
|
Krejci O, Wunderlich M, Geiger H, Chou FS,
Schleimer D, Jansen M, Andreassen PR and Mulloy JC: p53 signaling
in response to increased DNA damage sensitizes AML1-ETO cells to
stress-induced death. Blood. 111:2190–2199. 2008. View Article : Google Scholar
|
|
25
|
McCubrey JA, Steelman LS, Chappell WH,
Abrams SL, Montalto G, Cervello M, Nicoletti F, Fagone P, Malaponte
G, Mazzarino MC, et al: Mutations and deregulation of
Ras/Raf/MEK/ERK and PI3K/PTEN/Akt/mTOR cascades which alter therapy
response. Oncotarget. 3:954–987. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Hart JR and Vogt PK: Phosphorylation of
AKT: A mutational analysis. Oncotarget. 2:467–476. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Datta SR, Dudek H, Tao X, Masters S, Fu H,
Gotoh Y and Greenberg ME: Akt phosphorylation of BAD couples
survival signals to the cell-intrinsic death machinery. Cell.
91:231–241. 1997. View Article : Google Scholar : PubMed/NCBI
|