Introduction
Angiofibroma of soft tissue is a recently described
benign fibrovascular tumor of unknown cellular origin (1). It arises most commonly in the
extremities of middle-aged adults but displays a broad anatomic and
age distribution. Microscopically, it is characterized by bland,
uniform, probably fibroblastic spindle cell set in an abundant
fibromyxoid stroma, with a prominent and highly characteristic
vascular pattern composed of innumerable branching, thin-walled
blood vessels (1). Cytogenetic
knowledge about angiofibroma of soft tissue is based on the
analysis of six such tumors of which four showed a balanced
t(5;8)(p15;q12) translocation and a fifth tumor showed a three-way
t(5;8;8)(p15;q13;p11) (1).
Molecular analysis of four tumors carrying the t(5;8)(p15;q12)
showed in-frame AHRR-NCOA2 and
NCOA2-AHHR fusion transcripts in all of them
(2). A GTF2I-NCOA2
fusion gene was detected in a fifth tumor carrying a
t(7;8;14)(q11;q13;q31) as the sole chromosome change (3). To the best of our knowledge, the
above-mentioned tumors are the only angiofibromas of soft tissue
which have been investigated both cytogenetically and molecularly
for fusion genes. An additional angiofibroma of soft tissue with
t(5;8)(p15;q12) was also reported but without molecular analysis
(4). In three other studies,
fluorescence in situ hybridization (FISH) was performed with
probes for NCOA2 showing rearrangements of the NCOA2;
however, no further investigation of fusion genes was performed
(5–7).
We report here an angiofibroma of soft tissue which
had the chromosome translocations t(4;5)(q24;q31) and
t(5;8;17)(p15;q13;q21) and identified the fusion genes generated by
the two translocations. Our data show that, in addition to the
reported AHRR-NCOA2, the tumor carried also other
fusion genes resulting from the chromosomal aberrations that might
have contributed to tumorigenesis as well.
Materials and methods
Ethics statement
The study was approved by the regional Ethics
Committee (Regional komité for medisinsk forskningsetikk Sør-Øst,
Norge; http://helseforskning.etikkom.no), and written
informed consent was obtained from the patient to publication of
the case details. The Ethics Committee's approval included a review
of the consent procedure. All patient information has been
de-identified.
Case history
The patient was a 45-year-old male in whom MRI of
the abdomen and pelvis showed a 53-mm tumor in the right inguinal
region partially surrounding large vessels. The patient had been
aware of the lesion for several years. Surgery was performed with
removal of the entire tumor including part of the right deep
femoral artery with immediate reconstruction of the vessel. The
postoperative period was eventless and to date there is no sign of
tumor relapse.
The specimen (58×45×45 mm) showed an encapsuled,
well-circumscribed tumor with a homogenous gray/white cut surface.
There were no signs of necrosis or bleeding. Routine microscopy
showed a tumorous proliferation of small, spindled cells without
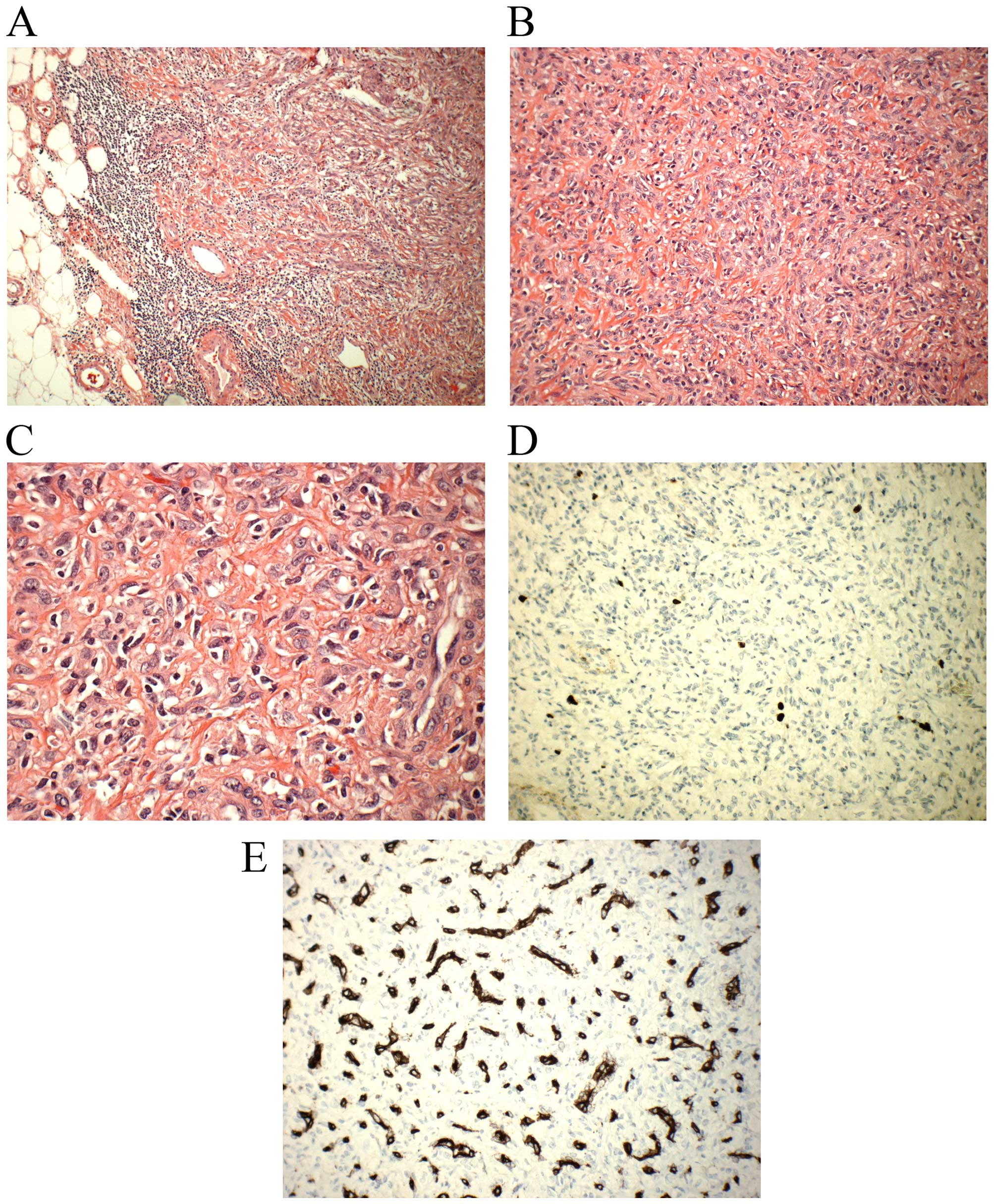
atypia or mitotic activity (Fig.
1A–C). There were a lot of small, thin-walled blood vessels in
the background (Fig. 1A–C).
Immunohistochemical examination showed low proliferative activity
(MIB1/Ki67 <5%) (Fig. 1D) and
the vessels highlighted by the endothelial marker CD34 (Fig. 1E). The clinical setting as well as
histopathological features fit well with a diagnosis of
angiofibroma of soft tissue (1).
G-banding and karyotyping
Fresh tissue from the tumor was processed for
cytogenetic analysis as part of our diagnostic routine. The sample
was disaggregated mechanically and enzymatically with collagenase
II (Worthington Biochemical Corp., Freehold, NJ, USA). The
resulting cells were cultured and harvested using standard
techniques. Chromosome preparations were G-banded with Wright stain
and examined. The karyotype was written according to the
International System for Human Cytogenetic Nomenclature (ISCN) 2013
guidelines (8).
High-throughput paired-end
RNA-sequencing
Total RNA was extracted using miRNeasy Mini Kit
according to the manufacturer's instructions (Qiagen, Hilden,
Germany). Tumor tissue was disrupted and homogenized in QIAzol
Lysis Reagent (Qiagen) using a 5-mm stainless steel bead and
TissueLyser II (Qiagen). Subsequently, total RNA was purified using
QIAcube (Qiagen). The RNA quality was evaluated using the Experion
Automated Electrophoresis System (Bio-Rad Laboratories, Oslo,
Norway). The RNA quality indicator (RQI) was 8.5. Total RNA (3
µg) was sent for high-throughput paired-end RNA-sequencing
at the Norwegian Sequencing Centre, Ullevål Hospital (http://www.sequencing.uio.no/). Detailed information
about the high-throughput paired-end RNA-sequencing was given
elsewhere (9). The software
FusionCatcher (10) (https://github.com/ndaniel/fusioncatcher) was used for
the discovery of fusion transcripts.
Molecular genetic analyses
The primers used for PCR amplification and
sequencing are listed in Table I.
The primer combinations, target fusion transcripts, and results of
PCR amplifications are shown in Table
II. cDNA was synthesized from 2 µg of total RNA in a
20-µl reaction volume using iScript Advanced cDNA Synthesis
Kit for RT-qPCR according to the manufacturer's instructions
(Bio-Rad Laboratories). cDNA was diluted to 100 µl and 2
µl were used as template in subsequent PCR assays. The
25-µl PCR volumes contained 12.5 µl of Premix Taq
(Takara Bio Europe SAS, Saint-Germain-en-laye, France), 1 µl
of diluted cDNA, and 0.4 µM of each of the forward and
reverse primers (Table II). The
quality of the cDNA synthesis was examined by amplification of a
cDNA fragment of the TBCK gene using the primers TBCK-2558F1
and TBCK-2908R1. The PCRs were run on a C1000 Thermal cycler
(Bio-Rad Laboratories) with the following cycling for the
amplifications: an initial denaturation at 94°C for 30 sec, 35
cycles of 7 sec at 98°C, 7 sec at 60°C, 1 min at 72°C, and a final
extension for 5 min at 72°C.
 | Table IPrimers used for PCR amplification
and Sanger sequencing analyses. |
Table I
Primers used for PCR amplification
and Sanger sequencing analyses.
| Name | Sequence
(5′→3′) | Position | Reference
sequence | Gene |
|---|
| TBCK-2908R1 |
TGGCGTGGATATGAAGAACTGTGC | 2931–2908 | NM_033115.4 | TBCK |
| TBCK-2558F1 |
CCTGGTGGTTGACATCCGGAATAG | 2558–2581 | NM_033115.4 | TBCK |
| P4HA2-785R1 |
AGCCAGGTAGCCCTCAGCATCAG | 807–785 | NM_004199.2 | P4HA2 |
| P4HA2-33F1 |
CCGCGGGAGGTTCTGGAAAC | 33–52 | NM_001142598.1 | P4HA2 |
|
NCOA2-intr14-R1 |
CACCATGTCGAGACTGCTGGCTC |
71106777–71106799 | NC_018919.2 | NCOA2 |
| NCOA2-3364R1 |
TCACTCGGAGACTCAGCTGCAGG | 3386–3364 | NM_006540.2 | NCOA2 |
| NCOA2-2858F1 |
CTGGACCTTTCCCACCAATCAGAA | 2858–2881 | NM_006540.2 | NCOA2 |
| ETV4-1496R1 |
GGGGCTCTCATCCAAGTGGGAC | 1517–1496 | NM_001986.2 | ETV4 |
| ETV4-863F1 |
TGGGGTCAATGGGCACAGGTAC | 863–884 | NM_001986.2 | ETV4 |
| AHRR-1932R1 |
TGCAGGGTGGAAAGGGGTCAG | 1952–1932 | NM_020731.4 | AHRR |
| AHRR-1503F1 |
AGCAGACCCATGCGGGATGTC | 1503–1523 | NM_020731.4 | AHRR |
| AHRR-1425F1 |
TGTGTCCAGGGCACTTTCAGGAA | 1425–1447 | NM_020731.4 | AHRR |
| EGFL7-353F1 |
ACCCCAAAGCCACATCTGTAGCC | 353–375 | NM_016215.4 | EGFL7 |
| MCF2l-3271R1 |
CGCCACGACCGTGTATTTACCTG | 3293–3271 | NM_024979.4 | MCF2L |
| CYP1B1-132F1 |
TCAACGCTGTGAGGAAACCTCGA | 132–154 | NM_000104.3 | CYP1B1 |
| CLU-1164R1 |
GACCTGGAGGGATTCGTCGAGC | 1185–1164 | NM_001831.3 | CLU |
 | Table IIPrimer combinations, target fusion
transcripts and results of PCR amplification. |
Table II
Primer combinations, target fusion
transcripts and results of PCR amplification.
| Primer
combination | Target fusion
transcripts | Results |
|---|
|
P4HA2-33F1/TBCK-2908R1 |
P4HA2-TBCK | Positive |
|
TBCK-2558F1/P4HA2-785R1 |
TBCK-P4HA2 | Positive |
|
AHRR-1503F1/NCOA2-intr14-R1 |
AHRR-NCOA2 | Positive |
|
AHRR-1425F1/NCOA2-3364R1 |
AHRR-NCOA2 | Positive |
|
ETV4-863F1/AHRR-1932R1 |
ETV4-AHRR | Positive |
|
NCOA2-2858F1/ETV4-1496R1 |
NCOA2-ETV4 | Positive |
|
EGFL7-353F1/MCF2L-3271R1 |
EGFL7-MCF2L | Negative |
|
CYP1B1-132F1/CLU-1164R1 |
CYP1B1-CLU | Negative |
The PCR products were analyzed on a QIAxcel Advanced
System according to the manufacturer's instructions (Qiagen). The
remaining PCR products were purified using the QIAquick PCR
Purification Kit or the QIAquick Gel Extraction Kit (both from
Qiagen) and direct sequenced using the dideoxy procedure with the
ABI Prism BigDye Terminator v1.1 Cycle Sequencing Kit (Applied
Biosystems, Foster City, CA, USA) on the Applied Biosystems 3500
Genetic Analyzer sequencing system. The BLAST software (http://www.ncbi.nlm.nih.gov/BLAST/) was used for
computer analysis of the sequence data.
Results
Cytogenetic analysis
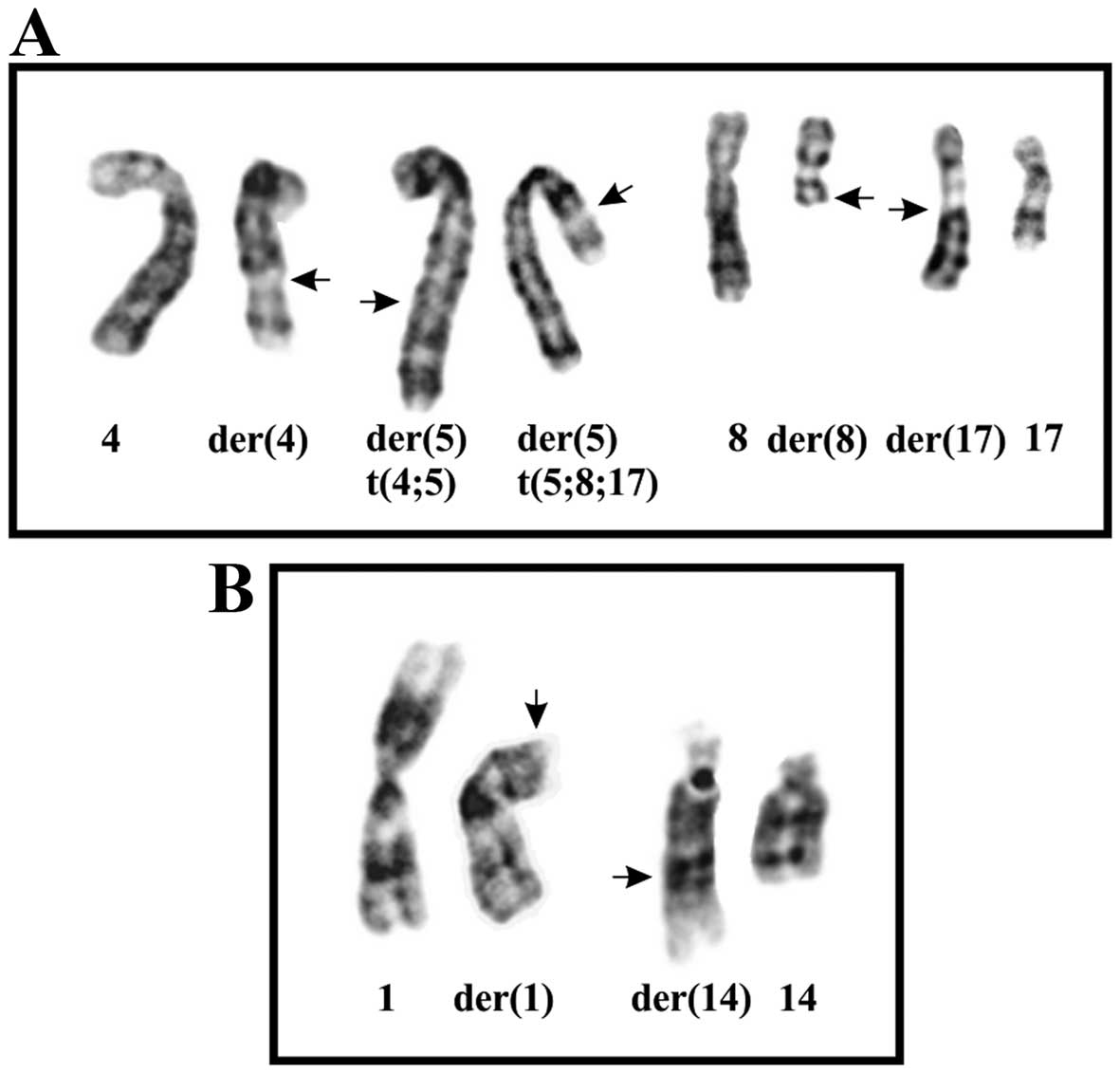
The G-banding analysis showed that the tumor had two
cytogenetically unrelated clones. The first clone, found in eight
metaphases, had the t(4;5)(q24;q31) and t(5;8;17)(p15;q13;q21)
chromosome aberrations (Fig. 2A).
The second, found in two metaphases, had the t(1;14)(p31;q32)
abnormality (Fig. 2B). This yielded
the following karyotype: 46,XY,t(4;5)(q24;q31),t(5;8;17)
(p15;q13;q21)[8]/46,XY,t(1;14)(p31;q32)[2]/46,XY[3].
High-throughput paired-end RNA-sequencing
analysis
Using the FusionCatcher software with the FASTQ
files obtained from the Norwegian Sequencing Centre, Ullevål
Hospital (http://www.sequencing.uio.no/), 39 potential fusions
were found: 28 fusions were described as readthrough short-distance
fusions and 5 as pseudogenes (Table
III). Among the other fusions, the program detected the
P4HA2-TBCK and the reciprocal
TBCK-P4HA2. According to the UCSC genome Browser on
Human, Feb. 2009, (GRCh37/hg19) assembly (http://genome-euro.ucsc.edu/cgi-bin/hggateway),
P4HA2 maps on chromosome subband 5q31.1 and TBCK on
band 4q24. Thus, the two fusions P4HA2-TBCK and the
reciprocal TBCK-P4HA2 most probably were the result
of the balanced chromosome translocation t(4;5)(q24;q31).
FusionCatcher also detected AHRR-NCOA2 and
ETV4-AHRR which correspond to the three-way
t(5;8;17)(p15;q13;q21) found in the tumor. The three genes
AHRR, NCOA2, and ETV4 map to chromosome
subbands 5p15.33, 8q13.3, and 17q21.31, respectively (https://genome.ucsc.edu/). In the three-way t(5;8;17),
the moving of 5p15 to 8q13 generated the AHRR-NCOA2
fusion whereas the translocation of 17q21 to 5p15 generated the
ETV4-AHRR. We assume that the moving of 8q13 to 17q21
would have generated an NCOA2-ETV4 fusion but no such
fusion was, for unknown reasons, detected by FusionCatcher. The
fusion transcrips EGFL7-MCF2L and a
CYP1B1-CLU were also detected by the analysis with
FusionCatcher, in all likelihood generated by t(9;13)(q34;q34) and
t(2;8)(p22.2;p21.1), respectively. No fusion gene corresponding to
the cytogenetically detected t(1;14)(p31;q32) was found.
 | Table IIIFusion transcripts detected using
FusionCatcher. |
Table III
Fusion transcripts detected using
FusionCatcher.
| 5′-Partner
gene | 3′-Partner
gene | Fusion
description | Fusion
sequence |
|---|
| PCDP1 | TMEM177 | Readthrough |
ATTCTAGAATGAAAGTCACCAGTAG*gaaagggaacatcacagaaaggtga |
|
MIR155HG | JAM2 | Readthrough |
CAAGGAGACGCTCCTGGCACTGCAG*atcataaggcctatgggttttctgc |
| GOLT1A | KISS1 | Readthrough |
ATGATCTCCATCACCGAATGGCAGA*cctcaaggcacttctaggacctgcc |
| SHISA9 |
U91319.1 | Readthrough |
AAGTACGCCTCCTTAAAGGCAGTCG*agctggaacacccttcttctcctgc |
| VPS45 | PLEKHO1 | Readthrough |
GCACCACAGTGCACAACACGAAAAG*ggacctcaggatggaaaccagcagc |
|
P4HA2 |
TBCK | |
AACGCCGGGAGCTGCGAGTGTCCAG*tttgcagctcaccttgtgaagatga |
|
TBCK |
P4HA2 | |
GCATGTGGCAAAACACACAGCTGAG*acacttccctctgtgaccatgaaac |
| ADCK4 | NUMBL | Readthrough |
TCCAGCCTCTCAGTGTGTTGGAGAG*acggggcgggcaccatgaacaagtt |
|
ETV4 |
AHRR | |
AAGGTCAGAGAAGTGACTGTTGATG*ggggacctgtgtggtccgacgctgc |
| FOSB | PPM1N | Readthrough |
TCCACCCACCGCCGCCGCCTCCCAG*aaggggcaggatggggctgggaagt |
| MFSD7 | ATP5I | Readthrough |
GGGGAGGATCCACTTGACTGGACAG*attacctaaaacctcgggcagaaga |
| DPY19L2 |
DPY19L2P2 | Pseudogene |
TTCTTCATCTTTGTTAATGACGTGG*ctaattcaaggtagtgcctggtggt |
|
DPY19L2P2 | DPY19L2 | Pseudogene |
TTCTTCATCTTTGTTAATGACATGG*ctaattcaaggtagtgcctggtggt |
| MATR3 | PAIP2 | Readthrough |
CCGCGTCCCGCTCGCTGGGAGAGAG*gttaaaaacgacaaccaacatcagc |
|
LINC00893 |
LINC00894 | Antisense |
AGGAAGCAGGAATGCTGGAGATGAG*acggagttttgctcttgttgcccag |
| PTPRG | C3ORF14 | Readthrough |
GAGGCCTGGAGTATTCACAGACATT*ggcaagcactttaaccttttaagcc |
| SIX3 |
AC012354.6 | Readthrough |
AGACACCGGCACCTCCATCCTCTCG*acaaggccacctacatcccaagcca |
| CTBS | GNG5 | Readthrough |
GCGGGCTCCTTATTATAACTATAAA*gtttcccaggcagctgcagacttga |
| CYP1B1 | CLU | Readthrough |
CGAGTGGGAGTTAAAGCTTCCAGTG*aaggcgacgatgaccggactgtgtg |
| ZBTB16 | NNMT | Readthrough |
CGGGACCCCCTCAGCCTCATTTCTG*aagggctgaactgatggaaggaatg |
|
KB-1507C5.4 |
ATP6V1C1 | Readthrough |
TCCATGTCGTAAGTTACACAAGAAG*aatctctcttgatttttgaggaaat |
| PPP1R21 | STON1 | Readthrough |
TGACACACTAAAGATGTCCAGTAAg*gagggagcgctctcccctcctctgg |
| SUZ12 | SUZ12P | Pseudogene |
GAAACTCCAGAACAAACATCAAAAG*cttgtcagctcatttgcagcttaca |
| SUZ12P | SUZ12 | Pseudogene |
AAATGACAGTATTTGATAAAAACAG*aggctgcctccattcgaaacatttt |
| TREM2 | TREML1 | Readthrough |
CTGCTCATCTTACTCTTTGTCACAG*catccccttgatctggggtgctgtg |
| TRIM2 | MND1 | Readthrough |
CGACTGGGGAAACAGCAGGATCCAG*tcaaagaaaaaaggactgagtgcag |
|
AC015977.6 | CIB4 | Readthrough |
GGTTCTGCCCAGAAGCCAGCTGCAG*gccctgaccttcctgaccagaaatga |
|
AHRR |
NCOA2 | |
GCAAGGTGTACCGATGCCTCCGGGG*ttcaacagaaaattatcttttggaa |
| CHD4 | NOP2 | Readthrough |
GGCACCCGAACCTACCCCACAGCAG*taccatggggcgcaagttggaccct |
| EGFL7 | MCF2L | |
GGGATGACTGATTCTCCTCCGCCAG*gttggagcaaaacgtcccactcact |
| GPR65 |
LINC01146 | Readthrough |
AAACACATCACCGGAAGAAATATGG*atgatgcatatcataaattattact |
| HERC3 |
FAM13A-AS1 | Readthrough |
AATTCTACATGATTAAAGAATCCAT*ccctttacagaaaacaactgaccaa |
|
KB-1572G7.2 |
AP000347.4 | Readthrough |
ACACCACTCTTCCTGTTGGCCCAAG*gtcagcccaagactaccccgtcggt |
| LCAT | PSMB10 | Readthrough |
TGAATAAAGACCTTCCTTTGCTACC*agtacccagtgagcagcacagaggg |
| LSP1 | TNNT3 | Short-distance |
CCGGCTCCCTAGGCGTCCCATCTCG*aaaccacccaccttcaccatgtctg |
| LTBP2 | NPC2 | Readthrough |
GATGCGGCCCACATGGCCTGCGTAG*gttctgtggatggagttataaagga |
| OSBPL2 | ADRM1 | Readthrough |
GGTTGCAAGCTGAGAACATCCAGAG*gaacccaagacagaccaggatgagg |
| PARL | MAP6D1 | Readthrough |
ATCTTGGGGGAGCTCTTTTTGGAAT*acaggaattccaggcttggactgga |
| PTPN22 | RSBN1 | Readthrough |
AACTCCAGCTCATTTCTGAATTTTG*aaacaccagatgaaaatggtaaaac |
We decided to investigate with molecular methods the
described fusion transcripts. No other fusions were examined.
Molecular genetic confirmation of
fusions
PCR with the primers TBCK-2558F1 and TBCK-2908R1
amplified a cDNA of the TBCK gene indicating that the
synthesized cDNA was of good quality.
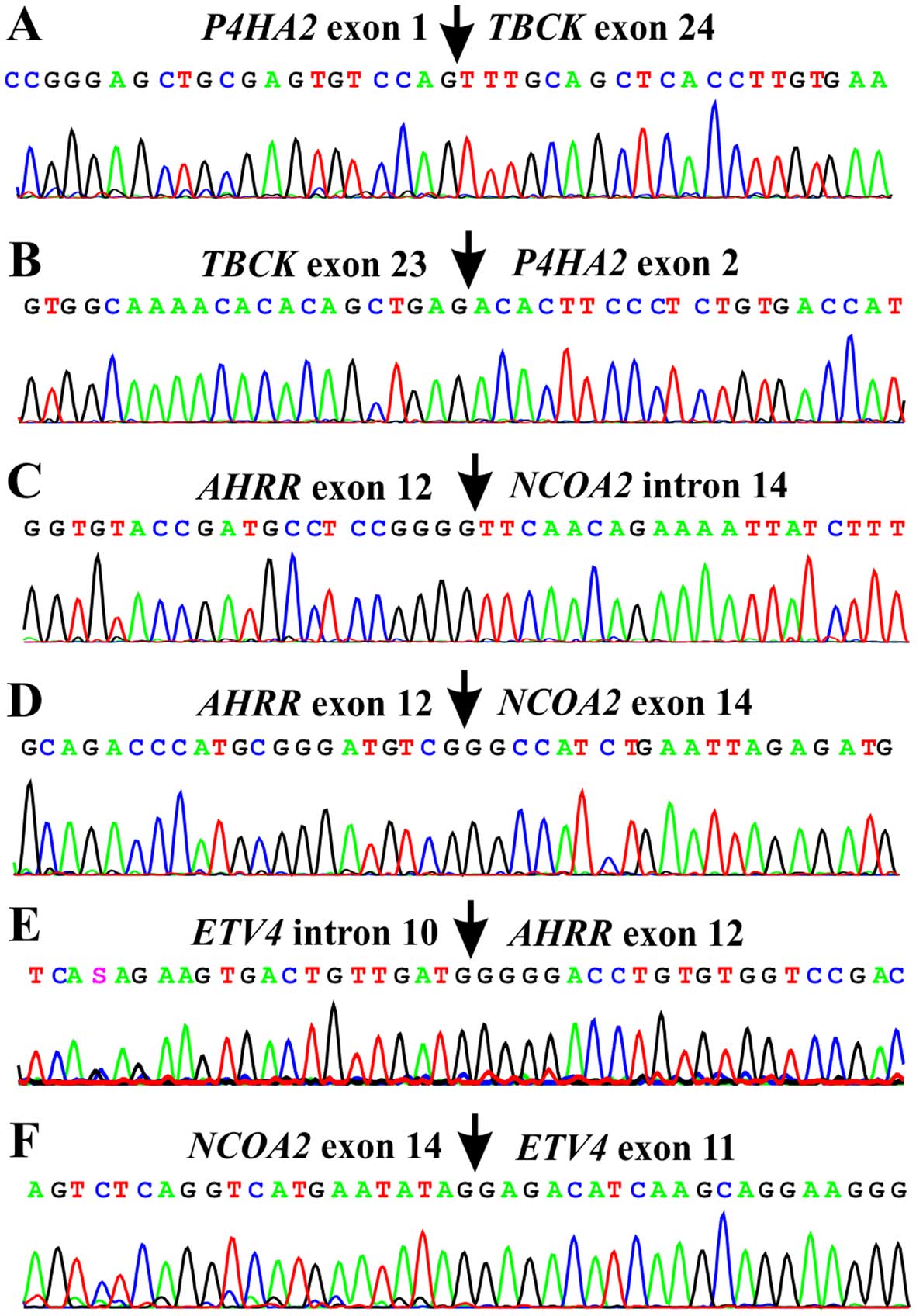
RT-PCR using cDNA from the tumor and subsequent
direct Sanger sequencing verified the presence of the
P4HA2-TBCK, TBCK-P4HA2,
AHRR-NCOA2, ETV4-AHRR, and
NCOA2-ETV4 fusion transcripts (Table II and Fig. 3). TBCK-P4HA2,
AHRR-NCOA2, and NCOA2-ETV4 were
in-frame fusions which would code for chimeric proteins. The
detected ETV4-AHRR fusion, on the other hand, was
out-of-frame and would not produce a chimeric protein, nor would
the P4HA2-TBCK code for any functional protein. No
EGFL7-MCF2L or CYP1B1-CLU fusion
transcript was found by RT-PCR amplification (Table II).
Discussion
The examined angiofibroma of soft tissue carried the
recurrent AHRR-NCOA2 fusion transcript but lacked the
reciprocal NCOA2-AHRR. This finding supports the
initial suggestion that AHRR-NCOA2 is the
pathogenetically significant fusion transcript in tumors carrying a
t(5;8)(p15;q12) (2,3). While we were examining the current
tumor, a report was published describing 13 cases of angiofibroma
of soft tissue with an AHRR-NCOA2 but with only eight
of them carrying the reciprocal NCOA2-AHRR (11). Current data therefore agree that the
AHRR-NCOA2 fusion gene is recurrent in angiofibroma
of soft tissue [(2,3,11),
present case] and indicate that this is the pathogenetically
crucial outcome of the t(5;8).
Using FISH on formalin-fixed, paraffin-embedded
specimens, Sugita et al (5)
found that 16–36% of the tumor cells showed NCOA2
rearrangement. A fairly small proportion of NCOA2 gene
rearrangement-positive cells (4–12 split signals per 50 tumor cell
nuclei) was recently reported also by Yamada et al (11). The split signals were mostly
detected in relatively large, spindle-shaped nuclei, indicating
that these were the ones belonging to the neoplastic parenchyma
(11).
The present tumor had two cytogenetically unrelated
clones: one (eight metaphases) with the translocations
t(4;5)(q24;q31) and t(5;8;17)(p15;q13;q21) and another (2 cells)
with t(1;14)(p31;q32) as the sole chromosome abnormality. Thus, our
data not only are in agreement with previous observations that only
a fraction of tumor cells carry the NCOA2 gene
rearrangement, but also demonstrate genetic heterogeneity of
uncertain pathogenetic significance within the tumor. Although no
fusion gene was found corresponding to t(1;14)(p31;q32), this
should not lead us to conclude that the translocation was
pathogenetically unimportant. The t(1;14)(p31;q32) chromosome
aberration may exert its influence through a position effect
causing deregulation of a gene in the proximity of the breakpoints.
Alternatively, the current methodology may be unable to detect a
fusion gene as has been demonstrated (9).
So far, three types of AHRR-NCOA2
fusion transcripts have been described: in the first type, exon 9
of AHRR is joined with exon 16 of NCOA2, the second
type shows exon 10 of AHRR being joined to exon 14 of
NCOA2, and in the third type there is an insertion of an
intronic sequence from the NCOA2 gene between exon 9 of
AHRR and exon 14 of NCOA2 (2,11). In
the present angiofibroma of soft tissue, two novel fusion
transcripts were found with different fusion positions from those
previously described: a fusion transcript in which nt 1670
(sequence with accession no. NM_020731) from exon 12 of the
AHRR gene was fused with a sequence from intron 14 of
NCOA2 and a transcript in which nt 1533 (also from exon 12)
of AHRR was fused to exon 15 of NCOA2 (sequence with
accession no. NM_006540.2). The resulting putative AHRR-NCOA2
protein would be similar to those reported (2) in as much as the C-terminal part of
AHRR is replaced by the C-terminal part of NCOA2.
The involvement of NCOA2 in neoplasia was
first reported in acute myeloid leukemia with the cytogenetic
inversion inv(8)(p11q13) which
resulted in a KAT6A-NCOA2, also known as
MOZ-TIF2 fusion gene (12,13).
Since then, NCOA2 has been implicated also in other
malignancies. A fusion between ETV6 (TEL) and
NCOA2 was reported in childhood leukemia with the recurrent
t(8;12)(q13;p13) (14). A
PAX3-NCOA2 gene was found as a rare variant fusion in
alveolar rhabdomyosarcoma; it was brought about by a
t(2;8)(q35;q13) translocation (15). A HEY1-NCOA2 fusion
gene was described in mesenchymal chondrosarcomas (16,17).
Recently, SRF-NCOA2, TEAD1-NCOA2, and
VGLL2-NCOA2 fusions were reported in
rhabdomyosarcomas (18,19). In all the above-mentioned fusions,
NCOA2 is the 3′-partner gene and all fusion proteins contain
the two C-terminal activation domains AD1/CID (activation domain
1/CREB-binding protein interacting domain) and AD2 (2,3,12–19).
The transforming activities of KAT6A-NCOA2 and
PAX3-NCOA2 have been demonstrated experimentally
(15,20). In addition,
KAT6A-NCOA2 was shown to induce acute myeloid
leukemia in transgenic fish (21).
Deguchi et al (20) showed
that the KAT6A-NCOA2 interaction with CREBBP through
AD1/CID is essential for transformation. Similarly, Sumegi et
al (15) showed that while
deletion of the AD2 portion of PAX3-NCOA2 fusion protein reduced
the transforming activity, deletion of the AD1/CID domain fully
abrogated the transforming activity of the chimeric protein. Thus,
the AD1/CID and AD2 domains of NCOA2 seem to be essential for the
transformation ability of the various fusion proteins.
The three-way translocation t(5;8;17)(p15;q13;q21)
of the present case not only generated an AHRR-NCOA2
resulting from the translocation of 5p15 to 8q13, but also two
additional fusion genes: an NCOA2-ETV4, stemming from
the moving of 8q13 to 17q21, and an ETV4-AHRR,
generated by the moving of 17q21 to 5p15. The detected
ETV4-AHRR fusion transcript is out-of-frame and so
cannot produce a chimeric protein. The NCOA2-ETV4
fusion transcript is in-frame coding for a chimeric NCOA2-ETV4
protein, the oncogenetic potential of which cannot be ruled out.
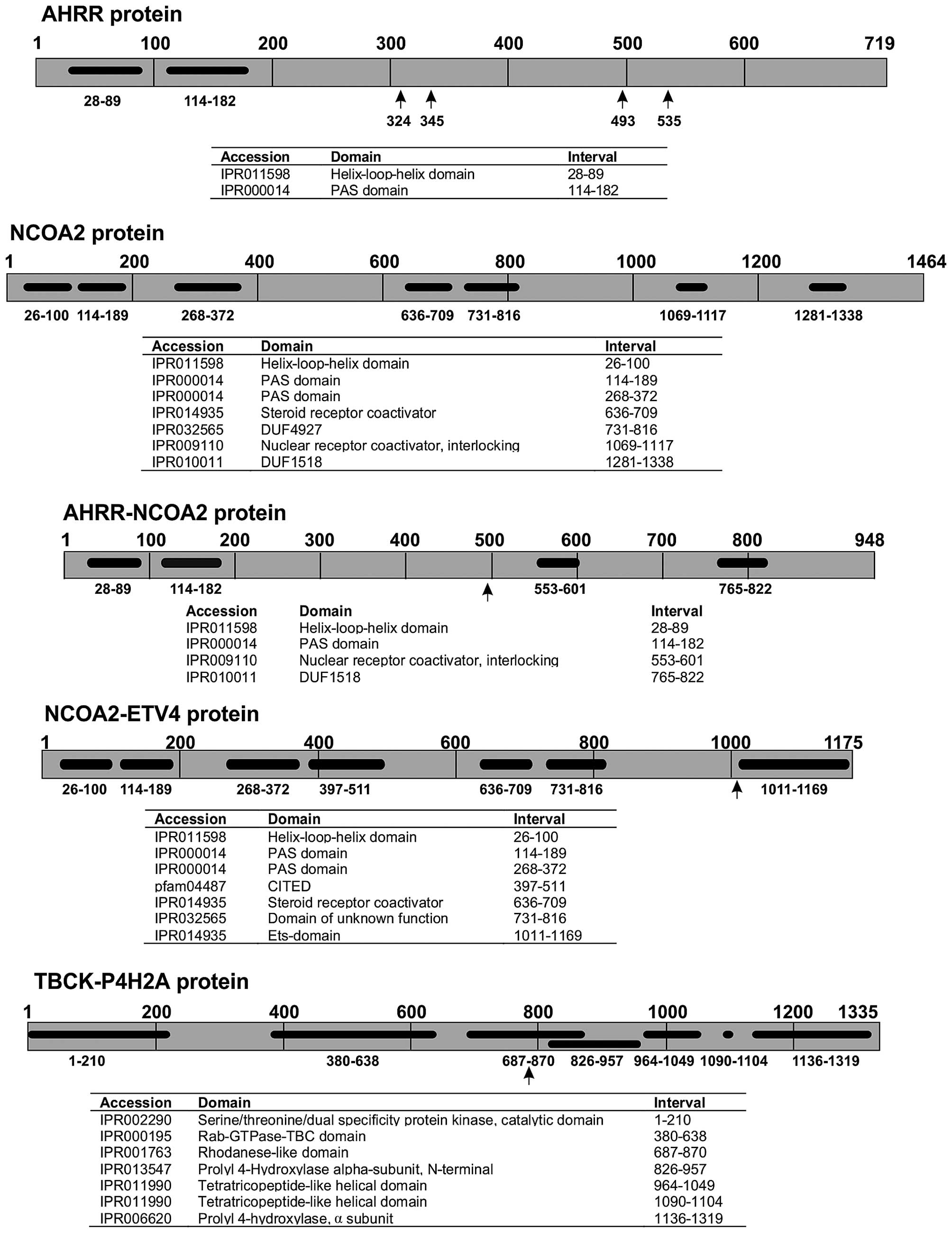
Based on the NCOA2 and ETV4 proteins with accession nos.
NP_006531.1 and NM_001986.2, respectively, the chimeric NCOA2-ETV4
would contain 1,175 amino acids. The NCOA2 N-terminal part of the
protein would contain the helix-loop-helix, PAS_9 and PAS_11, the
CITED, and the SRC-1 domains. The ETV4 C-terminal part would
contain the ETS DNA-binding domain of ETV4 (Fig. 4).
ETV4 was reported to contribute the 3′-part of the
oncogenic protein in the subset of Ewing's sarcomas characterized
by a t(17;22)(q12;q12) translocation (22,23).
The EWSR1-ETV4 protein, in which the N-terminal part of EWSR1 is
fused to the ETS DNA-binding domain of ETV4, has an oncogenetic
potential similar to that of the EWSR1-FLI1, EWSR1-ERg, EWSR1-FEV,
and EWSR1-ETV1 fusion proteins which may also be found in Ewing's
sarcoma (24). The ETV4 gene
was also described as the 3′-partner in fusion genes found in
prostate carcinoma (25–27). ETV4 was found to fuse with
the TMPRSS2, KLK2, CANT1, and DDX5
(25–27). All these fusions genes,
TMPRSS2-ETV4, KLK2-ETV4,
CANT1-ETV4, and DDX5-ETV4, contain
(like the present NCOA2-ETV4) the part of ETV4 coding
for the ETS DNA-binding domain.
The chromosome translocation t(4;5)(q24;q31)
generated the P4HA2-TBCK and TBCK-P4HA2
fusion transcripts. P4HA2-TBCK does not encode any
functional protein, whereas TBCK-P4HA2 encodes a
chimeric 1,335-amino acid protein. TBCK-P4HA2 would contain the
first 794 out of 830 amino acids of the TBCK protein (accession no.
NP_149106.2), 6 amino acids from the untranslated region of exon 2
of P4HA2 (accession no. NM_004199.2), and the entire 535 amino
acid-P4HA2 protein (NP_004190.1). The function of this putative
chimeric protein is difficult to predict since it would contain
both the protein kinase domain, the Rhodanese-like domain, and the
Tre-2/Bub2/Cdc16 (TBC) domain of TBCK together with the P4HA2
protein which is a component of the prolyl 4-hydroxylase. The TBCK
protein is thought to play a role in actin organization, cell
growth, and cell proliferation by regulating the mammalian target
of the rapamycin (mTOR) signaling pathway. This protein may also be
involved in the transcriptional regulation of the components of the
mTOR complex (http://www.ncbi.nlm.nih.gov/gene/93627). Depletion of
TBCK significantly inhibits cell proliferation, reduces cell size,
and disrupts the organization of actin but not microtubule.
Knockdown of TBCK induces a significant decrease in the protein
levels of components of mTOR complex (mTORC) and suppresses the
activity of mTOR signaling, but not the MAPK or PDK1/Akt pathway
(28).
The protein encoded by the P4HA2 gene is one
of several different types of α subunit of the prolyl 4-hydroxylase
and provides the major part of the catalytic site of the active
enzyme (http://www.ncbi.nlm.nih.gov/gene/8974). In collagen
and related proteins, prolyl 4-hydroxylase catalyzes the formation
of 4-hydroxyproline that is essential to the proper
three-dimensional folding of newly synthesized procollagen chains.
In breast cancer, P4HA2 was shown to promote progression and
metastasis by regulating collagen deposition (29). In squamous cell carcinoma of the
oral cavity, P4HA2 was identified as a metastasis associated
protein (30).
In spite of the now repeatedly documented
recurrence of AHRR-NCOA2 in angiofibroma of soft
tissue [present case, (2,11)], our findings indicate that also
additional genetic events, some of which lead to fusion genes, may
be important in tumor development. Worthy of mention is that of the
eight hitherto cytogenetically reported tumors, including the
present case, three had three-way translocations (1–3). What
lies behind this highly unusual feature is unknown. Obviously, more
such tumors must be studied cytogenetically and molecularly before
all important aspects of their pathogenesis are laid bare.
Acknowledgments
The authors would like to thank Hege Kilen Andersen
and Nina Øino for their excellent technical assistance. This study
was supported by grants from the Norwegian Radium Hospital Research
Foundation.
References
|
1
|
Mariño-Enríquez A and Fletcher CD:
Angiofibroma of soft tissue: Clinicopathologic characterization of
a distinctive benign fibrovascular neoplasm in a series of 37
cases. Am J Surg Pathol. 36:500–508. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jin Y, Möller E, Nord KH, Mandahl N, Von
Steyern FV, Domanski HA, Mariño-Enríquez A, Magnusson L, Nilsson J,
Sciot R, et al: Fusion of the AHRR and NCOA2 genes through a
recurrent translocation t(5;8)(p15;q13) in soft tissue angiofibroma
results in upregulation of aryl hydrocarbon receptor target genes.
Genes Chromosomes Cancer. 51:510–520. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Arbajian E, Magnusson L, Mertens F,
Domanski HA, Vult von Steyern F and Nord KH: A novel GTF2I/NCOA2
fusion gene emphasizes the role of NCOA2 in soft tissue
angiofibroma development. Genes Chromosomes Cancer. 52:330–331.
2013. View Article : Google Scholar
|
|
4
|
Schoolmeester JK, Sukov WR, Aubry MC and
Folpe AL: Angiofibroma of soft tissue: Core needle biopsy
diagnosis, with cytogenetic confirmation. Am J Surg Pathol.
36:1421–1423. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Sugita S, Aoyama T, Kondo K, Keira Y,
Ogino J, Nakanishi K, Kaya M, Emori M, Tsukahara T, Nakajima H, et
al: Diagnostic utility of NCOA2 fluorescence in situ hybridization
and Stat6 immunohistochemistry staining for soft tissue
angiofibroma and morphologically similar fibrovascular tumors. Hum
Pathol. 45:1588–1596. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Fukuda Y, Motoi T, Kato I, Ikegami M,
Funata N, Ohtomo R, Horiguchi S, Goto T and Hishima T: Angiofibroma
of soft tissue with fibrohistiocytic features and intratumor
genetic heterogeneity of NCOA2 gene rearrangement revealed by
chromogenic in situ hybridization: A case report. Pathol Int.
64:237–242. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Edgar MA, Lauer SR, Bridge JA and Rizzo M:
Soft tissue angiofibroma: Report of 2 cases of a recently described
tumor. Hum Pathol. 44:438–441. 2013. View Article : Google Scholar
|
|
8
|
Schaffer LG, McGowan-Jordan J and Schmid
M: ISCN 2013: An International System for Human Cytogenetic
Nomenclature (2013): Recommendations of the International Standing
Committee on Human Cytogenetic Nomenclature. 1st edition. S. Karger
AG; Basel: 2013
|
|
9
|
Panagopoulos I, Gorunova L, Bjerkehagen B
and Heim S: The ʻgrepʼ command but not FusionMap, FusionFinder or
ChimeraScan captures the CIC-DUX4 fusion gene from whole
transcriptome sequencing data on a small round cell tumor with
t(4;19)(q35;q13). PLoS One. 9:e994392014. View Article : Google Scholar
|
|
10
|
Nicorici D, Satalan M, Edgren H,
Kangaspeska S, Murumagi A, Kallioniemi O, Virtanen S and Kilkku O:
FusionCatcher - a tool for finding somatic fusion genes in
paired-end RNA-sequencing data. bioRxiv 011650. http://dx.doi.org/10.1101/011650.
|
|
11
|
Yamada Y, Yamamoto H, Kohashi K, Ishii T,
Iura K, Maekawa A, Bekki H, Otsuka H, Yamashita K, Tanaka H, et al:
Histological spectrum of angiofibroma of soft tissue: Histological
and genetic analysis of 13 cases. Histopathology. Feb 4–2016.Epub
ahead of print. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Carapeti M, Aguiar RC, Goldman JM and
Cross NC: A novel fusion between MOZ and the nuclear receptor
coactivator TIF2 in acute myeloid leukemia. Blood. 91:3127–3133.
1998.PubMed/NCBI
|
|
13
|
Carapeti M, Aguiar RC, Watmore AE, Goldman
JM and Cross NC: Consistent fusion of MOZ and TIF2 in AML with
inv(8)(p11q13). Cancer Genet Cytogenet. 113:70–72. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Strehl S, Nebral K, König M, Harbott J,
Strobl H, Ratei R, Struski S, Bielorai B, Lessard M, Zimmermann M,
et al: ETV6-NCOA2: A novel fusion gene in acute leukemia associated
with coexpression of T-lymphoid and myeloid markers and frequent
NOTCH1 mutations. Clin Cancer Res. 14:977–983. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Sumegi J, Streblow R, Frayer RW, Dal Cin
P, Rosenberg A, Meloni-Ehrig A and Bridge JA: Recurrent t(2;2) and
t(2;8) translocations in rhabdomyosarcoma without the canonical
PAX-FOXO1 fuse PAX3 to members of the nuclear receptor
transcriptional coactivator family. Genes Chromosomes Cancer.
49:224–236. 2010.
|
|
16
|
Panagopoulos I, Thorsen J, Gorunova L,
Micci F and Heim S: Sequential combination of karyotyping and
RNA-sequencing in the search for cancer-specific fusion genes. Int
J Biochem Cell Biol. 53:462–465. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang L, Motoi T, Khanin R, Olshen A,
Mertens F, Bridge J, Dal Cin P, Antonescu CR, Singer S, Hameed M,
et al: Identification of a novel, recurrent HEY1-NCOA2 fusion in
mesenchymal chondrosarcoma based on a genome-wide screen of
exon-level expression data. Genes Chromosomes Cancer. 51:127–139.
2012. View Article : Google Scholar
|
|
18
|
Alaggio R, Zhang L, Sung YS, Huang SC,
Chen CL, Bisogno G, Zin A, Agaram NP, LaQuaglia MP, Wexler LH, et
al: A molecular study of pediatric spindle and sclerosing
rhabdomyosarcoma: Identification of novel and recurrent
VGLL2-related fusions in infantile cases. Am J Surg Pathol.
40:224–235. 2016.
|
|
19
|
Mosquera JM, Sboner A, Zhang L,
Kitabayashi N, Chen CL, Sung YS, Wexler LH, LaQuaglia MP, Edelman
M, Sreekantaiah C, et al: Recurrent NCOA2 gene rearrangements in
congenital/infantile spindle cell rhabdomyosarcoma. Genes
Chromosomes Cancer. 52:538–550. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Deguchi K, Ayton PM, Carapeti M, Kutok JL,
Snyder CS, Williams IR, Cross NC, Glass CK, Cleary ML and Gilliland
DG: MOZ-TIF2-induced acute myeloid leukemia requires the MOZ
nucleosome binding motif and TIF2-mediated recruitment of CBP.
Cancer Cell. 3:259–271. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhuravleva J, Paggetti J, Martin L,
Hammann A, Solary E, Bastie JN and Delva L: MOZ/TIF2-induced acute
myeloid leukaemia in transgenic fish. Br J Haematol. 143:378–382.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kaneko Y, Yoshida K, Handa M, Toyoda Y,
Nishihira H, Tanaka Y, Sasaki Y, Ishida S, Higashino F and Fujinaga
K: Fusion of an ETS-family gene, EIAF, to EWS by t(17;22)(q12;q12)
chromosome translocation in an undifferentiated sarcoma of infancy.
Genes Chromosomes Cancer. 15:115–121. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Urano F, Umezawa A, Hong W, Kikuchi H and
Hata J: A novel chimera gene between EWS and E1A-F, encoding the
adenovirus E1A enhancer-binding protein, in extraosseous Ewing's
sarcoma. Biochem Biophys Res Commun. 219:608–612. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Braunreiter CL, Hancock JD, Coffin CM,
Boucher KM and Lessnick SL: Expression of EWS-ETS fusions in NIH3T3
cells reveals significant differences to Ewing's sarcoma. Cell
Cycle. 5:2753–2759. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Tomlins SA, Mehra R, Rhodes DR, Smith LR,
Roulston D, Helgeson BE, Cao X, Wei JT, Rubin MA, Shah RB, et al:
TMPRSS2:ETV4 gene fusions define a third molecular subtype of
prostate cancer. Cancer Res. 66:3396–3400. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Han B, Mehra R, Dhanasekaran SM, Yu J,
Menon A, Lonigro RJ, Wang X, Gong Y, Wang L, Shankar S, et al: A
fluorescence in situ hybridization screen for E26
transformation-specific aberrations: Identification of DDX5-ETV4
fusion protein in prostate cancer. Cancer Res. 68:7629–7637. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Hermans KG, Bressers AA, van der Korput
HA, Dits NF, Jenster G and Trapman J: Two unique novel
prostate-specific and androgen-regulated fusion partners of ETV4 in
prostate cancer. Cancer Res. 68:3094–3098. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Liu Y, Yan X and Zhou T: TBCK influences
cell proliferation, cell size and mTOR signaling pathway. PLoS One.
8:e713492013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Xiong G, Deng L, Zhu J, Rychahou PG and Xu
R: Prolyl-4-hydroxylase α subunit 2 promotes breast cancer
progression and metastasis by regulating collagen deposition. BMC
Cancer. 14:12014. View Article : Google Scholar
|
|
30
|
Chang KP, Yu JS, Chien KY, Lee CW, Liang
Y, Liao CT, Yen TC, Lee LY, Huang LL, Liu SC, et al: Identification
of PRDX4 and P4HA2 as metastasis-associated proteins in oral cavity
squamous cell carcinoma by comparative tissue proteomics of
microdissected specimens using iTRAQ technology. J Proteome Res.
10:4935–4947. 2011. View Article : Google Scholar : PubMed/NCBI
|