Introduction
Cancer is now accepted to be a genetic disease in
the sense that it arises due to acquired genetic abnormalities in
susceptible somatic cells (1).
Microscopic studies of cancer cells have shown that these
aberrations are often visible as balanced chromosomal changes, such
as translocations and inversions, as well as unbalanced anomalies,
such as deletions, monosomies, duplications, and trisomies
(1). Many hematologic malignancies,
including acute myeloid leukemia (AML) and acute lymphoblastic
leukemia (ALL), are characterized by the presence of acquired
chromosome translocations and inversions resulting in chimeric
genes of pathogenetic, diagnostic, and prognostic importance
(1). Whereas some genes, e.g.,
ABL, BCR, RUNX1T1, and PML, have only been reported
involved in one or a few translocations, other genes are
promiscuous, having numerous fusion partners in various
translocations and even in different types of malignancy suggesting
that the pathogenetic and phenotypic impact of the chimeras is
dependent on both genes participating in the fusion (1).
One such gene is RUNX1 at 21q22 (2) which codes for the alpha subunit of the
heterodimeric transcription factor named core binding factor (CBF)
that binds to the core element of many enhancers and promoters. To
date, RUNX1 (previously called AML1, CBFA2, PEBP2aB)
has been shown in both myeloid and lymphoblastic acute leukemias to
fuse with more than 30 different partner genes encoding a
heterogeneous group of structurally diverse proteins (1). Recently, RUNX1 fusions were
also found in adenocarcinoma of breast and lung as well as in
squamous cell carcinoma of the oral cavity (3). Some of the fusions are common, such as
ETV6-RUNX1 [t(12;21)(p13;q22)] in pre-B-ALL,
RUNX1-RUNX1T1 [t(8;21) (q22;q22)] in AML, and
RUNX1/MECOM [t(3;21)(q26;q22)] in myelodysplasia (MDS), AML,
and chronic myeloid leukemia in blastic phase, whereas others have
been reported in single cases, i.e., they have not yet been shown
to be recurrent (2,4). The prognostic impact of the common
RUNX1 fusions is well known (5–8).
Corresponding knowledge for the infrequent RUNX1 chimeras is
lacking (9).
Acquired point mutations distributed throughout
RUNX1 are also frequently found in both de novo and
secondary (therapy-related) MDS/AML (10,11).
They are not found together with RUNX1 chromosomal
translocations or complex abnormal karyotypes, and they are
associated with poor outcome in MDS (12–16).
The mutation spectrum includes missense, nonsense, frameshift,
in-frame insertion/deletion mutations, as well as exon-skipping
mutations (15). Nonsense mutations
in RUNX1 account for 11% of the total and generate a
repertoire of truncated RUNX1 proteins which to varying degree show
lack of the C-terminal region. Most of them affect the
transactivation domain (15).
Although less frequent, truncated RUNX1 proteins can
also be the result of a chromosomal translocation which generates a
premature stop codon in the RUNX1 open reading frame,
leading to expression of C-terminal truncated forms. These
chromosome translocations can be divided into two categories: in
the first, the translocations produce only out-of-frame fusion
transcripts (17–25) whereas, in the second category, they
generate both in-frame and out-of-frame fusion transcripts
(26–31).
The generation of C-terminally truncated RUNX1
proteins via different mechanisms suggests that their expression is
important in leukemogenesis. Truncated RUNX1 protein was shown to
reduce the transactivation capacity of CBF on specific myeloid
promoters that function as inhibitors of normal RUNX1 (18–20).
Recently, the truncated RUNX1 protein resulting from the
t(1;21)(p32;q22) chromosomal translocation was shown to impair
proliferation and differentiation of human hematopoietic
progenitors (25).
Since acute leukemia treatment protocols are in part
based on the presence of certain genetic changes, it is of clinical
interest to obtain more information also about rare RUNX1
fusions, even in disease subgroups that so far cannot be treated
with medications specifically directed against the leukemogenic
defect. It is important to underscore that this may be the case
also for infrequent pathogenetic mechanisms where information is
gathered by the addition of single case reports, as recently
exemplified by the story of the rare RUNX1-USP42 fusion and
5q deletion in AML (9,32–35).
For this reason, we here present the molecular
genetic and clinical features of a case of AML with a cryptic
t(6;21)(q25;q22) which resulted in the generation of a truncated
RUNX1.
Patient and methods
Ethics statement
The study was approved by the regional ethics
committee (Regional komité for medisinsk forskningsetikk Sør-Øst,
Norge, http://helseforskning.etikkom.no), and written
informed consent was obtained from the patient's parents to
publication of the case details. The ethics committee's approval
included a review of the consent procedure. All patient information
has been de-identified.
Case report
A 7-year-old girl was admitted to the Children's
Hospital because of petechiae. Prior to admission she had a one
week history of fever, throat and abdominal pain and had been
prescribed antibiotics on the suspicion of tonsillitis. On clinical
examination, the girl was pale and had petechiae on the extremities
and trunk, as well as a few hematomas on the legs. The peripheral
blood values were hemoglobin 88 g/l, leukocytes
369.0×109/l, platelets 59×109/l, lactate
dehydrogenase 1886 U/L, and C-reactive protein 71 mg/l. She had
continuous epistaxis despite sustained platelet counts of
60×109 cells/l, normal international normalized ratio
(INR), and activated partial thromboplastin time (APTT). There was
no central nervous system involvement. Leucocytes gradually
increased to 480.0×109 cells/l before start of the
treatment.
Morphology and immunophenotypic findings were in
keeping with the diagnosis acute myeloid leukemia with minimal
differentiation (AML M0). Normal hematopoiesis was completely
replaced by large blasts without conspicuous granulation or Auer
rods and with lacy chromatin and prominent nucleoli. The blasts
were positive for CD34, CD71, CD117, CD123, HLA-DR antigens, and
the common myeloid markers CD13, CD33, and CD15. Less than 10% of
the blasts were positive for cytoplasmic myeloperoxidase. Of
interest, partial expression of Tdt and aberrant expression of CD7
and CD9 were demonstrated. The blasts were negative for B-cell,
T/NK-cell as well as for monocytic, erythroid, and megakaryocytic
lineage markers.
The bone marrow karyotype was 47,XX,+4[15] (see
below). In addition, a FLT3 ITD mutation was detected, but no
mutations in the nucleophosmin 1 gene. Upon induction treatment
according to the NOPHO-AML 2004 protocol (NOPHO: Nordic Pediatric
Hematology and Oncology) (36),
morphologic remission (<5% blasts) was obtained. Due to the
presence of a FLT3-ITD mutation, the patient became eligible for
allogeneic stem cell transplantation (SCT). However, because a
suitable donor was not found, consolidation therapy was completed
with chemotherapy only. Four months after completed therapy, the
patient had a bone marrow relapse. She went into a second remission
on a clofarabin-based regimen and was transplanted with stem cells
from her 7-month-old matching sibling. Unfortunately, she relapsed
again 6 months after SCT and died one month later.
G-banding analysis
Bone marrow cells were cytogenetically investigated
by standard methods. Chromosome preparations were made from
metaphase cells of a 24-h culture, G-banded using Leishman stain,
and karyotyped according to the ISCN 2009 guidelines (37).
Fluorescence in situ hybridization
(FISH)
As part of our standard cytogenetic diagnosis,
initial interphase FISH analyses of bone marrow cells were
performed with the Cytocell multiprobe ALL panel (Cytocell,
http://www.cytocell.co.uk/) looking for
MYC rearrangements, CDKN2A (P16) deletion, TCF3
(E2A) rearrangements, ETV6-RUNX1 fusion, hyperdiploidy,
MLL rearrangements, BCR-ABL1 fusion, and IGH
rearrangements. On the basis of findings made using the above
panel, further FISH was performed on metaphase spreads and
interphase nuclei using the Vysis LSI TEL/AML1 ES Dual Color
Translocation Probe (Abbott Molecular, http://www.abbottmolecular.com). This is a mixture of
the LSI TEL probe labeled with SpectrumGreen and the LSI AML1 probe
labeled with SpectrumOrange. Fluorescent signals were captured and
analyzed using the CytoVision system (Leica Biosystems, Newcastle,
UK).
RNA sequencing
Total RNA (3 μg) extracted from the patient's
bone marrow at the time of diagnosis was sent to the Norwegian
Sequencing Centre at Ullevål Hospital (http://www.sequencing.uio.no/) for high-throughput
paired-end RNA-sequencing. The Illumina software pipeline was used
to process image data into raw sequencing data. Only sequence reads
marked as 'passed filtering' were used in the downstream data
analysis. A total of 103 million reads were obtained. The FASTQC
software was used for quality control of the raw sequence data
(http://www.bioinformatics.babraham.ac.uk/projects/fastqc/).
The software deFuse was used for the discovery of fusion
transcripts (38) (http://compbio.bccrc.ca/software/defuse/).
In addition, the 'grep' command (http://en.wikipedia.org/wiki/Grep) was used to
search the fastq files of the sequence data (http://en.wikipedia.org/wiki/FASTQ_format) for
RUNX1 fusion sequences (NM_001754 version 4). To confirm the
RUNX1 fusion identified by the deFuse program (see below),
the 'expression' used was 'CAGATGCAGGAAGACTTTTG' which is a
sequence of 20 nucleotides (nt) at the fusion point: 10 bases
upstream (5′-end of RUNX1 gene, CAGATGCAGG), and 10 bases
downstream from the junction (3′-end of the 6q25 intergenic
sequence, AAGACTTTTG). The sequences obtained by 'grep' were
blasted against the human genomic plus transcript database
(http://blast.ncbi.nlm.nih.gov/Blast.cgi) as well as
the reference sequences NM_001754 version 4 (RUNX1) and
NC_000006.12 (chromosome 6).
PCR analysis
For reverse transcriptase-Polymerase Chain Reaction
(RT-PCR), 1 μg of total RNA was reverse-transcribed in a 20
μl reaction volume using iScript Advanced cDNA Synthesis kit
for RT-qPCR according to the manufacturer's instructions (Bio-Rad
Laboratories, Oslo, Norway). The cDNA was diluted to 50 μl
of which 1 μl was used as templates in subsequent PCR
assays. The 25 μl PCR volume contained 12.5 μl Premix
Ex Taq™ DNA Polymerase Hot Start Version (Takara Bio, AH
diagnostics, Oslo, Norway), cDNA, and 0.4 μM of each of the
forward and reverse primers. For detection of the RUNX1
fusion transcript, the forward RUNX1-809N-F1 (CGG CAG AAA CTA GAT
GAT CAG ACC A) and reverse 6q25-R1 (TCC TTC AAG CAG CAA AAT CTG TGA
G) primers were used. The PCR was run on a C-1000 Thermal cycler
(Bio-Rad) with an initial denaturation at 94°C for 30 sec, followed
by 35 cycles of 7 sec at 98°C, 30 sec at 60°C, 1 min at 72°C, and a
final extension for 5 min at 72°C. PCR products (3 μl) were
stained with GelRed (Biotium, Hayward, CA, USA), analyzed by
electrophoresis through 1.0% agarose gel, and photographed. DNA gel
electrophoresis was performed using lithium borate buffer (39). The remaining PCR products were
purified using the GeneJET PCR Purification kit (Thermo Fisher
Scientific, Oslo, Norway) and sequenced at GATC Biotech (Germany,
http://www.gatc-biotech.com/en/home.html). The BLAST
software (http://blast.ncbi.nlm.nih.gov/Blast.cgi) was used for
computer analysis of sequence data.
Results
Cytogenetics
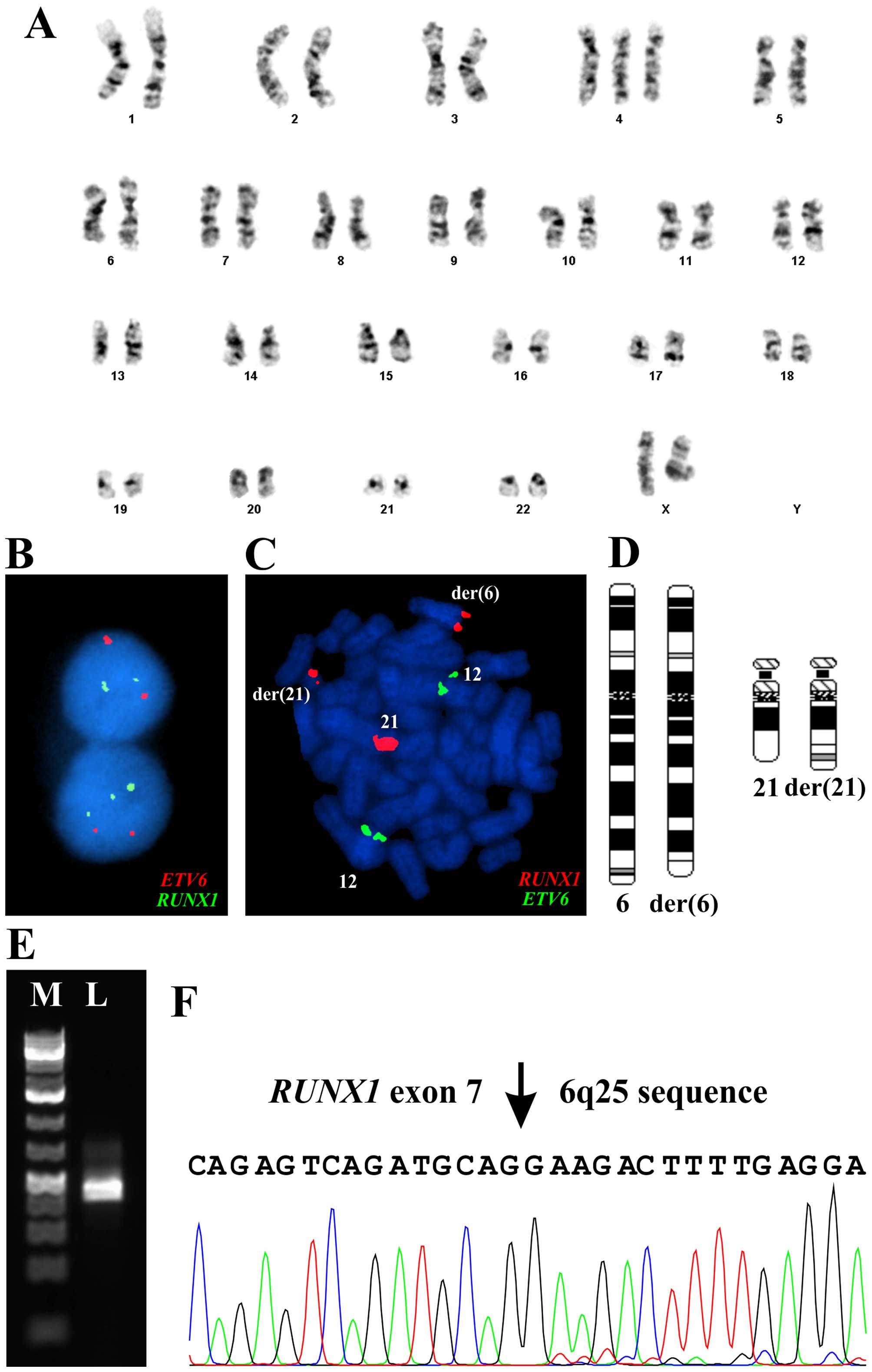
The G-banding analysis at diagnosis showed trisomy 4
in all 15 cells analyzed (Fig. 1A).
The ETV6-RUNX1 probe showed abnormal signals with splitting
of the RUNX1 probe in 203 out of 233 interphase nuclei
examined in spite of no cytogenetically visible rearrangement of
chromosome arm 21q (Fig. 1B). In
the same experiment, 10 metaphase cells were examined in which part
of the RUNX1 probe was unexpectedly seen to be located on
the distal part of 6q (Fig. 1C).
The data showed a novel cryptic t(6;21)(q25-27;q22) chromosome
translocation (Fig. 1D). Other FISH
analyses detected no rearrangements of MYC, TCF3, MLL, and
IGH, no CDKN2A (P16) deletion, no hyperdiploidy, and
none of the fusions ETV6-RUNX1 and BCR-ABL1.
Therefore, the whole karyotype was: 47,XX,+4[15].nuc
ish(ETV6x2,AML1x3) [209/233].ish
t(6;21)(q25-27;q22)(AML1+;AML1+)[10] (Fig. 1A–D).
Analysis of RNA-sequencing with
defuse
Using deFuse on the raw sequencing data, 39
potential fusion transcripts were found (data not shown), among
them a fusion between RUNX1 and a sequence mapping close to
the MIR1202 locus which corresponds well to the 6q breakpoint of
the t(6;21) (q25-27;q22) suggested by combined G-banding and FISH.
In order to verify the fusion obtained with the deFuse software, we
used the 'grep' command utility to search for expressions composed
of 10 nt of RUNX1 and 10 nt of 6q25 upstream and downstream
of the fusion point (Table I).
Using the expression 'CAGATGCAGGAAGACTTTTG', 9 sequences were
retrieved which corresponded to the fusion RUNX1-transcript
found by defuse (Table I).
 | Table ISequences, obtained with the grep
command using the expression 'CAGATGCAGGAAGACTTTTG'. which
show the fusion of exon 7 of RUNX1 (NM_001754.4) with
sequence from chromosome band 6q25. |
Table I
Sequences, obtained with the grep
command using the expression 'CAGATGCAGGAAGACTTTTG'. which
show the fusion of exon 7 of RUNX1 (NM_001754.4) with
sequence from chromosome band 6q25.
| Sequence | NM_001754.4 BP | NC_000006.12 |
|---|
|
CTTTAACCCTCAGCCTCAGAGTCAGATGCAGGAAGACTTTTGAGGATAAAGAAAGGATGAAAATTCTCCAAGAAAATGGAGCACCAAGACTGATGTTGCAC | 996 | 155903358 |
|
CTCGTGCCTCCCTGAACCACTCCACTGCCTTTAACCCTCAGCCTCAGAGTCAGATGCAGGAAGACTTTTGAGGATAAAGAAAGGATGAAAATTCTCCAAGA | 996 | 155903358 |
|
CTCGTGCCTCCCTGAACCACTCCACTGCCTTTAACCCTCAGCCTCAGAGTCAGATGCAGGAAGACTTTTGAGGATAAAGAAAGGATGAAAATTCTCCAAGA | 996 | 155903358 |
|
CAGCCTCAGAGTCAGATGCAGGAAGACTTTTGAGGATAAAGAAAGGATGAAAATTCTCCAAGAAAATGGAGCACCAAGACTGATGTTGCACGAAATGCCAA | 996 | 155903358 |
|
CAGCCTCAGAGTCAGATGCAGGAAGACTTTTGAGGATAAAGAAAGGATGAAAATTCTCCAAGAAAATGGAGCACCAAGACTGATGTTGCACAGATCGGAAG | 996 | 155903358 |
|
CTCAGCCTCAGAGTCAGATGCAGGAAGACTTTTGAGGATAAAGAAAGGATGAAAATTCTCCAAGAAAATGGAGCACCAAGACTGATGTTGCACGAAATGCA | 996 | 155903358 |
|
CCAACCCTCGTGCCTCCCTGAACCACTCCACTGCCTTTAACCCTCAGCCTCAGAGTCAGATGCAGGAAGACTTTTGAGGATAAAGAAAGGATGAAAATTCT | 996 | 155903358 |
|
CTCGTGCCTCCCTGAACCACTCCACTGCCTTTAACCCTCAGCCTCAGAGTCAGATGCAGGAAGACTTTTGAGGATAAAGAAAGGATGAAAATTCTCAGATC | 996 | 155903358 |
|
CCCAGCCCCCACGCCCAACCCTCGTGCCTCCCTGAACCACTCCACTGCCTTTAACCCTCAGCCTCAGAGTCAGATGCAGGAAGACTTTTGAGGATAAAGAA | 996 | 155903358 |
Molecular confirmation of the
RUNX1-fusions
PCR with the RUNX1-809N-F1/6q25-R1 primer
combination amplified a 358 bp cDNA fragment (Fig. 1E). Direct sequencing of the
amplified fragment verified the presence of the RUNX1-fusion
transcript. The fusion point was identical to that found with
deFuse (Fig. 1F). Therefore, the
final karyotype after G-banding, FISH, and molecular examination
could be written 47,XX,+4,t(6;21)(q25;q22)[10] (Fig. 1A and D).
Discussion
We present herein a case of childhood AML in which
the leukemic cells had trisomy 4, a novel cryptic t(6;21)(q25;q22)
chromosome translocation, and FLT3-ITD mutation. The molecular
analysis of the translocation showed fusion of the RUNX1
gene with an intergenic sequence from 6q25 resulting in a putative
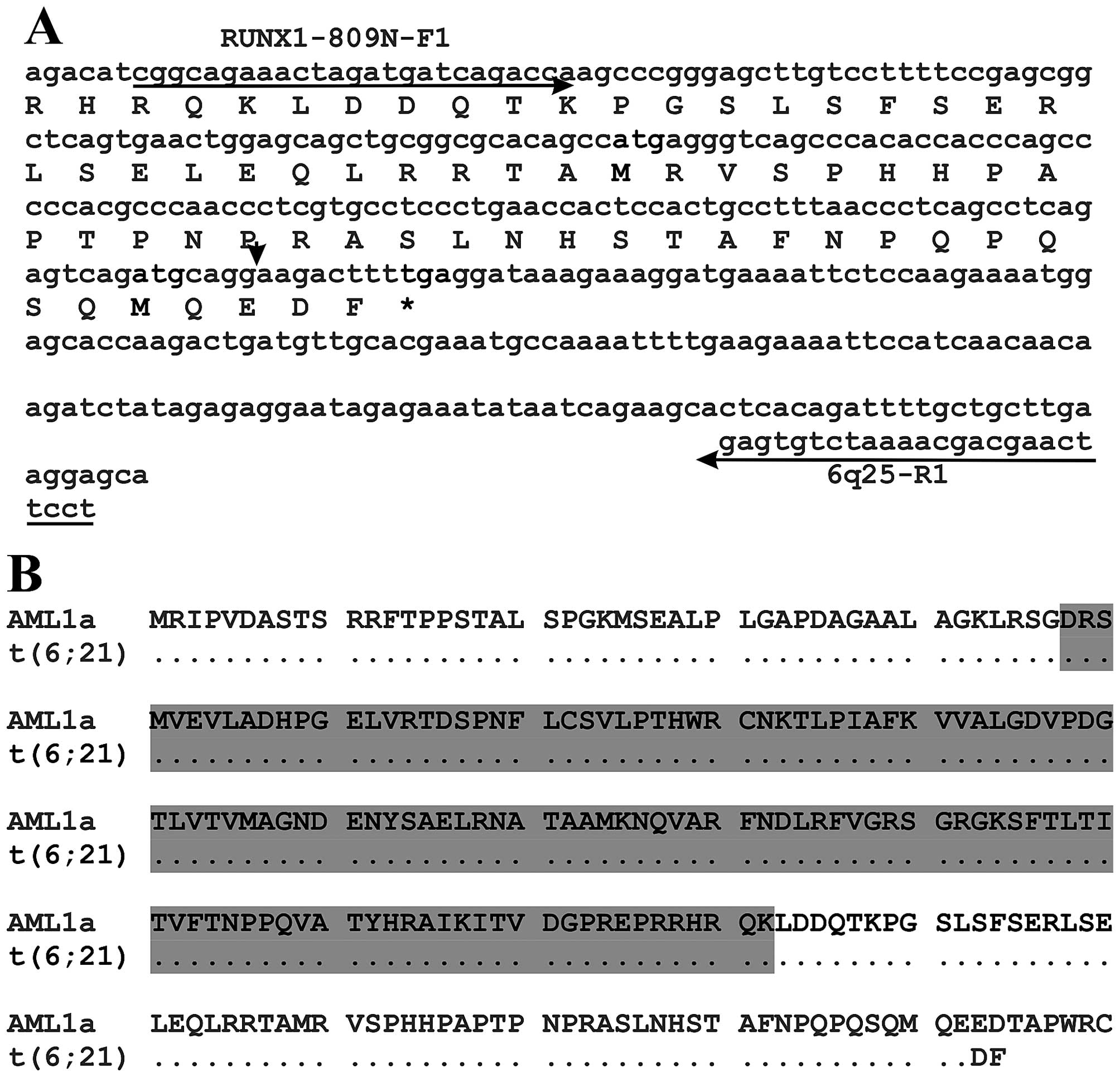
RUNX1 truncated protein (Fig. 2A and
B). The predicted truncated protein would contain the Runt
homology domain (RHD) which is responsible for both
heterodimerization with CBFB and DNA binding (40). Functionally, the truncated RUNX1
would be similar to the isoform AML1a of the RUNX1 protein
(Fig. 2B, protein with accession
number NP_001116079) (41–43). The isoform AML1a is a 250 amino acid
RUNX1 protein which contains the RHD but lacks the proline-,
serine-, and threonine-rich (PST) region which is the
transcriptional activation domain at the C terminal end (41–43).
AML1a does not itself have any transactivation function, but it
inhibits the transcriptional activity of AML1b by competing for the
DNA sequence of target genes with higher affinity (43). Overexpression of AML1a was shown to
suppress granulocytic differentiation and to stimulate cell
proliferation in 32Dcl3 murine myeloid cells treated with
granulocyte colony-stimulating factor (43). AML1a was found to inhibit erythroid
differentiation induced by sodium butyrate and enhance the
megakaryocytic differentiation of K562 leukemia cells (44). AML1a also enhanced hematopoietic
lineage commitment from human embryonic stem cells and inducible
pluripotent stem cells (45). AML1a
was reported to be highly abundant in the primitive stem/progenitor
compartment of human cord blood, and forced expression of AML1a in
these cells enhanced maintenance of primitive potential both in
vitro and in vivo (46).
Overexpression of AML1a was reported in patients with acute
lymphoblastic leukemia and AML-M2 patients (47). In the same study, AML1a was found to
repress transcription of promoter of macrophage colony-stimulating
factor receptor mediated by AML1b (47). When murine bone marrow mononuclear
cells were transduced with AML1a and then transplanted into
lethally irradiated mice, the mice developed lymphoblastic leukemia
after transplantation (47). Thus,
AML1a seems to be an important contributing factor to
leukemogenesis.
Truncated RUNX1 proteins generated by chromosomal
translocations were shown to have functions similar to those of the
AML1a isoform. In a patient with secondary AML carrying a
t(19;21)(q13;q22), RUNX1 was fused out-of-frame to
chromosome 19 sequences resulting in a truncated AML protein
bearing the DNA binding domain but not the transcriptional
activation domain. The fusion AML1 protein functioned as an
inhibitor of the normal RUNX1 protein (19). The RUNX1-RPL22P1 (also known
as AML1-EAP) fusion gene which is the result of the
t(3;21)(q26;q22) chromosome translocation in AML, codes for a
truncated RUNX1 protein which acts as an inhibitor of AML1b
(17,18). The fusion of RUNX1 to
CPNE8 in an AML with t(12;21)(q12;q22) also resulted in a
truncated inhibitory RUNX1 protein (20). Recently, in vitro analysis of
transduced human hematopoietic/progenitor stem cells showed that
truncated RUNX1 proteins generated by a t(1;21)(p32;q22)
chromosomal translocation increased proliferation and self-renewal
and disrupted the differentiation program by interfering with AML1b
(25). In a mouse model, truncated
RUNX1 protein resulting from a point mutation induced pancytopenia
with erythroid dysplasia, followed by progression to MDS-RAEB or
MDS/AML (48). Dowdy et al
studied the RUNX1 C-terminus in a mouse model by introducing a
premature translational stop codon after amino acid 307
(Runx1Q307X) which mimicked RUNX1 mutations found in
MDS/AML and CMML patients (49).
They found that Runx1Q307X homozygous mice exhibited
embryonic lethality at E12.5 due to central nervous system
hemorrhage and a complete lack of hematopoietic stem cell function
(49). They also showed that while
the RUNX1 truncated protein was capable of binding to DNA, it was
unable to associate with the nuclear matrix and failed to activate
target gene promoters (49).
Taking all the above-mentioned data into
consideration, it appears that the truncated RUNX1 protein (or
absence from it of the C terminal part which contains subnuclear
targeting and transactivation domains) is at least a contributing
factor in leukemogenesis.
The patient described here also had, apart from the
t(6;21)-RUNX1 rearrangement, trisomy 4 and FLT3-ITD
mutation. The molecular genetic consequences of trisomy 4 are, as
for numerical chromosome changes in general, unknown. Possible
mechanisms could be global gene expression alterations because of
gene dosage effect generated by the trisomy and duplication of any
rearranged or mutated genes on chromosome 4. The prognosis for
AML-patients with trisomy 4 is unclear, but based on a review of 30
such patients, Gupta et al (50) concluded that the outcome is poor
compared to that of other cytogenetic subsets within the
intermediate risk group. More importantly, a recent international
collaborative study on pediatric t(8;21)-AML showed that gain of
chromosome 4 in addition to t(8;21) represents a prognostically
unfavorable feature (51).
FLT3-ITD mutation has been shown to be a prognostic
factor although its impact has to be interpreted against the
overall genetic background of the leukemic cells (52). In adult patients with a normal
karyotype, FLT3-ITD is associated with poor prognosis (53,54).
In core-binding factor (CBF) AML, higher mutant levels of FLT3-ITD
were an adverse factor for overall survival (55). However, a recent report on adult
patients with CBF AML stated that MRD levels, rather than the
FLT3-ITD mutations, were significant prognostic markers for outcome
(56). In pediatric patients, FLT3
mutations have been associated with poor prognosis (57,58).
Reports on the significance of FLT3 mutations in pediatric CBF AML
are lacking.
All in all, we cannot say which genetic event (+4,
FLT3-ITD, or t(6;21)-RUNX1 truncation) was more pathogenetically or
prognostically important. The case nevertheless illustrates that
submicroscopic chromosomal rearrangements may accompany visible
numerical changes and perhaps should be actively sought for
whenever a single trisomy is found. To what extent and at which
frequency such submicroscopic changes target the RUNX1 gene
remains unknown. An active search for them may provide both
pathogenetic and prognostic novel information in the future.
Acknowledgments
This study was supported by grants from the
Norwegian Radium Hospital Foundation.
References
|
1
|
Heim S and Mitelman F: Cancer
Cytogenetics: Chromosomal and Molecular Genetic Abberations of
Tumor Cells. Forth Edition. Wiley-Blackwell; 2015, http://dx.doi.org/10.1002/9781118795569.
View Article : Google Scholar
|
|
2
|
De Braekeleer E, Douet-Guilbert N, Morel
F, Le Bris MJ, Férec C and De Braekeleer M: RUNX1 translocations
and fusion genes in malignant hemopathies. Future Oncol. 7:77–91.
2011. View Article : Google Scholar
|
|
3
|
Yoshihara K, Wang Q, Torres-Garcia W,
Zheng S, Vegesna R, Kim H and Verhaak RG: The landscape and
therapeutic relevance of cancer-associated transcript fusions.
Oncogene. 34:4845–4854. 2015. View Article : Google Scholar :
|
|
4
|
Abe A, Katsumi A, Kobayashi M, Okamoto A,
Tokuda M, Kanie T, Yamamoto Y, Naoe T and Emi N: A novel
RUNX1-C11orf41 fusion gene in a case of acute myeloid leukemia with
a t(11;21)(p14;q22). Cancer Genet. 205:608–611. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bhojwani D, Pei D, Sandlund JT, Jeha S,
Ribeiro RC, Rubnitz JE, Raimondi SC, Shurtleff S, Onciu M, Cheng C,
et al: ETV6-RUNX1-positive childhood acute lymphoblastic leukemia:
Improved outcome with contemporary therapy. Leukemia. 26:265–270.
2012. View Article : Google Scholar :
|
|
6
|
Cho EK, Bang SM, Ahn JY, Yoo SM, Park PW,
Seo YH, Shin DB and Lee JH: Prognostic value of AML 1/ETO fusion
transcripts in patients with acute myelogenous leukemia. Korean J
Intern Med. 18:13–20. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Gandemer V, Chevret S, Petit A, Vermylen
C, Leblanc T, Michel G, Schmitt C, Lejars O, Schneider P, Demeocq
F, et al FRALLE Group: Excellent prognosis of late relapses of
ETV6/RUNX1-positive childhood acute lymphoblastic leukemia: Lessons
from the FRALLE 93 protocol. Haematologica. 97:1743–1750. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Sangle NA and Perkins SL: Core-binding
factor acute myeloid leukemia. Arch Pathol Lab Med. 135:1504–1509.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ji J, Loo E, Pullarkat S, Yang L and
Tirado CA: Acute myeloid leukemia with t(7;21)(p22;q22) and 5q
deletion: A case report and literature review. Exp Hematol Oncol.
3:82014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Osato M: Point mutations in the RUNX1/AML1
gene: Another actor in RUNX leukemia. Oncogene. 23:4284–4296. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Silva FP, Swagemakers SM,
Erpelinck-Verschueren C, Wouters BJ, Delwel R, Vrieling H, van der
Spek P, Valk PJ and Giphart-Gassler M: Gene expression profiling of
minimally differentiated acute myeloid leukemia: M0 is a distinct
entity subdivided by RUNX1 mutation status. Blood. 114:3001–3007.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Dicker F, Haferlach C, Kern W, Haferlach T
and Schnittger S: Trisomy 13 is strongly associated with AML1/RUNX1
mutations and increased FLT3 expression in acute myeloid leukemia.
Blood. 110:1308–1316. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Harada H, Harada Y, Niimi H, Kyo T, Kimura
A and Inaba T: High incidence of somatic mutations in the
AML1/RUNX1 gene in myelodysplastic syndrome and low blast
percentage myeloid leukemia with myelodysplasia. Blood.
103:2316–2324. 2004. View Article : Google Scholar
|
|
14
|
Mendler JH, Maharry K, Radmacher MD,
Mrózek K, Becker H, Metzeler KH, Schwind S, Whitman SP, Khalife J,
Kohlschmidt J, et al: RUNX1 mutations are associated with poor
outcome in younger and older patients with cytogenetically normal
acute myeloid leukemia and with distinct gene and MicroRNA
expression signatures. J Clin Oncol. 30:3109–3118. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Schnittger S, Dicker F, Kern W, Wendland
N, Sundermann J, Alpermann T, Haferlach C and Haferlach T: RUNX1
mutations are frequent in de novo AML with noncomplex karyotype and
confer an unfavorable prognosis. Blood. 117:2348–2357. 2011.
View Article : Google Scholar
|
|
16
|
Silva FP, Lind A, Brouwer-Mandema G, Valk
PJ and Giphart-Gassler M: Trisomy 13 correlates with RUNX1 mutation
and increased FLT3 expression in AML-M0 patients. Haematologica.
92:1123–1126. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Nucifora G, Begy CR, Erickson P, Drabkin
HA and Rowley JD: The 3;21 translocation in myelodysplasia results
in a fusion transcript between the AML1 gene and the gene for EAP,
a highly conserved protein associated with the Epstein-Barr virus
small RNA EBER 1. Proc Natl Acad Sci USA. 90:7784–7788. 1993.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zent CS, Mathieu C, Claxton DF, Zhang DE,
Tenen DG, Rowley JD and Nucifora G: The chimeric genes AML1/MDS1
and AML1/EAP inhibit AML1B activation at the CSF1R promoter, but
only AML1/MDS1 has tumor-promoter properties. Proc Natl Acad Sci
USA. 93:1044–1048. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hromas R, Busse T, Carroll A, Mack D,
Shopnick R, Zhang DE, Nakshatri H and Richkind K: Fusion AML1
transcript in a radiation-associated leukemia results in a
truncated inhibitory AML1 protein. Blood. 97:2168–2170. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ramsey H, Zhang DE, Richkind K,
Burcoglu-O'Ral A and Hromas R: Fusion of AML1/Runx1 to copine VIII,
a novel member of the copine family, in an aggressive acute
myelogenous leukemia with t(12;21) translocation. Leukemia.
17:1665–1666. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Mikhail FM, Coignet L, Hatem N, Mourad ZI,
Farawela HM, El Kaffash DM, Farahat N and Nucifora G: A novel gene,
FGA7, is fused to RUNX1/AML1 in a t(4;21)(q28;q22) in a patient
with T-cell acute lymphoblastic leukemia. Genes Chromosomes Cancer.
39:110–118. 2004. View Article : Google Scholar
|
|
22
|
Ågerstam H, Lilljebjörn H, Lassen C,
Swedin A, Richter J, Vandenberghe P, Johansson B and Fioretos T:
Fusion gene-mediated truncation of RUNX1 as a potential mechanism
underlying disease progression in the 8p11 myeloproliferative
syndrome. Genes Chromosomes Cancer. 46:635–643. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Giguère A and Hébert J: CLCA2, a novel
RUNX1 partner gene in a therapy-related leukemia with
t(1;21)(p22;q22). Cancer Genet Cytogenet. 202:94–100. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Giguère A and Hébert J: Identification of
a novel fusion gene involving RUNX1 and the antisense strand of
SV2B in a BCR-ABL1-positive acute leukemia. Genes Chromosomes
Cancer. 52:1114–1122. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Rodriguez-Perales S, Torres-Ruiz R, Suela
J, Acquadro F, Martin MC, Yebra E, Ramirez JC, Alvarez S and
Cigudosa JC: Truncated RUNX1 protein generated by a novel
t(1;21)(p32;q22) chromosomal translocation impairs the
proliferation and differentiation of human hematopoietic
progenitors. Oncogene. 35:125–134. 2016. View Article : Google Scholar
|
|
26
|
Chinen Y, Taki T, Nishida K, Shimizu D,
Okuda T, Yoshida N, Kobayashi C, Koike K, Tsuchida M, Hayashi Y, et
al: Identification of the novel AML1 fusion partner gene, LAF4, a
fusion partner of MLL, in childhood T-cell acute lymphoblastic
leukemia with t(2;21)(q11;q22) by bubble PCR method for cDNA.
Oncogene. 27:2249–2256. 2008. View Article : Google Scholar
|
|
27
|
Dai HP, Xue YQ, Zhou JW, Li AP, Wu YF, Pan
JL, Wang Y and Zhang J: LPXN, a member of the paxillin superfamily,
is fused to RUNX1 in an acute myeloid leukemia patient with a
t(11;21)(q12;q22) translocation. Genes Chromosomes Cancer.
48:1027–1036. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hazourli S, Chagnon P, Sauvageau M, Fetni
R, Busque L and Hébert J: Overexpression of PRDM16 in the presence
and absence of the RUNX1/PRDM16 fusion gene in myeloid leukemias.
Genes Chromosomes Cancer. 45:1072–1076. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
LaFiura KM, Edwards H, Taub JW, Matherly
LH, Fontana JA, Mohamed AN, Ravindranath Y and Ge Y; Children's
Oncology Group: Identification and characterization of novel
AML1-ETO fusion transcripts in pediatric t(8;21) acute myeloid
leukemia: A report from the Children's Oncology Group. Oncogene.
27:4933–4942. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Sakai I, Tamura T, Narumi H, Uchida N,
Yakushijin Y, Hato T, Fujita S and Yasukawa M: Novel RUNX1-PRDM16
fusion transcripts in a patient with acute myeloid leukemia showing
t(1;21)(p36;q22). Genes Chromosomes Cancer. 44:265–270. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Stevens-Kroef MJ, Schoenmakers EF, van
Kraaij M, Huys E, Vermeulen S, van der Reijden B and van Kessel AG:
Identification of truncated RUNX1 and RUNX1-PRDM16 fusion
transcripts in a case of t(1;21)(p36;q22)-positive therapy-related
AML. Leukemia. 20:1187–1189. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Panagopoulos I, Gorunova L, Brandal P,
Garnes M, Tierens A and Heim S: Myeloid leukemia with
t(7;21)(p22;q22) and 5q deletion. Oncol Rep. 30:1549–1552.
2013.PubMed/NCBI
|
|
33
|
Jeandidier E, Gervais C, Radford-Weiss I,
Zink E, Gangneux C, Eischen A, Galoisy AC, Helias C, Dano L,
Cammarata O, et al: A cytogenetic study of 397 consecutive acute
myeloid leukemia cases identified three with a t(7;21) associated
with 5q abnormalities and exhibiting similar clinical and
biological features, suggesting a new, rare acute myeloid leukemia
entity. Cancer Genet. 205:365–372. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Foster N, Paulsson K, Sales M, Cunningham
J, Groves M, O'Connor N, Begum S, Stubbs T, McMullan DJ, Griffiths
M, et al: Molecular characterisation of a recurrent, semi-cryptic
RUNX1 translocation t(7;21) in myelodysplastic syndrome and acute
myeloid leukaemia. Br J Haematol. 148:938–943. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Paulsson K, Békássy AN, Olofsson T,
Mitelman F, Johansson B and Panagopoulos I: A novel and
cytogenetically cryptic t(7;21) (p22;q22) in acute myeloid leukemia
results in fusion of RUNX1 with the ubiquitin-specific protease
gene USP42. Leukemia. 20:224–229. 2006. View Article : Google Scholar
|
|
36
|
Hasle H, Abrahamsson J, Forestier E, Ha
SY, Heldrup J, Jahnukainen K, Jónsson OG, Lausen B, Palle J and
Zeller B; Nordic Society of Paediatric Haematology Oncology
(NOPHO): Gemtuzumab ozogamicin as postconsolidation therapy does
not prevent relapse in children with AML: Results from NOPHO-AML
2004. Blood. 120:978–984. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Schaffer LG, Slovak ML and Campbell LJ:
ISCN 2009: An International System for Human Cytogenetic
Nomenclature. Karger S: Basel: 2009
|
|
38
|
McPherson A, Hormozdiari F, Zayed A,
Giuliany R, Ha G, Sun MG, Griffith M, Heravi Moussavi A, Senz J,
Melnyk N, et al: deFuse: An algorithm for gene fusion discovery in
tumor RNA-Seq data. PLOS Comput Biol. 7:e10011382011. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Singhal H, Ren YR and Kern SE: Improved
DNA electrophoresis in conditions favoring polyborates and lewis
acid complexation. PLoS One. 5:e113182010. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Ogawa E, Maruyama M, Kagoshima H, Inuzuka
M, Lu J, Satake M, Shigesada K and Ito Y: PEBP2/PEA2 represents a
family of transcription factors homologous to the products of the
Drosophila runt gene and the human AML1 gene. Proc Natl Acad Sci
USA. 90:6859–6863. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Miyoshi H, Shimizu K, Kozu T, Maseki N,
Kaneko Y and Ohki M: t(8;21) breakpoints on chromosome 21 in acute
myeloid leukemia are clustered within a limited region of a single
gene, AML1. Proc Natl Acad Sci USA. 88:10431–10434. 1991.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Miyoshi H, Ohira M, Shimizu K, Mitani K,
Hirai H, Imai T, Yokoyama K, Soeda E and Ohki M: Alternative
splicing and genomic structure of the AML1 gene involved in acute
myeloid leukemia. Nucleic Acids Res. 23:2762–2769. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Tanaka T, Tanaka K, Ogawa S, Kurokawa M,
Mitani K, Nishida J, Shibata Y, Yazaki Y and Hirai H: An acute
myeloid leukemia gene, AML1, regulates hemopoietic myeloid cell
differentiation and transcriptional activation antagonistically by
two alternative spliced forms. EMBO J. 14:341–350. 1995.PubMed/NCBI
|
|
44
|
Niitsu N, Yamamoto-Yamaguchi Y, Miyoshi H,
Shimizu K, Ohki M, Umeda M and Honma Y: AML1a but not AML1b
inhibits erythroid differentiation induced by sodium butyrate and
enhances the megakaryocytic differentiation of K562 leukemia cells.
Cell Growth Differ. 8:319–326. 1997.PubMed/NCBI
|
|
45
|
Ran D, Shia WJ, Lo MC, Fan JB, Knorr DA,
Ferrell PI, Ye Z, Yan M, Cheng L, Kaufman DS, et al: RUNX1a
enhances hematopoietic lineage commitment from human embryonic stem
cells and inducible pluripotent stem cells. Blood. 121:2882–2890.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Tsuzuki S, Hong D, Gupta R, Matsuo K, Seto
M and Enver T: Isoform-specific potentiation of stem and progenitor
cell engraftment by AML1/RUNX1. PLoS Med. 4:e1722007. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Liu X, Zhang Q, Zhang DE, Zhou C, Xing H,
Tian Z, Rao Q, Wang M and Wang J: Overexpression of an isoform of
AML1 in acute leukemia and its potential role in leukemogenesis.
Leukemia. 23:739–745. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Watanabe-Okochi N, Kitaura J, Ono R,
Harada H, Harada Y, Komeno Y, Nakajima H, Nosaka T, Inaba T and
Kitamura T: AML1 mutations induced MDS and MDS/AML in a mouse BMT
model. Blood. 111:4297–4308. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Dowdy CR, Xie R, Frederick D, Hussain S,
Zaidi SK, Vradii D, Javed A, Li X, Jones SN, Lian JB, et al:
Definitive hematopoiesis requires Runx1 C-terminal-mediated
subnuclear targeting and transactivation. Hum Mol Genet.
19:1048–1057. 2010. View Article : Google Scholar :
|
|
50
|
Gupta V, Minden MD, Yi QL, Brandwein J and
Chun K: Prognostic significance of trisomy 4 as the sole
cytogenetic abnormality in acute myeloid leukemia. Leuk Res.
27:983–991. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Klein K, Kaspers G, Harrison CJ, Beverloo
HB, Reedijk A, Bongers M, Cloos J, Pession A, Reinhardt D,
Zimmerman M, et al: Clinical impact of additional cytogenetic
aberrations, cKIT- and RAS mutations and other factors in pediatric
t(8;21)-AML: Results from an International Retrospective Study by
the International Berlin-Frankfutrt-Munster Study Group. J Clin
Oncol. 33:4247–4258. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Levis M: FLT3 mutations in acute myeloid
leukemia: What is the best approach in 2013? Hematology Am Soc
Hematol Educ Program. 2013:220–226. 2013.PubMed/NCBI
|
|
53
|
Döhner H, Estey EH, Amadori S, Appelbaum
FR, Büchner T, Burnett AK, Dombret H, Fenaux P, Grimwade D, Larson
RA, et al European LeukemiaNet: Diagnosis and management of acute
myeloid leukemia in adults: Recommendations from an international
expert panel, on behalf of the European LeukemiaNet. Blood.
115:453–474. 2010. View Article : Google Scholar
|
|
54
|
Estey EH: Acute myeloid leukemia: 2014
update on risk-stratification and management. Am J Hematol.
89:1063–1081. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Allen C, Hills RK, Lamb K, Evans C,
Tinsley S, Sellar R, O'Brien M, Yin JL, Burnett AK, Linch DC, et
al: The importance of relative mutant level for evaluating impact
on outcome of KIT, FLT3 and CBL mutations in core-binding factor
acute myeloid leukemia. Leukemia. 27:1891–1901. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Jourdan E, Boissel N, Chevret S, Delabesse
E, Renneville A, Cornillet P, Blanchet O, Cayuela JM, Recher C,
Raffoux E, et al French AML Intergroup: Prospective evaluation of
gene mutations and minimal residual disease in patients with core
binding factor acute myeloid leukemia. Blood. 121:2213–2223. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Meshinchi S, Alonzo TA, Stirewalt DL,
Zwaan M, Zimmerman M, Reinhardt D, Kaspers GJ, Heerema NA, Gerbing
R, Lange BJ, et al: Clinical implications of FLT3 mutations in
pediatric AML. Blood. 108:3654–3661. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Zwaan CM, Meshinchi S, Radich JP, Veerman
AJ, Huismans DR, Munske L, Podleschny M, Hählen K, Pieters R,
Zimmermann M, et al: FLT3 internal tandem duplication in 234
children with acute myeloid leukemia: Prognostic significance and
relation to cellular drug resistance. Blood. 102:2387–2394. 2003.
View Article : Google Scholar : PubMed/NCBI
|