Introduction
Acute lymphoblastic leukemia (ALL) is an aggressive
hematologic malignancy arising from the hematopoietic precursors of
lymphoid. It is most common in childhood. T-cell acute
lymphoblastic leukemia (T-ALL) represents 10–15% of pediatric and
25% of adult ALL cases (1),
resulting from the malignant transformation of T cell progenitors.
Although the treatment for T-ALL has gradually improved, T-ALL
patients with primary resistant or relapsed leukemia have poor
prognosis (2–4).
MicroRNAs (miRs) are short non-coding RNAs which
negatively regulate protein expression via binding to the
complementary sequences within the 3′-untranslated region (UTR) of
target mRNA (5,6). Several studies have indicated the
importance of microRNAs in the pathogenesis of human leukemia
(7–9).
miR-101 is reported as a putative tumor suppressor
in several types of cancer, including gastric, prostate cancer,
renal cell carcinoma and melanoma (10–13).
Recently, increasing studies revealed that miR-101 is also
associated with the development of hematological malignancies
(14–16). Correia et al (17) have demonstrated that miR-101 is
downregulated in T-ALL patient specimens and T-ALL cell lines.
However, the exact role of miR-101 in T-ALL progression and
chemoresistance remains unclear. Notch1 is a transmembrane receptor
that regulates cell growth, differentiation, angiogenesis and
metastasis (18–20). Notch1 signaling activation plays key
roles in the majority of hematological malignancies including T-ALL
(21,22).
In the present study, we detected the expression of
miR-101 in the blood samples of patients with T-ALL. The in
vitro functional studies were performed on Jurkat cell line to
elucidate the effect of miR-101 on cell proliferation, apoptosis,
invasion and chemoresistance. Furthermore, whether miR-101 exerts
its effect on T-ALL by targeting Notch1 was identified.
Materials and methods
Clinical samples
The study was approved by the Ethics Committee of
The Second Affiliated Hospital of Xi'an Jiaotong University, and
all the participants signed a written informed consent for
participation in this study. The blood samples were obtained from
25 T-ALL patients and 30 healthy controls.
Cell culture and transfection
The Jurkat and HEK293 cell lines were purchased from
the American Type Culture Collection (ATCC; Rockville, MD, USA),
and cultured in RPMI-1640 medium (Gibco, Grand Island, NY, USA)
supplemented with 10% fetal bovine serum (FBS; Gibco) at 37°C in a
humidified atmosphere with 5% CO2. Adriamycin (ADM) was
obtained from Sangon Biotech Co., Ltd., (Shanghai, China) and
dissolved in phosphate-buffered saline (PBS). Cells were treated
with 5 µg/ml adriamycin for 24 h. The cells were transfected
with miR-negative control (NC), miR-101 mimic, miR-101 inhibitor,
Notch1-pcDNA3.1 or pcDNA3.1 empty vector (Shanghai GenePharma, Co.,
Ltd., Shanghai, China) using Lipofectamine 2000 (Invitrogen,
Waltham, MA, USA) following the manufacturer's protocols. After 6
h, the medium was replaced with fresh medium for further
experiments.
Cell proliferation assay
The cells were plated in 96-well plates at
5×103 cells/well and allowed to grow for 1–4 days.
Subsequently, the cells were incubated with 10 µl CCK-8
(Beyotime Institute of Biotechnology, Shanghai, China) at 37°C for
4 h. Absorbance was measured at 450 nm using a microplate reader
(DNM-9606; Perlong Medical Equipment Co., Ltd., Beijing,
China).
Cell apoptosis assay
Cell apoptosis rate was examined using the Annexin
V-FITC and propidium iodide (PI) apoptosis kit (Nanjing KeyGen
Biotech, Co., Ltd., Nanjing, China) following the manufacturer's
protocols. Briefly, the cells were washed with PBS, and
dual-stained with Annexin V and PI. The apoptosis cells were
detected by a FACSCalibur flow cytometer (Becton-Dickinson, Sparks,
MD, USA).
Cell invasion assay
Invasiveness of Jurkat cells were performed using
Transwell inserts (5 µm pore size; Corning Inc., Corning,
NY, USA) coated with Matrigel (BD Biosciences, Bedford, MA, USA).
The matrix solutions were loaded into the upper well of Transwell
chambers, and incubated at 37°C for 1 h. Each group of cells were
resuspended in FBS-free medium and placed into the upper Transwell
chambers. The lower chambers were filled with RPMI-1640 medium
containing 10% FBS. After incubation at 37°C for 24 h, the
non-invading cells on the top of the membrane were removed by
wiping. The invading cells on the lower face of the membrane were
fixed with 3.7% paraformaldehyde, and stained with crystal violet
staining solution (Beyotime Institute of Biotechnology). Cells in
the lower compartments were also counted.
Luciferase assay
The wild-type Notch1 3′ untranslated region (UTR)
carrying a putative miR-101 binding site, and the mutant Notch1
3′UTR were inserted into psiCHECK-2 vector (Promega, Madison, WI,
USA). HEK293 cells were co-transfected with miR-NC/miR-101 and
wild-type/mutant Notch1-3′UTR using Lipofectamine 2000. The
Renilla luciferase reporter vector was transfected as an
internal control in each assay. Luciferase activity was measured 24
h after transfection using the Luciferase Reporter assay system
(Promega).
Reverse transcription quantitative
polymerase chain reaction (RTqPCR)
Total RNA was extracted using the RNeasy/miRNeasy
Mini kit (Qiagen, Limburg, The Netherlands) according to the
manufacturer's protocols. Total RNA (5 ng) was used for reverse
transcription, using the RevertAid™ First Strand cDNA Synthesis kit
(Fermentas, Vilnius, Lithuania). The primers for miR-101 were the
exact sequence of mature miR-101. They were purchased from
GenScript (Nanjing, China). PCR was performed with the SYBR-Green
PCR Master Mix (Applied Biosystems, Foster City, CA, USA) on the
ABI PRISM 7700 Sequence detection system (Applied Biosystems). The
relative expression of miR-101 was calculated by the
2−ΔΔCt method that was normalized to the U6 internal
control.
Western blot analysis
Whole cell lysates were prepared using ice-cold RIPA
buffer supplemented with the protease inhibitor (Beyotime Institute
of Biotechnology). The protein concentration was determined using
the Bradford reagent (Pierce, Rockford, IL, USA). An equal amount
of protein (20 µg) was resolved by 10% SDS-PAGE, and then
transferred to nitrocellulose membranes (Bio-Rad Laboratories,
Inc., Hercules, CA, USA). The membranes were blocked with 5%
non-fat milk at room temperature for 2 h, and then incubated with
the specific antibodies at 4°C overnight, including rabbit
polyclonal to Notch1 (1:600; cat. no. 3881-100; BioVision Incorp.,
Milpitas, CA, USA) and rabbit polyclonal to β-actin (1:1,000; cat.
no. C1836; Applygen Technologies, Inc., Beijing, China). After
washing with TBST, the membranes were further incubated with
HRP-labelled goat anti-rabbit IgG (1:1,000; cat. no. C2226;
Applygen Technologies) at 37°C for 1 h. The immunoreactive bands
were visualized using the chemiluminescent substrate (Pierce).
Statistical analysis
All data represent at least three independent
experiments and are expressed as the means ± SD. Student's t-test
or one-way ANOVA test was used to compare the differences between
groups. P<0.05 was considered statistically significant.
Results
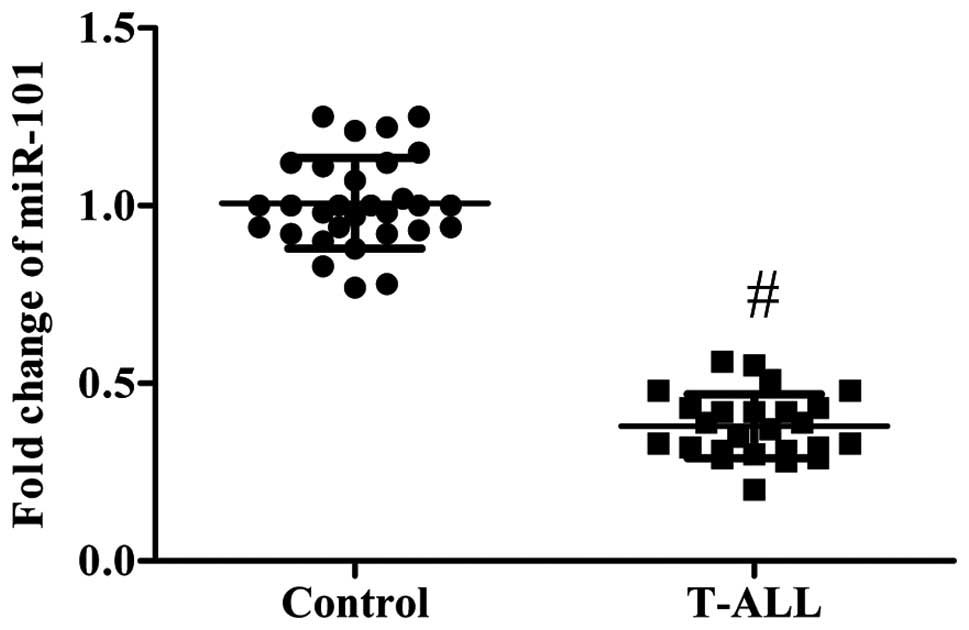
Expression of miR-101 in the blood
samples of patients with T-ALL
RT-qPCR was performed to detect miR-101 expression
in the blood samples of 25 T-ALL patients and 30 healthy controls.
Compared with the healthy controls, the expression of miR-101 was
significantly downregulated in the blood samples of patients with
T-ALL (P<0.01; Fig. 1).
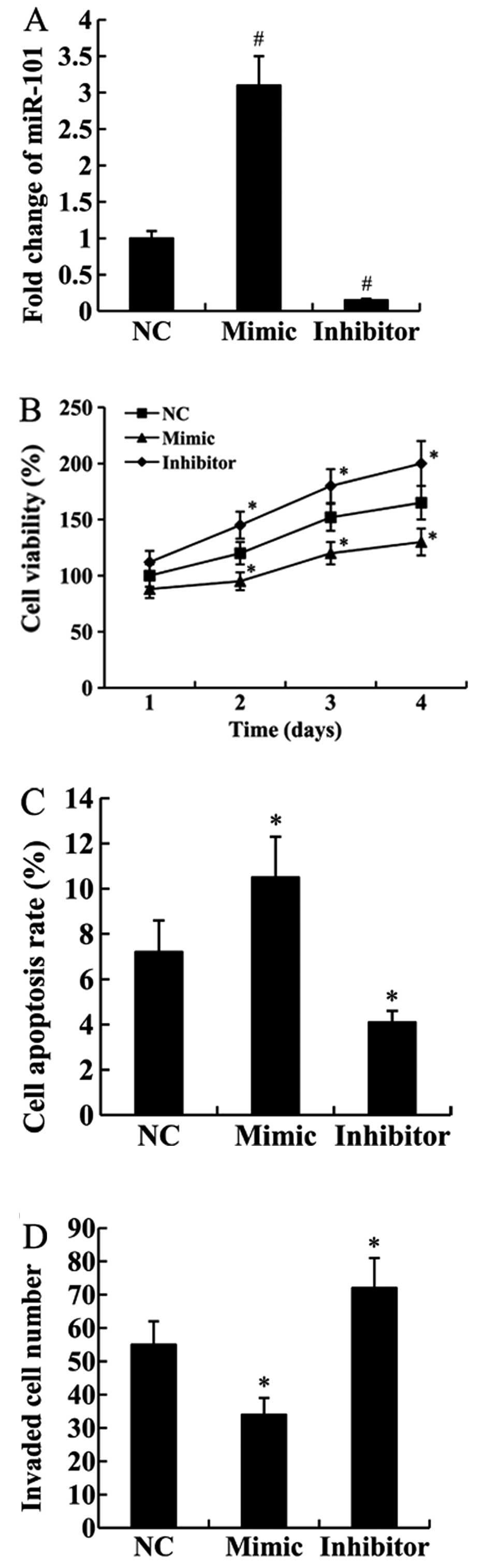
Effect of miR-101 on cell proliferation,
apoptosis and invasion of Jurkat cells
The Jurkat cells were transfected with the miR-NC,
miR-101 mimic or miR-101 inhibitor, and then RT-qPCR analysis was
performed to detect miR-101 expression. The results showed that
compared with the miR-NC-transfected cells, the expression of
miR-101 was significantly upregulated in miR-101 mimic-transfected
cells but downregulated in miR-101 inhibitor-transfected cells
(P<0.01; Fig. 2A).
To elucidate the effect of miR-101 on T-ALL
progression, the in vitro functional assays were performed
on Jurkat cells. As shown in Fig.
2B, the proliferation ability of Jurkat cells transfected with
the miR-101 mimic was significantly weaker than those transfected
with the miR-NC (P<0.05). In addition, the cell proliferation
ability was enhanced in miR-101 inhibitor transfected cells
compared with the control cells (P<0.05).
We examined whether miR-101 could affect cell
apoptosis using FCM analysis. We found that compared with the
miR-NC-transfected cells, cell apoptosis rate was significantly
increased in the cells transfected with the miR-101 mimic but
decreased in the cells transfected with the miR-101 inhibitor
(P<0.05; Fig. 2C).
Cell invasion assay confirmed that the invasive
ability of Jurkat cells was inhibited by transfection with the
miR-101 mimic (P<0.05). By contrast, transfection with the
miR-101 inhibitor significantly increased cell invasive ability
(P<0.05; Fig. 2D).
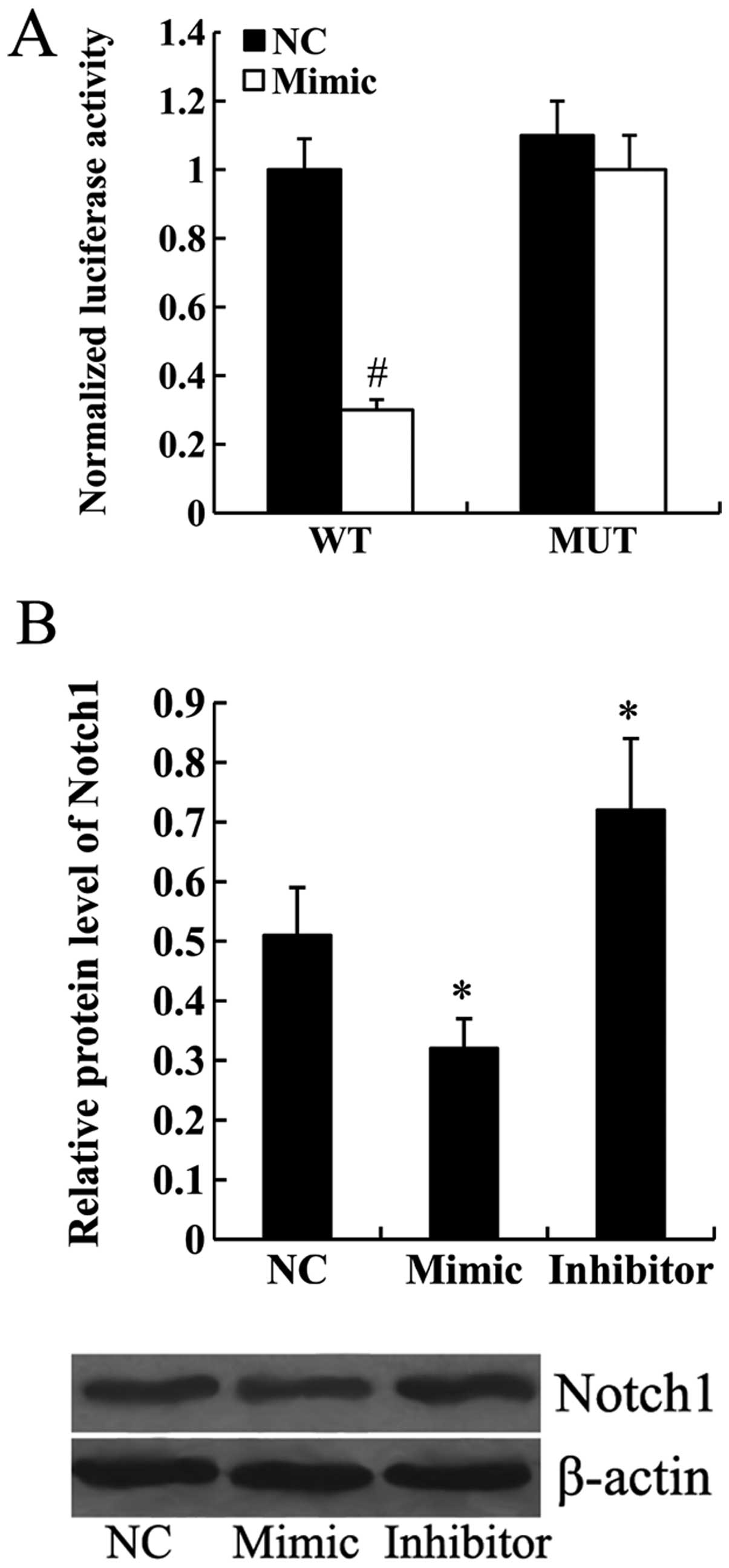
Notch1 is a direct target of miR-101
To determine whether Notch1 was a target gene of
miR-101, wild-type or mutant Notch1-3′UTR was transfected into the
HEK293 cells along with the miR-NC or miR-101 mimic. Luciferase
assay demonstrated that miR-101 mimic significantly inhibited the
transcription activity of wild-type Notch1-3′UTR (P<0.01).
However, the transcription activity of mutant Notch1-3′UTR was not
affected by the transfection of miR-101 mimic (Fig. 3A). Furthermore, the results from
western blot analysis revealed that the expression of Notch1
protein was significantly downregulated in the miR-101
mimic-transfected cells and upregulated in the miR-101
inhibitor-transfected cells (P<0.05; Fig. 3B).
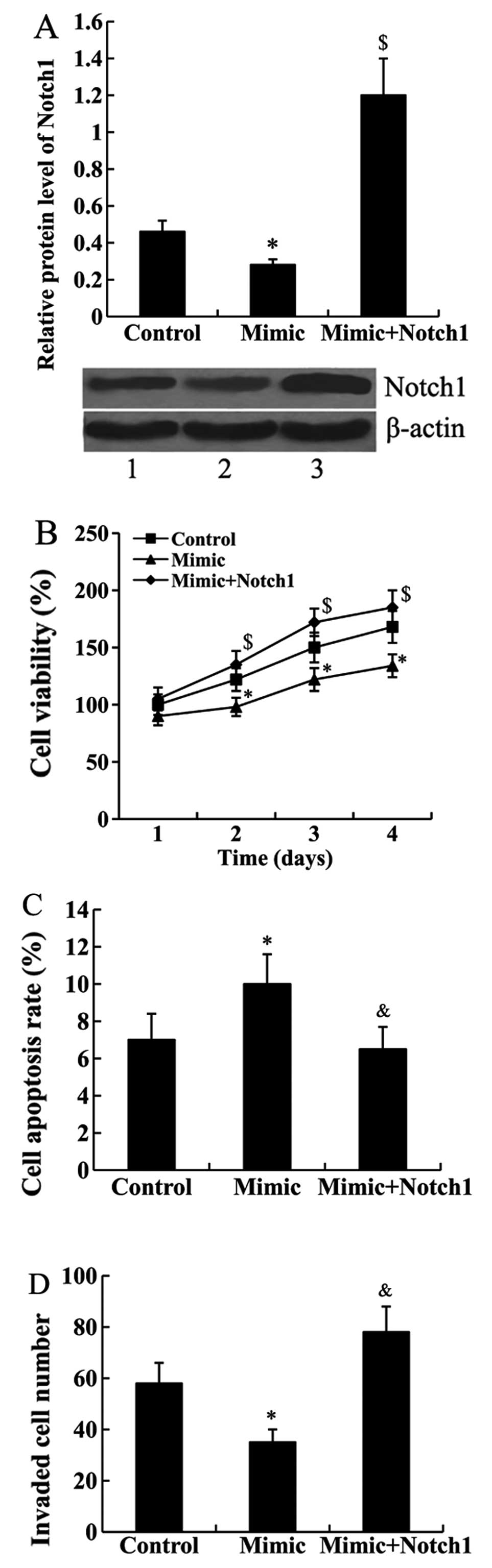
Notch1 attenuates the effect of miR-101
on cell proliferation, apoptosis and invasion
Notch1-pcDNA3.1 was transfected into the Jurkat
cells to overexpress Notch 1. As shown in Fig. 4A, the results from the western blot
analysis confirmed that the relative protein level of Notch1 was
significantly upregulated in the cells transfected with the miR-101
mimic and Notch1-pcDNA3.1 compared with the cells transected with
the miR-101 mimic only (P<0.01).
Subsequently, CCK-8, FCM and cell invasion assays
were performed to determine whether Notch1 mediates the effect of
miR-101 on cell proliferation, apoptosis and invasion. We found
that the suppressed cell proliferation and invasion abilities by
miR-101 mimic transfection were attenuated by Notch1 overexpression
in Jurkat cells. In addition, miR-101 mimic-induced cell apoptosis
rate was inhibited by Notch1-pcDNA3.1 transfection (Fig. 4B–D).
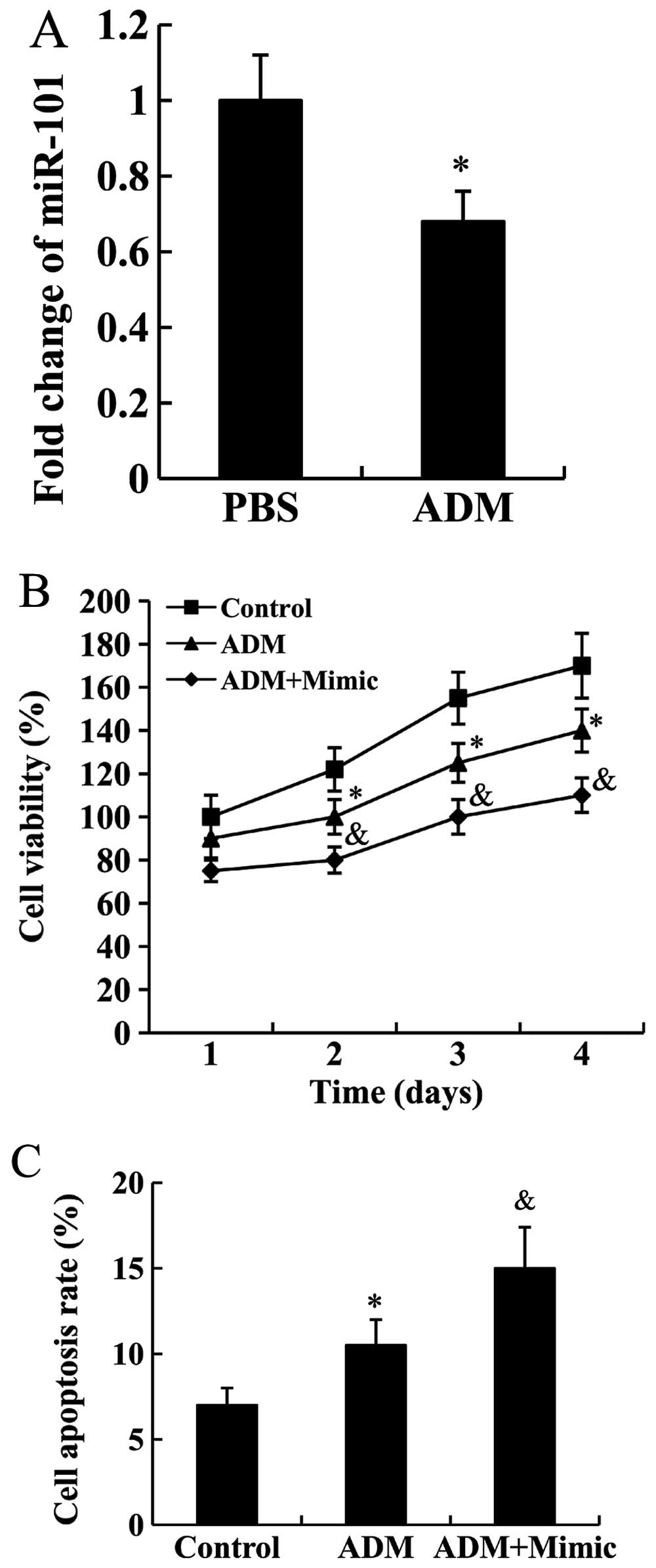
miR-101 enhanced drug sensitivity of
Jurkat cells
Jurkat cells were treated with 5 µg/ml
adriamycin for 24 h, and the expression of miR-101 was subsequently
detected. We found that miR-101 expression was significantly
decreased in Jurkat cells following treatment with adriamycin
(P<0.05; Fig. 5A).
Jurkat cells transfected with miR-101 mimic were
subjected to adriamycin treatment, then cell proliferation and
apoptosis rate were analyzed. As shown in Fig. 5B and C, adriamycin was able to
inhibit cell proliferation and promote cell apoptosis (P<0.05),
these effects were enhanced by miR-101 mimic (P<0.05).
Discussion
The expression of miR-101 has been extensively
studied in hematological malignancies. It was reported that miR-101
was downregulated in samples of Burkitt lymphoma (14) and adult ALL cases (23). Fallah et al (24) revealed that miR-101 was upregulated
in patients with newly diagnosed chronic myeloid leukemia in
chronic phase. Recently, Correia et al (17) demonstrated that miR-101 is
downregulated in T-ALL patient specimens and T-ALL cell lines, and
it may be involved in the development of T-cell acute lymphoblastic
leukemia. In the present study, we detected miR-101 expression in
the blood samples of patients with T-ALL. We found that compared
with the healthy controls, the expression of miR-101 was
significantly downregulated in T-ALL patients. This finding was
consistent with the previous study on T-ALL patient specimens and
T-ALL cell lines (17).
miR-101 has been reported to be downregulated in
various types of cancer, and it can repress cell proliferation and
metastatic ability in gallbladder, liver, breast and ovarian
cancers (25–28). Thus, miR-101 is widely recognized as
a tumor suppressor. miR-101 is also downregulated in T-ALL,
however, its function in T-ALL has not been reported. We further
performed in vitro studies to determine the effect of
miR-101 on Jurkat cell proliferation, apoptosis and invasion. Both
the gain- and loss-of-function experiments revealed that miR-101
could inhibit cell proliferation and invasion, and increase
apoptosis of Jurkat cells, suggesting the tumor-suppressive role of
miR-101 in T-ALL. Furthermore, the present study provided the first
evidence that miR-101 could enhance the drug sensitivity of Jurkat
cells to adriamycin.
Notch1 is a well-known regulator that plays an
oncogenic role in many malignancies (29). Notch1 mutation is present in over
50% of T-cell acute lymphoblastic leukemias (T-ALL) (30). Constitutive activation of Notch1
signaling can induce T-ALL in murine models. Suppression of Notch1
signaling leads to the decreased cell proliferation and increased
cell apoptosis in the context of T-ALL (31,32).
Furthermore, Notch1 is related to chemoresistance (33,34), a
major cause of poor prognosis in T-ALL. Using miRanda (http://www.microrna.org), Notch1 was predicted to be a
target of miR-101. In the present study, we confirmed that miR-101
could inhibit the transcription activity of Notch1-3′UTR using
luciferase assay. In addition, the expression of Notch1 protein was
downregulated by miR-101. These results identified Notch1 as a
direct target gene of miR-101. Furthermore, we found that Notch1
overexpression could attenuate the effect of miR-101 on cell
proliferation, apoptosis and invasion of Jurkat cells. These
findings suggested that miR-101 exerts its effect on T-ALL at least
partially by downregulating Notch1.
In conclusion, the present study revealed the
tumor-suppressive role of miR-101 in T-ALL by directly targeting
Notch1. In addition, miR-101 was able to enhance adriamycin
sensitivity in Jurkat cells. miR-101 could be potentially of value
in T-ALL therapy.
Acknowledgments
The present study was supported by the foundation of
Xi'an Municipal Bureau of Science and Technology [no.
SF1317(2)].
References
|
1
|
Ferrando AA, Neuberg DS, Staunton J, Loh
ML, Huard C, Raimondi SC, Behm FG, Pui CH, Downing JR, Gilliland
DG, et al: Gene expression signatures define novel oncogenic
pathways in T cell acute lymphoblastic leukemia. Cancer Cell.
1:75–87. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Pui CH, Robison LL and Look AT: Acute
lymphoblastic leukaemia. Lancet. 371:1030–1043. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Goldberg JM, Silverman LB, Levy DE, Dalton
VK, Gelber RD, Lehmann L, Cohen HJ, Sallan SE and Asselin BL:
Childhood T-cell acute lymphoblastic leukemia: The Dana-Farber
Cancer Institute acute lymphoblastic leukemia consortium
experience. J Clin Oncol. 21:3616–3622. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Oudot C, Auclerc MF, Levy V, Porcher R,
Piguet C, Perel Y, Gandemer V, Debre M, Vermylen C, Pautard B, et
al: Prognostic factors for leukemic induction failure in children
with acute lymphoblastic leukemia and outcome after salvage
therapy: The FRALLE 93 study. J Clin Oncol. 26:1496–1503. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Seca H, Almeida GM, Guimarães JE and
Vasconcelos MH: miR signatures and the role of miRs in acute
myeloid leukaemia. Eur J Cancer. 46:1520–1527. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Giza DE and Calin GA: microRNA and chronic
lymphocytic leukemia. Adv Exp Med Biol. 889:23–40. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yendamuri S and Calin GA: The role of
microRNA in human leukemia: A review. Leukemia. 23:1257–1263. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhou X, Xia Y, Li L and Zhang G: MiR-101
inhibits cell growth and tumorigenesis of Helicobacter pylori
related gastric cancer by repression of SOCS2. Cancer Biol Ther.
16:160–169. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hao Y, Gu X, Zhao Y, Greene S, Sha W,
Smoot DT, Califano J, Wu TC and Pang X: Enforced expression of
miR-101 inhibits prostate cancer cell growth by modulating the
COX-2 pathway in vivo. Cancer Prev Res (Phila). 4:1073–1083. 2011.
View Article : Google Scholar
|
|
12
|
Sakurai T, Bilim VN, Ugolkov AV, Yuuki K,
Tsukigi M, Motoyama T and Tomita Y: The enhancer of zeste homolog 2
(EZH2), a potential therapeutic target, is regulated by miR-101 in
renal cancer cells. Biochem Biophys Res Commun. 422:607–614. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Luo C, Merz PR, Chen Y, Dickes E, Pscherer
A, Schadendorf D and Eichmüller SB: MiR-101 inhibits melanoma cell
invasion and proliferation by targeting MITF and EZH2. Cancer Lett.
341:240–247. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Robertus JL, Kluiver J, Weggemans C, Harms
G, Reijmers RM, Swart Y, Kok K, Rosati S, Schuuring E, van Imhoff
G, et al: MiRNA profiling in B non-Hodgkin lymphoma: A MYC-related
miRNA profile characterizes Burkitt lymphoma. Br J Haematol.
149:896–899. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Cheng J, Guo S, Chen S, Mastriano SJ, Liu
C, D'Alessio AC, Hysolli E, Guo Y, Yao H, Megyola CM, et al: An
extensive network of TET2-targeting MicroRNAs regulates malignant
hematopoiesis. Cell Rep. 5:471–481. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Papakonstantinou N, Ntoufa S,
Chartomatsidou E, Papadopoulos G, Hatzigeorgiou A, Anagnostopoulos
A, Chlichlia K, Ghia P, Muzio M, Belessi C, et al: Differential
microRNA profiles and their functional implications in different
immunogenetic subsets of chronic lymphocytic leukemia. Mol Med.
19:115–123. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Correia NC, Melão A, Póvoa V, Sarmento L,
Gómez de Cedrón M, Malumbres M, Enguita FJ and Barata JT: microRNAs
regulate TAL1 expression in T-cell acute lymphoblastic leukemia.
Oncotarget. 7:8268–8281. 2016.PubMed/NCBI
|
|
18
|
Sethi N and Kang Y: Notch signalling in
cancer progression and bone metastasis. Br J Cancer. 105:1805–1810.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Roy M, Pear WS and Aster JC: The
multifaceted role of Notch in cancer. Curr Opin Genet Dev.
17:52–59. 2007. View Article : Google Scholar
|
|
20
|
Sahlgren C, Gustafsson MV, Jin S,
Poellinger L and Lendahl U: Notch signaling mediates
hypoxia-induced tumor cell migration and invasion. Proc Natl Acad
Sci USA. 105:6392–6397. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Liu N, Zhang J and Ji C: The emerging
roles of Notch signaling in leukemia and stem cells. Biomark Res.
1:232013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zou J, Li P, Lu F, Liu N, Dai J, Ye J, Qu
X, Sun X, Ma D, Park J, et al: Notch1 is required for
hypoxia-induced proliferation, invasion and chemoresistance of
T-cell acute lymphoblastic leukemia cells. J Hematol Oncol.
6:32013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ninomiya S, Tyybäkinoja A, Borze I, Räty
R, Saarinen-Pihkala UM, Usvasalo A, Elonen E and Knuutila S:
Integrated analysis of gene copy number, copy neutral LOH, and
microRNA profiles in adult acute lymphoblastic leukemia. Cytogenet
Genome Res. 136:246–255. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Fallah P, Amirizadeh N, Poopak B, Toogeh
G, Arefian E, Kohram F, Hosseini Rad SM, Kohram M, Teimori Naghadeh
H and Soleimani M: Expression pattern of key microRNAs in patients
with newly diagnosed chronic myeloid leukemia in chronic phase. Int
J Lab Hematol. 37:560–568. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Su H, Yang JR, Xu T, Huang J, Xu L, Yuan Y
and Zhuang SM: MicroRNA-101, down-regulated in hepatocellular
carcinoma, promotes apoptosis and suppresses tumorigenicity. Cancer
Res. 69:1135–1142. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Liu XY, Liu ZJ, He H, Zhang C and Wang YL:
MicroRNA-101-3p suppresses cell proliferation, invasion and
enhances chemotherapeutic sensitivity in salivary gland adenoid
cystic carcinoma by targeting Pim-1. Am J Cancer Res. 5:3015–3029.
2015.PubMed/NCBI
|
|
27
|
Li JT, Jia LT, Liu NN, Zhu XS, Liu QQ,
Wang XL, Yu F, Liu YL, Yang AG and Gao CF: MiRNA-101 inhibits
breast cancer growth and metastasis by targeting CX chemokine
receptor 7. Oncotarget. 6:30818–30830. 2015.PubMed/NCBI
|
|
28
|
Zheng HB, Zheng XG and Liu BP: miRNA-101
inhibits ovarian cancer cells proliferation and invasion by
down-regulating expression of SOCS-2. Int J Clin Exp Med.
8:20263–20270. 2015.
|
|
29
|
Capaccione KM and Pine SR: The Notch
signaling pathway as a mediator of tumor survival. Carcinogenesis.
34:1420–1430. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
O'Neil J, Calvo J, McKenna K,
Krishnamoorthy V, Aster JC, Bassing CH, Alt FW, Kelliher M and Look
AT: Activating Notch1 mutations in mouse models of T-ALL. Blood.
107:781–785. 2006. View Article : Google Scholar
|
|
31
|
Chan SM, Weng AP, Tibshirani R, Aster JC
and Utz PJ: Notch signals positively regulate activity of the mTOR
pathway in T-cell acute lymphoblastic leukemia. Blood. 110:278–286.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Real PJ, Tosello V, Palomero T, Castillo
M, Hernando E, de Stanchina E, Sulis ML, Barnes K, Sawai C,
Homminga I, et al: Gamma-secretase inhibitors reverse
glucocorticoid resistance in T cell acute lymphoblastic leukemia.
Nat Med. 15:50–58. 2009. View
Article : Google Scholar
|
|
33
|
Nefedova Y, Cheng P, Alsina M, Dalton WS
and Gabrilovich DI: Involvement of Notch-1 signaling in bone marrow
stromamediated de novo drug resistance of myeloma and other
malignant lymphoid cell lines. Blood. 103:3503–3510. 2004.
View Article : Google Scholar
|
|
34
|
Nefedova Y, Sullivan DM, Bolick SC, Dalton
WS and Gabrilovich DI: Inhibition of Notch signaling induces
apoptosis of myeloma cells and enhances sensitivity to
chemotherapy. Blood. 111:2220–2229. 2008. View Article : Google Scholar
|