Introduction
Hepatocellular carcinoma (HCC) is one of the most
common cancers worldwide and the third leading cause of
cancer-related death globally (1).
Despite considerable advances in treatment modalities, the
long-term survival of HCC patients remains poor because of its high
recurrence and early metastasis (2). However, the underlying mechanism
responsible for the development and progression of HCC has not been
fully elucidated (3). Therefore, it
is urgent to develop a novel therapeutic target involved in
HCC.
Runt-related transcription factor 2 (RUNX2), which
belongs to RUNX family, are distinctive by a highly conserved 128
amino acid DNA binding/protein-protein interaction domain (4). It functions as a major regulator for
osteoblast differentiation and regulates endochondral bone
formation physiological progression (5–7).
Recent studies suggested that RUNX2 are involved in many types of
human cancer development, progression and metastasis (8–10).
In vitro studies showed that RUNX2 promoted the migration
and invasion capacity of prostate cancer cells (11,12).
In breast cancer, RUNX2 promoted tumorsphere formation by
regulating soluble E-cadherin expression associated with the TAZ
transcriptional co-activator (13).
Moreover, it was recently reported that RUNX2 played a critical
role in the process of epithelial to mesenchymal transition (EMT)
whose characteristics include increased migration, invasion and
metastasis potential by upregulating the transcription factors,
such as SOX9 and SMAD3 (12). In
addition, RUNX2 was essential for cellular movement and
cytoskeleton remodeling. In non-small cell lung cancer, the
increased RUNX2 presents resistance to cisplatin chemosensitivity.
Furthermore, RUNX2 promoted migration and invasion potential of
thyroid tumor cells by activating the expression levels of MMPs
(14). These studies suggest that
RUNX2 probably functions as an oncogene for tumorigenesis and
metastasis. However, the precise function of RUNX2 and the
underlying mechanisms in HCC remain unclear.
In the present study, we demonstrated that RUNX2
expression is upregulated in HCC. Clinical analysis reveals that
the increased RUNX2 expression was associated with poor prognostic
features and was an independent prognostic marker for predicting
survival of HCC patients. RUNX2 promotes cell migration and
invasion by regulating MMP9 expression in HCC cells.
Mechanistically, the pro-metastatic effect of RUNX2 could be
abrogated by inhibiting MMP9 expression in vitro. Our data
suggest that RUNX2 probably promotes MMP9 expression and thus,
induces the metastasis of HCC cells.
Materials and methods
Clinical tissues and data
Ninety-six HCC tissues and matched tumor-adjacent
tissues were obtained from patients including 82 males and 14
females, who underwent curative resection surgery in the Department
of Hepato-Biliary-Pancreatic Surgery, The Affiliated Cancer
Hospital of Zhengzhou University from January 2006 to December
2009. All tissues were used after obtaining informed consent.
Patients did not receive preoperative chemotherapy or embolization.
The demographic and clinicopathological data were obtained through
medical records. The experimental protocols were approved by the
Zhengzhou University Ethics Committee according to the Declaration
of Helsinki.
Cell lines and transfection
The human HCC cell lines (HepG2, MHCC-97L, Hep3B,
SMMC-7721, MHCC-97H and HCCLM3) and immortalized normal hepatic
cell line LO2 were purchased from the Institute of Biochemistry and
Cell Biology (Chinese Academy of Sciences, Shanghai, China). The
cells were cultured in complete Dulbecco's modified Eagle's medium
(DMEM; Invitrogen, Carlsbad, CA, USA) supplemented with 10% fetal
bovine serum (FBS; Gibco, Grand Island, NY, USA) at 37°C in a
humidified incubator in 5% CO2.
Retroviral vectors pMMP-RUNX2 and pMMP-MMP9 were
generated by inserting the cDNA into pMMP. The specific siRNA
against RUNX2 (5′-UAACAGCAGAGGCAUUUCGUAGCUC-3′), MMP9
(5′-CUAUGGUCCUCGCCCUGAA-3′) and scramble siRNA
(5′-UUCUCCGAACGUGUCACGUUUGUGC-3′) were synthesized by Shanghai
GenePharma Co., Ltd. (Shanghai, China). Cells were transfected with
the siRNAs mentioned above using Lipofectamine 2000 (Invitrogen)
according to the manufacturer's instructions.
Real-time quantitative reverse
transcription polymerase chain reaction (qRT-PCR)
Total RNA was extracted from respective HCC cells
and clinical samples using TRIzol reagent (Invitrogen). The first
strand cDNA was reverse transcribed with miRNA assay kit (Applied
Biosystems, Foster City, CA, USA) and quantified by a
SYBR® Premix Ex Taq™ II (Perfect Real-Time) kit (Takara
Bio, Inc., Shiga, Japan) and performed on the ABI PRISM 7300
Sequence Detection System (Applied Biosystems). qPCR primer against
RUNX2 (HQP016478) and GAPDH (HQP006940) were purchased from
Genecopoeia (Guangzhou, China).
Western blot analysis
The HCC cells and clinical tissues were collected
and lysed, then the protein concentration was quantified using the
BCA reagent (Pierce, Rockford, IL, USA). A total of 30 µg
protein was separated by 10% SDS-PAGE and transferred onto a PVDF
membrane (Bio-Rad Laboratories, Hercules, CA, USA). The membranes
were probed with the following primary antibodies: RUNX2 (1:1,000;
Cell Signaling Technology, Inc., Danvers, MA, USA), MMP9 (1:1,000;
Santa Cruz Biotechnology, Santa Cruz, CA, USA), GAPDH (G8140; US
Biological, Salem, MA, USA) overnight. The membranes were incubated
with appropriate HRP-conjugated secondary antibody (ZSGB-BIO,
Beijing, China). Protein bands were visualized using an enhanced
chemiluminescence kit (Amersham, Little Chalfont, UK).
Immunohistochemical staining
Samples were fixed in formalin and embedded in
paraffin and sections were cut at 4 µm thickness. RUNX2 and
MMP9 (1:400; Cell Signaling Technology) antibody was performed in
immunohistochemistry according to a standard
streptavidin-peroxidase-conjugated (SP-IHC) procedures. The
staining results for the RUNX2 and MMP9 protein were
semi-quantitatively evaluated by the staining intensity and the
percentage of positive staining cells. The percentage of positive
cells was divided into four grades: 0 for <5%; 1 for 6–25%; 2
for 26–50%; 3 for 51–75% and 4 for >75%. Staining intensity was
assessed by four degrees: 0, negative; 1, weak; 2, moderate; and 3,
strong. Each section was assayed for ten independent high
magnification (×400) fields to get the average scores.
Cell migration and invasion assays
Transwell cell migration and invasion assays were
carried out by the 8 µM pore-sized Transwell inserts (Nalge
Nunc International Corp., Naperville, IL, USA). Transfected cells
were seeded at 2.5×105/ml in 200 µl serum-free
DMEM medium into the upper chamber, and 750 µl DMEM medium
containing 10% FBS was placed in the lower chamber. After 24-h
incubation, cells were fixed in 4% paraformaldehyde for 20 min and
stained with 0.1% crystal violet dye for 15 min. The cells on the
inner layer were softly removed with a cotton swab; 1:6 dilution
Matrigel invasion chamber (BD Biosciences, San Jose, CA, USA) was
performed for invasion assays and the following assays were same as
before.
Statistical analysis
Results are shown as mean ± standard deviation. The
SPSS 13 (SPSS, Inc., Chicago, IL, USA) and GraphPad Prism 5
software (GraphPad Software, Inc., San Diego, CA, USA) were used
for Pearson Chi-squared test and the multivariate Cox regression
analysis. Two-tailed Student's t-test, a Kaplan-Meier plot, a
log-rank test, a Spearman's rank correlation coefficient or an
ANOVA were used to evaluate the statistical significance.
Difference was defined as P<0.05.
Results
The expression level of RUNX2 in HCC
tissues and cells
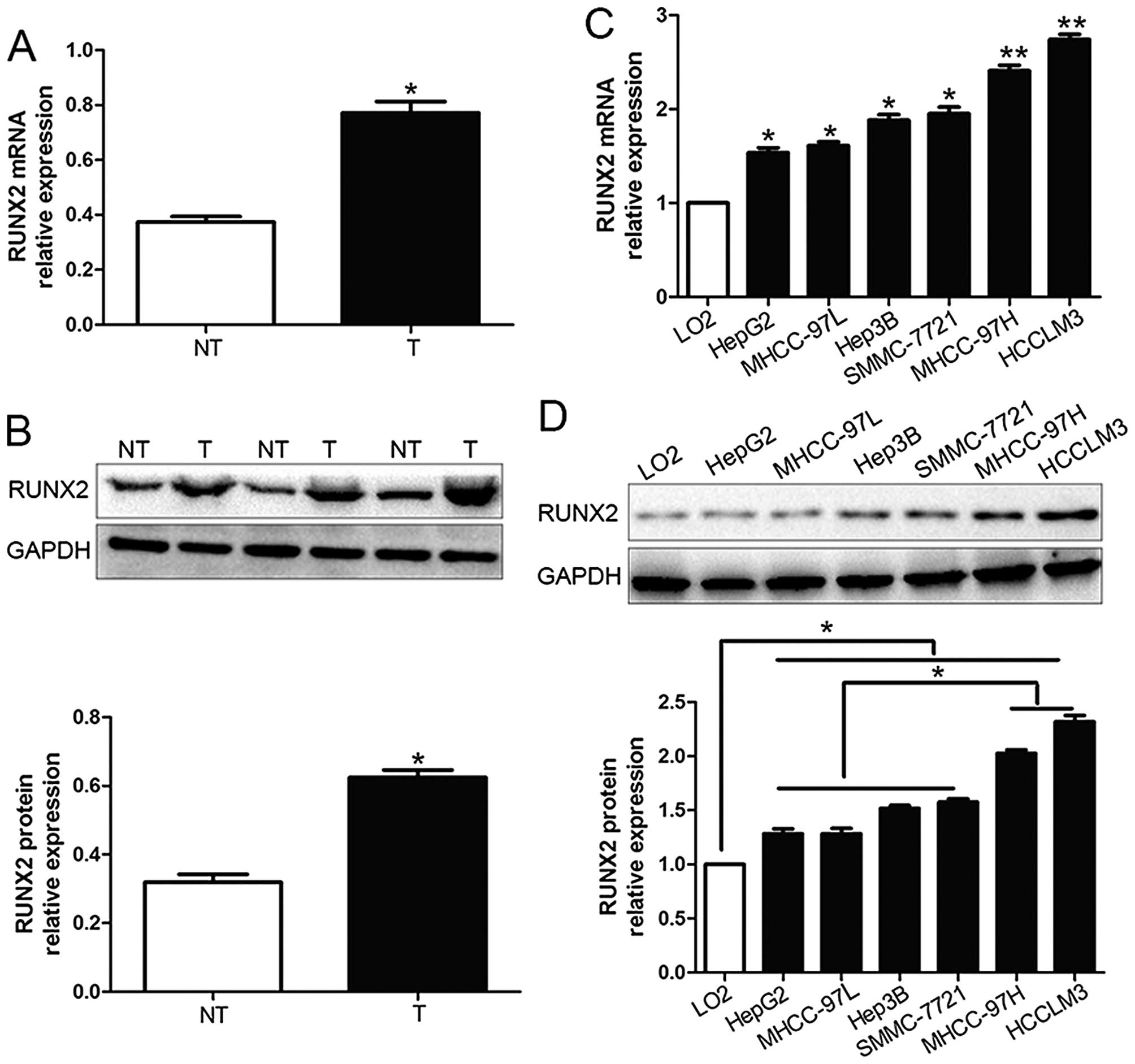
We investigated the expression level of RUNX2 in 96
pairs of HCC tissues and corresponding adjacent non-tumor tissues
by qRT-PCR and western blot analysis. We found that RUNX2 mRNA and
protein expression in HCC tissues were both obviously higher than
those in corresponding adjacent non-tumor tissues (P<0.05;
Fig. 1A and B). Moreover, RUNX2
mRNA levels were increased in HCC cell lines (HepG2, Hep3B,
SMMC-7721, MHCC-97L, HCCLM3 and MHCC-97H) compared to the normal
hepatocyte cell line, LO2 (P<0.05; Fig. 1C). In addition, the levels of RUNX2
protein showed the same result with mRNA levels in all cell lines
by western blot analysis (P<0.05; Fig. 1D). Furthermore, RUNX2 expression in
MHCC-97H and HCCLM3, which was considered as highly metastasis were
prominently higher than those in the low metastasis HCC cell lines
(HepG2, Hep3B, SMMC-7721 and MHCC-97L) (P<0.05; Fig. 1D). Thus, these results suggest that
elevated RUNX2 expression probably plays a critical role in the
development of HCC.
Correlation between RUNX2 expression and
clinicopathological features
We defined the mean level of RUNX2 protein as a
cut-off value to distinguish the RUNX2 expression level. As shown
in Table I, the high expression of
RUNX2 protein was significantly associated with multiple tumor
nodes (P=0.006), venous infiltration (P=0.009), high
Edmondson-Steiner grading (P=0.001) and advanced
tumor-node-metastasis (TNM) stage (P= 0.004). Hence, these data
suggest that the increased expression of RUNX2 is correlated with
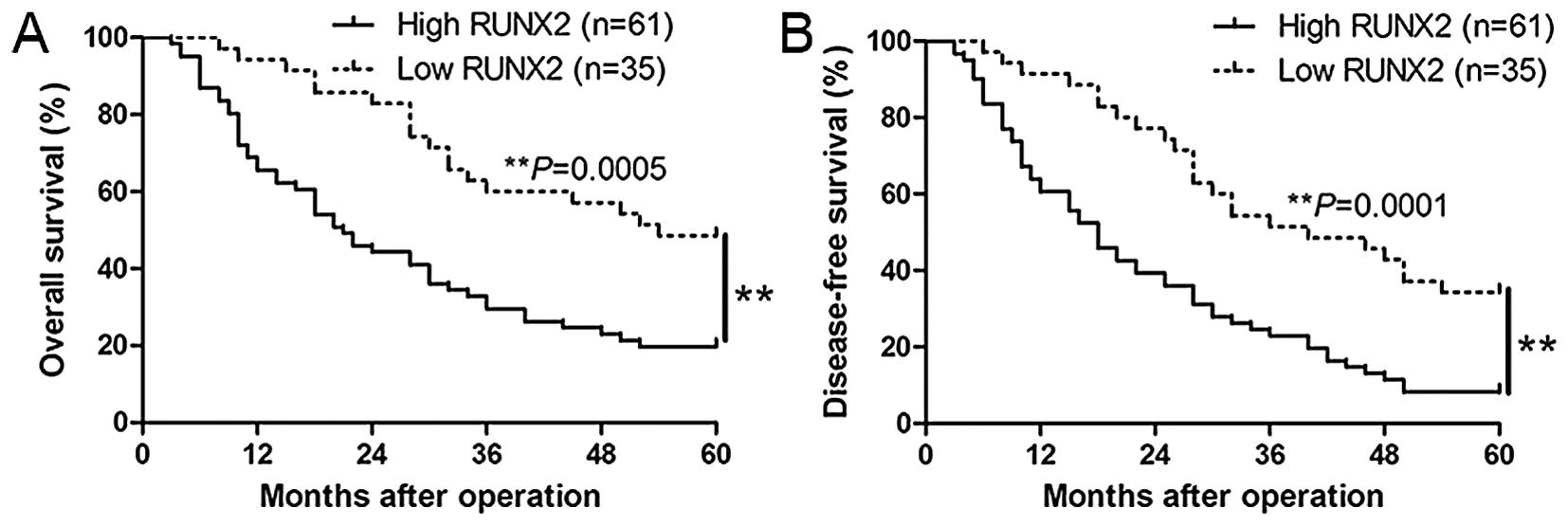
adverse prognostic features of HCC. In addition, Kaplan-Meier
analysis revealed that high RUNX2 expression was correlated with
worse overall survival (P=0.0005; Fig.
2A) and disease-free survival (P=0.0001, Fig. 2B) of HCC patients. In addition,
RUNX2 expression was a novel independent factor for predicting both
5-year overall and disease-free survival in HCC patients (P=0.021
and 0.039, respectively, Table
II). Taken together, these results highlight the potential
value of RUNX2 for the outcome of HCC.
 | Table IClinical correlation of RUNX2
expression in HCC (n=96). |
Table I
Clinical correlation of RUNX2
expression in HCC (n=96).
| Clinical
parameters | Cases
(n) | Expression level
| P-value
(aP<0.05) |
|---|
RUNX2high
(n=61) |
RUNX2low
(n=35) |
|---|
| Age (years) | | | | |
| <65 | 28 | 18 | 10 | 0.923 |
| ≥65 | 68 | 43 | 25 | |
| Gender | | | | |
| Male | 82 | 52 | 30 | 0.950 |
| Female | 14 | 9 | 5 | |
| Tumor size (cm) | | | | 0.128 |
| <5 | 37 | 27 | 10 | |
| ≥5 | 59 | 34 | 25 | |
| Tumor number | | | |
0.006a |
| Solitary | 76 | 43 | 33 | |
| Multiple | 20 | 18 | 2 | |
| Edmondson | | | |
0.001a |
| I+II | 32 | 13 | 19 | |
| III+IV | 64 | 48 | 16 | |
| TNM stage | | | |
0.004a |
| I+II | 65 | 35 | 30 | |
| III+IV | 31 | 26 | 5 | |
| Venous
infiltration | | | |
0.009a |
| Present | 26 | 22 | 4 | |
| Absent | 70 | 39 | 31 | |
| AFP (ng/ml) | | | | 0.619 |
| <400 | 38 | 23 | 15 | |
| ≥400 | 58 | 38 | 20 | |
| HBsAg | | | | 0.810 |
| Positive | 84 | 53 | 31 | |
| Negative | 12 | 8 | 4 | |
 | Table IIMultivariate Cox regression analysis
of 5-year overall and disease-free survival of 96 HCC patients. |
Table II
Multivariate Cox regression analysis
of 5-year overall and disease-free survival of 96 HCC patients.
| Variables | Overall survival
| Disease-free
survival
|
|---|
| HR | 95% CI | P-value | HR | 95% CI | P-value |
|---|
| RUNX2 | 2.179 | 1.107–4.658 | 0.021a | 3.648 | 1.084–5.103 | 0.039a |
| Edmondson
grade | 2.148 | 0.846–5.082 | 0.068 | 1.981 | 0.834–4.596 | 0.135 |
| TNM stage | 1.134 | 1.013–1.948 | 0.023a | 1.023 | 1.004–1.768 | 0.003a |
| No. of tumor
nodule | 1.712 | 0.684–4.249 | 0.214 | 1.132 | 0.954–3.821 | 0.873 |
| Venous
infiltration | 1.078 | 0.987–1.247 | 0.057 | 1.023 | 0.658–1.784 | 0.856 |
RUNX2 promotes HCC cell migration and
invasion
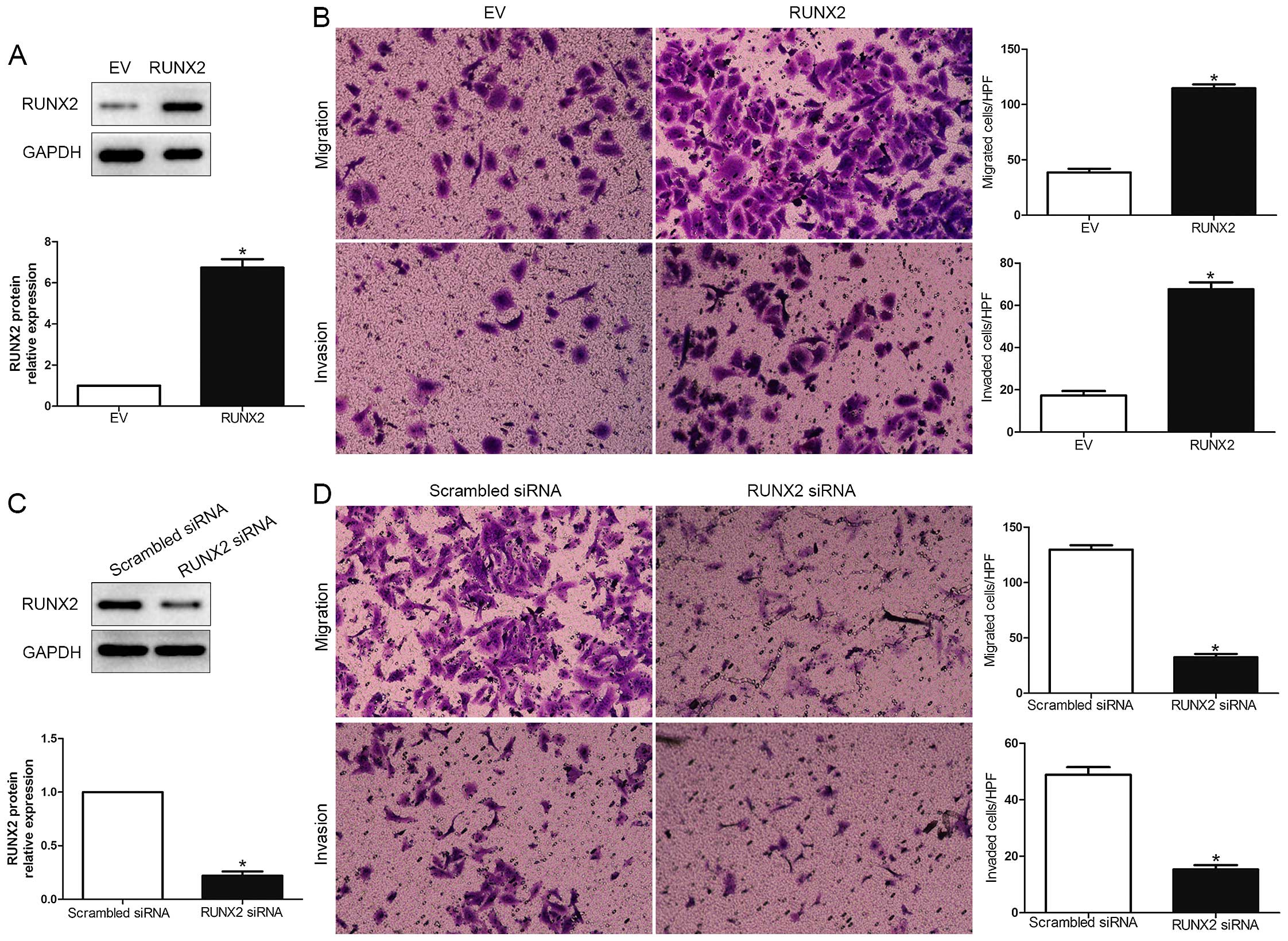
To explore the biological function of RUNX2 in HCC,
we transfected HCC cell line MHCC-97L with empty vector (EV) or
RUNX2 retroviruses (P<0.01; Fig.
3A). We demonstrated that RUNX2 overexpression prominently
promoted HCC cell migration and invasion in MHCC-97L (P<0.05;
Fig. 3B). Moreover, RUNX2 was
knocked down using a specific siRNA in HCCLM3 cells (P<0.05;
Fig. 3C). As expected,
downregulated RUNX2 led to a significant reduction of cell
migration and invasion (P<0.05; Fig.
3D). These data suggest that RUNX2 regulates the migration and
invasion of HCC cells.
RUNX2 positively regulates MMP9 in
HCC
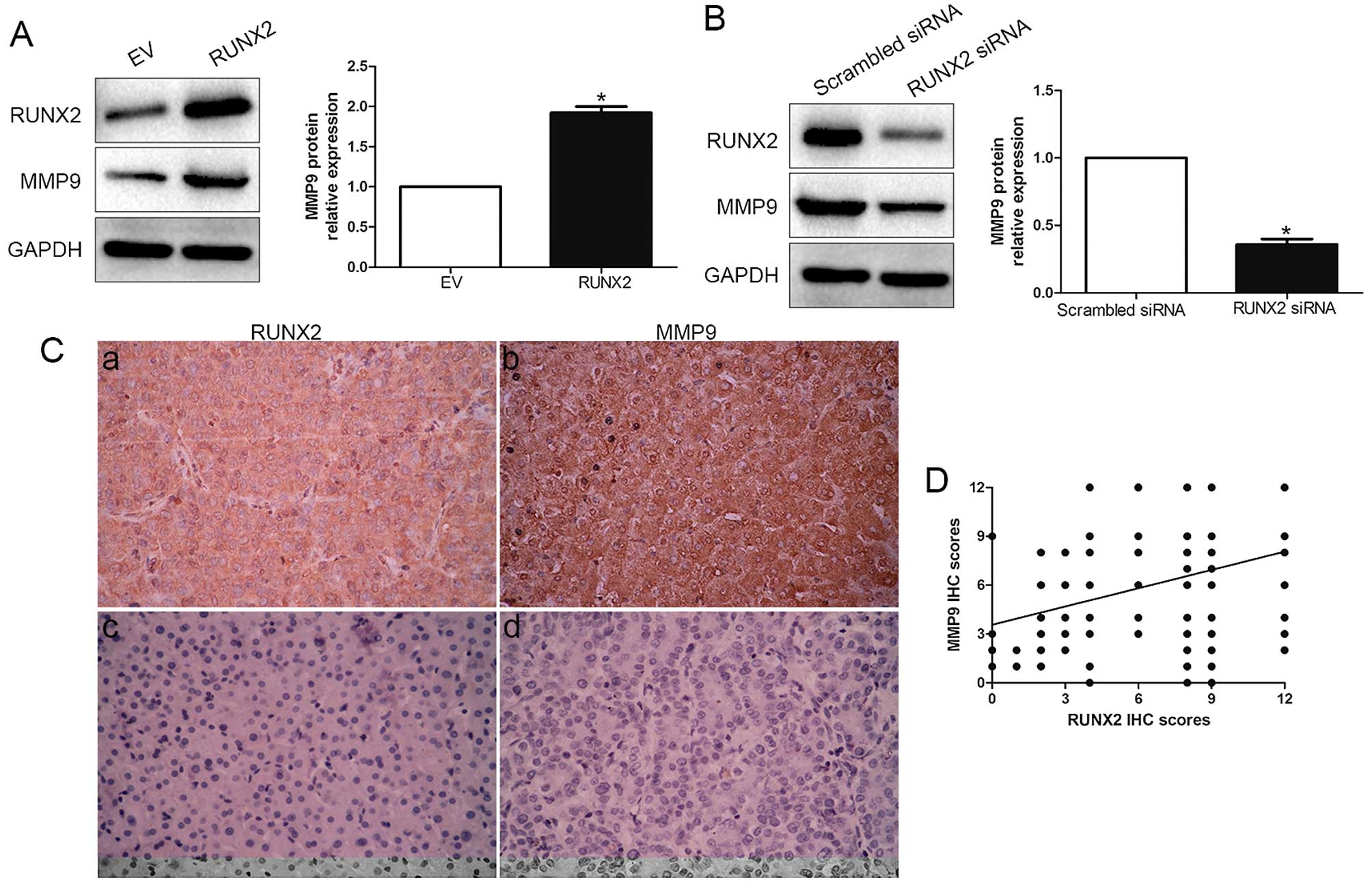
The positive correlation between RUNX2 and MMP9
expression was reported previously in breast cancer (15–17),
thus, we evaluated the effect of RUNX2 on MMP9. Amazingly, our
results showed that RUNX2 overexpression obviously increased the
expression level of MMP9 in MHCC-97L cells (P<0.05; Fig. 4A). Moreover, RUNX2 knockdown
significantly decreased the level of MMP9 in HCCLM3 cells
(P<0.05; Fig. 4B). We then
analysed RUNX2 and MMP9 expression by immunostaining in HCC
samples. Both RUNX2 and MMP9 expression in cancer tissues were
significantly higher than those in paired non-cancerous tissues
(P<0.05; Fig. 4C). Moreover, IHC
scores were evaluated for semi-quantitative analysis, we found a
strong positive correlation between RUNX2 and MMP9 (r=0.534,
P<0.001; Fig. 4D). These data
revealed that RUNX2 exerts its biological promotive function
through elevating expression of MMP9.
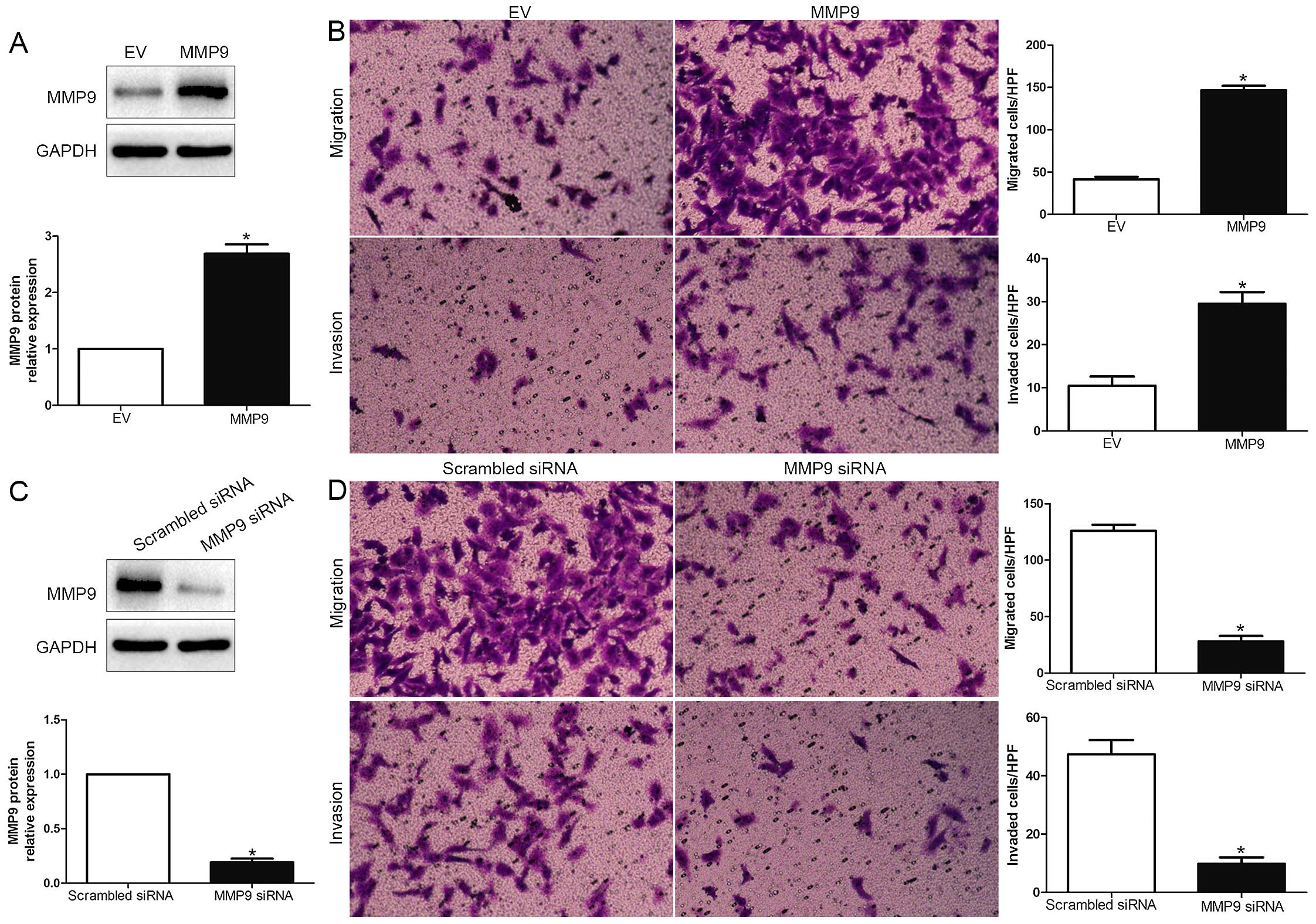
RUNX2 promotes HCC cell migration and
invasion by increasing MMP9
To explore whether MMP9 participates in RUNX2
mediated promotion of HCC cell migration and invasion, MMP9
expression was significantly increased by MMP9 retroviruses in
MHCC-97L cells (P<0.05; Fig. 5A)
and significantly knocked down using a specific siRNA in HCCLM3
cells (P<0.05; Fig. 5C). As
shown in Fig. 5B and 5D, MMP9 overexpression obviously promoted,
while MMP9 knockdown obviously blocked HCC cell migration and
invasion (P<0.05; respectively). In addition, RUNX2
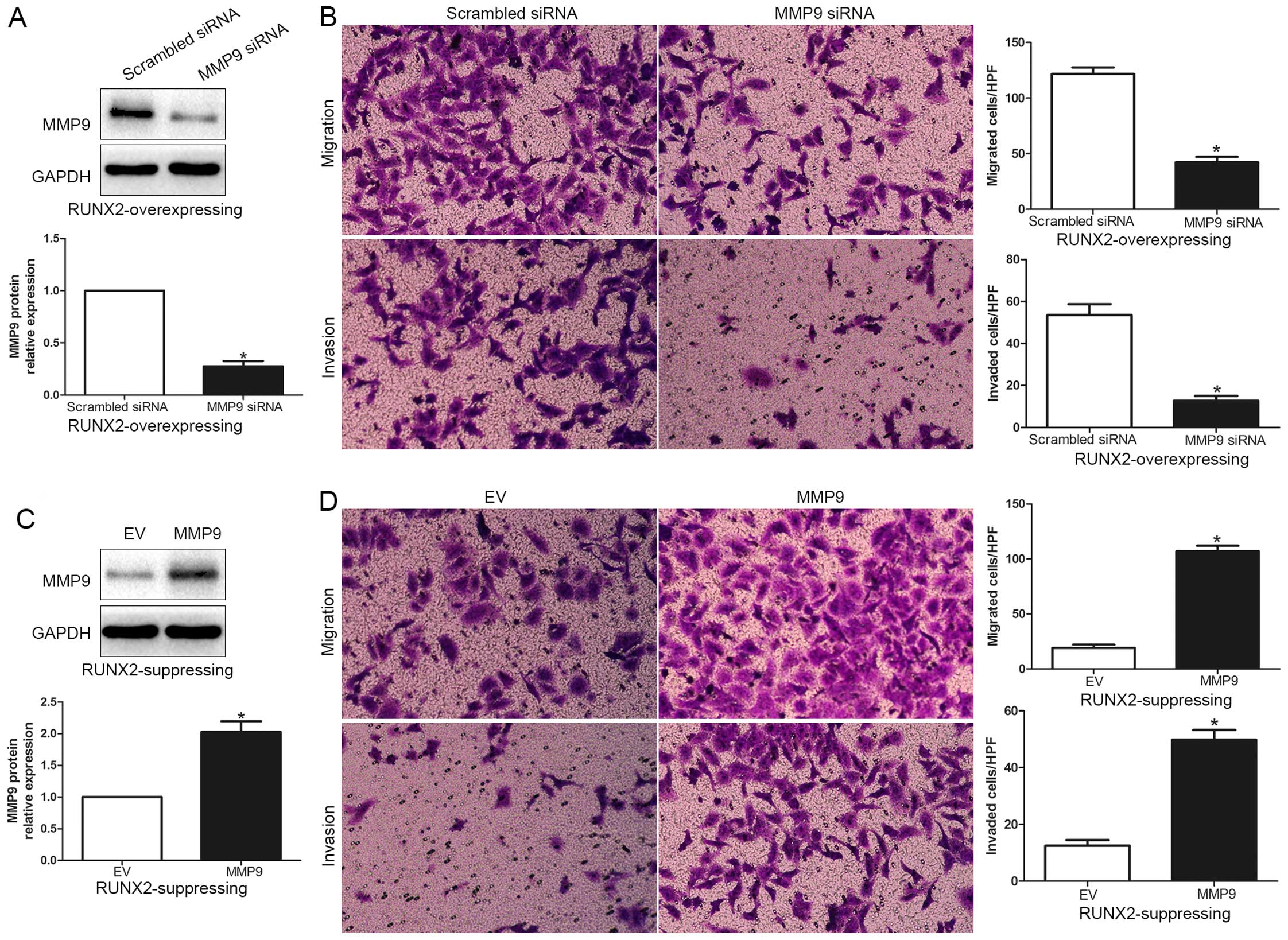
overexpressing MHCC-97L cells were subsequently transfected with
MMP9 siRNA (P<0.05; Fig. 6A).
MMP9 knockdown markedly abrogated the function of exogenous RUNX2
overexpression, causing a significant decrease in the number of
migrated and invaded cells (P<0.05; Fig. 6B). Similarly, MMP9 overexpression
promoted HCC cell migration and invasion in RUNX2-knockdown HCCLM3
cells (P<0.05, respectively; Fig. 6C
and D). In conclusion, these results suggest that MMP9 may
function as a downstream factor in RUNX2 mediated promotion of HCC
cell migration and invasion.
Discussion
In the present study, we initially checked RUNX2
expression level in 96 specimens of HCC tissues. Our results show
that the RUNX2 expression level in HCC was obviously higher than
that in non-tumor tissues. Increased expression levels of RUNX2
mRNA and protein were also confirmed in different HCC cell lines,
especially in the highly metastatic cell lines. Moreover, elevated
expression of RUNX2 was significantly correlated with multiple
tumor nodes, high histological grade, TNM stage and venous
infiltration. These data suggest that the increased RUNX2
expression is correlated with adverse prognostic characters in HCC.
Furthermore, patients with higher RUNX2 had a worse prognosis of
HCC patients, which was consistent with that in acute lymphoblastic
leukemia patients (18).
Multivariate Cox repression analysis demonstrated that RUNX2 was a
novel independent prognostic factor for predicting survival of HCC
patients. Taken together, these results indicate that RUNX2
expression is critical for prognosis of HCC patients.
Previous studies suggest that RUNX2 functions as an
oncogene in breast cancer development through Wnt and TGF-β
signaling pathways (19–21). Moreover, downregulated RUNX2
inhibits the invasion of osteosarcoma (22). In the present study, we found that
RUNX2 overexpression prominently promoted MHCC-97L cell migration
and invasion and RUNX2 knockdown significantly reduced the migrated
and invaded HCCLM3 cells. These data suggest that RUNX2 exerts its
biological function on HCC cell migration and invasion. Peng et
al (23) reported that RUNX2
promoted trophoblast invasion via MMP9 expression. Our data
demonstrated that RUNX2 overexpression upregulated MMP9 in MHCC-97L
cells and RUNX2 knockdown led to MMP9 reduction in HCCLM3 cells. In
addition, we demonstrated that the MMP9 expression level in high
RUNX2-expressed HCC tissues was prominently higher than that from
low RUNX2-expressed group. Spearman correlation analysis suggest a
positive correlation between RUNX2 and MMP9 expression in HCC
tissues. Taken together, these data indicate that RUNX2 positively
regulate MMP9 accumulation in HCC. Furthermore, we confirmed that
MMP9 overexpression obviously promoted HCC cell migration and
invasion in MHCC-97L cells, while MMP9 knockdown obviously
inhibited migration and invasion in HCCLM3 cells, which was
consistent with previous reports. Moreover, the promotion in HCC
migration and invasion by RUNX2 overexpression could be abolished
by MMP9 knockdown, and the suppression in HCC cell migration and
invasion by RUNX2 knockdown could be reverted by restoring MMP9
expression. Taken together, these data indicate that RUNX2 promotes
HCC cell migration and invasion by increasing MMP9 expression. In
osteoblast, RUNX2 exerts as a regulator in metastasis by
interaction with PI3K/AKT signaling (24,25).
Similarly, in breast cancer, RUNX2 directly regulates the
expression of MMP9 and MMP13. Furthermore, RUNX2 overexpression
upregulates transcription factors (SOX9, SMAD3 and SNAI2)
implicated in the process of epithelial to mesenchymal transition
(12,26–29).
Therefore, we need further investigation to explore the molecular
mechanisms between RUNX2 and MMP9 in HCC.
In conclusion, the data show that the expression of
RUNX2 is elevated in HCC tissues and cell lines and its high
expression is associated with malignant clinicopathological
features. We confirm that RUNX2 is an independent prognostic marker
for predicting 5-year survival of HCC patients. We demonstrate that
RUNX2 promotes HCC cell migration and invasion in vitro.
Mechanistically, we suggest that RUNX2 may promote HCC invasion and
metastasis by increasing MMP9. Taken together, we consider that
RUNX2 may potentially act as a clinical biomarker, and may also be
a therapeutic target in HCC.
Acknowledgments
The present study was supported by a grant from the
Scientific Research Foundation of Henan (no. 092102310090).
References
|
1
|
El-Serag HB and Rudolph KL: Hepatocellular
carcinoma: Epidemiology and molecular carcinogenesis.
Gastroenterology. 132:2557–2576. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Talwalkar JA and Gores GJ: Diagnosis and
staging of hepatocellular carcinoma. Gastroenterology. 127(Suppl
1): S126–S132. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ito Y: Oncogenic potential of the RUNX
gene family: 'overview'. Oncogene. 23:4198–4208. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Li J, Hao L, Wu J, Zhang J and Su J:
Linarin promotes osteogenic differentiation by activating the
BMP-2/RUNX2 pathway via protein kinase A signaling. Int J Mol Med.
37:901–910. 2016.PubMed/NCBI
|
|
6
|
Ozaki T, Nakamura M and Shimozato O: Novel
implications of DNA damage response in drug resistance of malignant
cancers obtained from the functional interaction between p53 family
and RUNX2. Biomolecules. 5:2854–2876. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sugimoto H, Nakamura M, Yoda H, Hiraoka K,
Shinohara K, Sang M, Fujiwara K, Shimozato O, Nagase H and Ozaki T:
Silencing of RUNX2 enhances gemcitabine sensitivity of
p53-deficient human pancreatic cancer AsPC-1 cells through the
stimulation of TAp63-mediated cell death. Cell Death Dis.
6:e19142015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wysokinski D, Blasiak J and Pawlowska E:
Role of RUNX2 in breast carcinogenesis. Int J Mol Sci.
16:20969–20993. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Shrivats AR, McDermott MC, Klimak M,
Averick SE, Pan H, Matyjaszewski K, Mishina Y and Hollinger JO:
Nanogel-mediated RNAi against Runx2 and Osx inhibits osteogenic
differentiation in constitutively active BMPR1A osteoblasts. ACS
Biomater Sci Eng. 1:1139–1150. 2015. View Article : Google Scholar
|
|
10
|
Fujita T, Azuma Y, Fukuyama R, Hattori Y,
Yoshida C, Koida M, Ogita K and Komori T: Runx2 induces osteoblast
and chondrocyte differentiation and enhances their migration by
coupling with PI3K-Akt signaling. J Cell Biol. 166:85–95. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Akech J, Wixted JJ, Bedard K, van der Deen
M, Hussain S, Guise TA, van Wijnen AJ, Stein JL, Languino LR,
Altieri DC, et al: Runx2 association with progression of prostate
cancer in patients: Mechanisms mediating bone osteolysis and
osteoblastic metastatic lesions. Oncogene. 29:811–821. 2010.
View Article : Google Scholar :
|
|
12
|
Baniwal SK, Khalid O, Gabet Y, Shah RR,
Purcell DJ, Mav D, Kohn-Gabet AE, Shi Y, Coetzee GA and Frenkel B:
Runx2 transcriptome of prostate cancer cells: Insights into
invasiveness and bone metastasis. Mol Cancer. 9:2582010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Brusgard JL, Choe M, Chumsri S, Renoud K,
MacKerell AD Jr, Sudol M and Passaniti A: RUNX2 and TAZ-dependent
signaling pathways regulate soluble E-cadherin levels and
tumorsphere formation in breast cancer cells. Oncotarget.
6:28132–28150. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Sancisi V, Borettini G, Maramotti S,
Ragazzi M, Tamagnini I, Nicoli D, Piana S and Ciarrocchi A: Runx2
isoform I controls a panel of proinvasive genes driving
aggressiveness of papillary thyroid carcinomas. J Clin Endocrinol
Metab. 97:E2006–E2015. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Mendoza-Villanueva D, Deng W,
Lopez-Camacho C and Shore P: The Runx transcriptional co-activator,
CBFbeta, is essential for invasion of breast cancer cells. Mol
Cancer. 9:1712010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Pratap J, Javed A, Languino LR, van Wijnen
AJ, Stein JL, Stein GS and Lian JB: The Runx2 osteogenic
transcription factor regulates matrix metalloproteinase 9 in bone
metastatic cancer cells and controls cell invasion. Mol Cell Biol.
25:8581–8591. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Selvamurugan N, Kwok S and Partridge NC:
Smad3 interacts with JunB and Cbfa1/Runx2 for transforming growth
factor-beta1-stimulated collagenase-3 expression in human breast
cancer cells. J Biol Chem. 279:27764–27773. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Edwards H, Xie C, LaFiura KM, Dombkowski
AA, Buck SA, Boerner JL, Taub JW, Matherly LH and Ge Y: RUNX1
regulates phosphoinositide 3-kinase/AKT pathway: Role in
chemotherapy sensitivity in acute megakaryocytic leukemia. Blood.
114:2744–2752. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Tandon M, Chen Z, Othman AH and Pratap J:
Role of Runx2 in IGF-1Rβ/Akt- and AMPK/Erk-dependent growth,
survival and sensitivity towards metformin in breast cancer bone
metastasis. Oncogene. Jan 25–2016.Epub ahead of print. View Article : Google Scholar
|
|
20
|
Sancisi V, Gandolfi G, Ragazzi M, Nicoli
D, Tamagnini I, Piana S and Ciarrocchi A: Cadherin 6 is a new RUNX2
target in TgF-β signalling pathway. PLoS One. 8:e754892013.
View Article : Google Scholar
|
|
21
|
van der Deen M, Akech J, Wang T,
Fitzgerald TJ, Altieri DC, Languino LR, Lian JB, van Wijnen AJ,
Stein JL and Stein GS: The cancer-related Runx2 protein enhances
cell growth and responses to androgen and TGFbeta in prostate
cancer cells. J Cell Biochem. 109:828–837. 2010.PubMed/NCBI
|
|
22
|
Zeng H and Xu X: RUNX2 RNA interference
inhibits the invasion of osteosarcoma. Oncol Lett. 9:2455–2458.
2015.PubMed/NCBI
|
|
23
|
Peng B, Zhu H, Klausen C, Ma L, Wang YL
and Leung PC: GnRH regulates trophoblast invasion via
RUNX2-mediated MMP2/9 expression. Mol Hum Reprod. 22:119–129. 2016.
View Article : Google Scholar
|
|
24
|
Cohen-Solal KA, Boregowda RK and Lasfar A:
RUNX2 and the PI3K/AKT axis reciprocal activation as a driving
force for tumor progression. Mol Cancer. 14:1372015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Tandon M, Chen Z and Pratap J: Runx2
activates PI3K/Akt signaling via mTORC2 regulation in invasive
breast cancer cells. Breast Cancer Res. 16:R162014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Niu DF, Kondo T, Nakazawa T, Oishi N,
Kawasaki T, Mochizuki K, Yamane T and Katoh R: Transcription factor
Runx2 is a regulator of epithelial-mesenchymal transition and
invasion in thyroid carcinomas. Lab Invest. 92:1181–1190. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhang X, Akech J, Browne G, Russell S,
Wixted JJ, Stein JL, Stein GS and Lian JB: Runx2-Smad signaling
impacts the progression of tumor-induced bone disease. Int J
Cancer. 136:1321–1332. 2015. View Article : Google Scholar :
|
|
28
|
Chimge NO, Baniwal SK, Little GH, Chen YB,
Kahn M, Tripathy D, Borok Z and Frenkel B: Regulation of breast
cancer metastasis by Runx2 and estrogen signaling: The role of
SNAI2. Breast Cancer Res. 13:R1272011. View
Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zaidi SK, Sullivan AJ, van Wijnen AJ,
Stein JL, Stein GS and Lian JB: Integration of Runx and Smad
regulatory signals at transcriptionally active subnuclear sites.
Proc Natl Acad Sci USA. 99:8048–8053. 2002. View Article : Google Scholar : PubMed/NCBI
|