Introduction
Mammalian genomes generate thousands of regulatory
RNAs that are either long non-coding RNAs (lncRNAs) or microRNAs
(miRNAs) (1,2). lncRNAs are more than 200 nucleotides,
and synthesized by RNA polymerase II, spliced and sometimes
polyadenylated (3). They are
pervasively transcribed, and exhibit spatially and temporally
regulated expression patterns (4).
Unlike small ncRNAs, lncRNAs can fold into complex secondary and
higher order structures to provide greater potential and
versatility for both protein and target recognition (5). lncRNAs have been found to play crucial
regulatory roles in a diverse range of cellular processes and
biological pathways, including genomic imprinting, chromosome
inactivation, differentiation and development of many human
diseases (6). lncRNAs are emerging
as new players in the cancer biology paradigms and their
dysfunction are correlated with tumorigenesis and malignancy
transformation in various types of cancers (7,8).
miRNAs, the most well characterized ncRNAs, are
short endogenous molecules, approximately 22 nucleotides in length,
that are processed by the RNase III enzymes Drosha and Dcr. miRNAs
post-transcriptionally regulate the gene expression through
interaction between the 5′ end and the 3′ untranslated region
(3′UTR) of mRNA. miRNA can guide the RNA-induced silencing complex
(RISC) to miRNA response element (MRE) on target transcript,
usually resulting in degradation of the transcript or inhibition of
its translation (9). Dysregulation
of miRNA expression is involved in various diseases (10). Accumulating evidence highlights the
role of miRNA-mediated regulation in cell growth, differentiation,
proliferation and apoptosis. Alterations in the miRNA balance in
the cell can lead to dysregulation of tumor suppressor genes and/or
oncogenes regulated by aberrantly expressed miRNAs, leading to
cancer (11,12).
Recent studies have described a complicated
interplay among diverse RNA species, including coding and
non-coding RNAs. These RNAs inclusive of mRNA, pseudogene, lncRNA
or circular RNA, interact and co-regulate with each other by acting
as competing endogenous RNAs (ceRNAs). ceRNAs have MRE, and serve
as miRNA sponges to control miRNAs available to their target RNAs.
ceRNA can sequester miRNAs, thereby protecting their target RNAs
from repression (13).
Understanding this novel RNA interaction will lead to significant
insight into gene regulatory networks in human development and
disease. Although lacking 3′UTRs, lncRNAs have been reported to be
downregulated by miRNAs and work as ceRNAs. The experimental
evidence is already emerging of lncRNAs as competitive platforms
for both miRNAs and mRNAs (14,15).
Glioblastoma multiforme (GBM) is the most common and
malignant brain tumor with poor prognosis. According to the 2007
World Health Organization classification, gliomas are classified
into 4 histopathological grades based on malignancy degree, and GBM
is the highest-grade glioma (grade IV) (16). Patients with newly diagnosed GBM
exhibit a median survival of approximately 15 months (17). Despite maximal surgical,
radiological and chemotherapeutic interventions, these figures have
changed little in the past two decades (18). New therapeutic strategies will
likely evolve from a better understanding of GBM biology.
Efforts have been made to study the relationship
between the lncRNA expression and the GBM pathogenesis (8,19–21),
but many more lncRNAs playing crucial roles in GBM remain to be
determined. The aberrant miRNA expression has features of GBM
(22). Nevertheless, the
miRNA-lncRNA-mRNA regulation networks in the GBM, as well as the
potential roles of ceRNAs in the biogenesis and development of GBM
have not been explored.
In this study, we aimed at profiling the miRNA,
lncRNA and mRNA expression signature, and constructing
miRNA-lncRNA-mRNA crosstalk by analyzing a cohort of sample-matched
exon and miRNA expression microarrays from the Cancer Genome Atlas
(TCGA), and predicting the functions of lncRNAs acting as ceRNAs in
GBM. The identified sets of lncRNA, miRNA and mRNA specific to GBM
were subsequently confirmed by quantitative reverse
transcription-polymerase chain reaction (qRT-PCR) in GBM
samples.
Materials and methods
Data-set characteristics
The sample matched whole-transcript and miRNA
expression profiling upon GBM were obtained from the TCGA database
(https://tcga-data.nci.nih.gov/tcga/).
To compare the miRNA, lncRNA and mRNA expression signatures in GBM,
we selected 16 data-sets that included 8 GBM and 8 non-tumoral
brain samples. Two panels of data-sets were included in our study:
Affymetrix Human Exon 1.0 array and Agilent Human MicroRNA array
8×15K.
Data analysis
Two-class differential was used to determine the
differentially expressed miRNA, lncRNA and mRNA between the normal
and GBM groups. The random variance model (RVM) t-test was applied
to filter the differentially expressed genes for it can effectively
increase the degrees of freedom in cases of small samples. The
false discovery rate (FDR) was calculated to correct the P-value.
We selected the differentially expressed miRNAs, lncRNAs and mRNAs
according to the P-value and FDR. P-values <0.05 and FDR
<0.05 were considered significant.
The differentially expressed probe sets were
imported into Cluster and TreeView (Stanford University) to perform
hierarchical cluster analysis (HCA) (23).
Gene Ontology (GO) and pathway
analysis
A GO analysis was applied to analyze the main
functions of the differentially expressed mRNAs (24). Specifically, a two-sided Fisher's
exact test and a χ2 test were used to classify the GO
category. We computed P-values of the GO for each differential
gene. Enrichment provides a measure for the significant function:
As the enrichment increases, the corresponding function is more
specific. Within the significant category, the enrichment Re was
given as follows: Re = (nf/n)/(Nf/N), where
nf is the number of flagged genes within the particular
category, n is the total number of genes within the same category,
Nf is the number of flagged genes in the entire
microarray, and N is the total number of genes in the
microarray.
Pathway analysis was used to identify the
significant pathway of the differential mRNAs according to KEGG,
BioCarta and Reactome. We used Fisher's exact test and the
χ2 test to select the significant pathway, and the
threshold of significance was defined by P-value and FDR. The
enrichment Re was calculated as described above (25).
Construction of lncRNA-mRNA co-expression
network
The lncRNA-mRNA networks were built according to the
normalized signal intensity of specific mRNA and lncRNA expression
in microarray. For each pair of mRNA-lncRNA, mRNA-mRNA or
lncRNA-lncRNA, we calculated the Pearson correlation and chose the
significant correlation pairs to construct the network (26). In a network analysis, degree is the
most important measure of an mRNA or lncRNA centrality within a
network. Degree centrality is defined as the link numbers one node
has to the other (27). The
clustering coefficient represents the density of each gene with the
adjacent gene, and the larger the clustering coefficient, the
greater importance the gene has in regulating the network.
Patient samples
GBM specimens were derived from patients with GBM
who underwent surgical treatment at Beijing Tian Tan Hospital. All
histologic diagnoses were made on formalin fixed, paraffin-embedded
H&E sections and were reviewed blinded to the original
diagnosis according to the 2007 World Health Organization
classification. Normal brain tissues were obtained from severe head
trauma patients for whom partial resection of normal brain was
required during surgery at Beijing Tian Tan Hospital. Samples were
collected immediately after surgical resection, snap-frozen and
stored in liquid nitrogen. The study was approved by the
institutional review board of Beijing Tian Tan Hospital.
RNA preparation and qRT-PCR
Total RNA from tissue specimens was extracted using
the TRIzol reagent (Invitrogen Life Technologies, Carlsbad, CA,
USA). RNA integrity was analyzed on a 1.2% agarose gel. RNA
quantity was determined using a NanoDrop 2000c Spectrophotometer
(Thermo Fisher Scientific, Inc., Waltham, MA, USA). RNA (1
μg) was reverse transcribed with a PrimeScript™ RT reagent
kit (Takara Biotechnology Co., Ltd., Dalian, China) for cDNA
synthesis and genomic DNA removal. For miRNA detection, total RNA
was reverse transcribed using miRNA specific primers. qPCRs were
performed according to the instructions of the SYBR Premix Ex Taq™
II kit (Takara Biotechnology Co., Ltd.) and carried out in the
Takara real-time PCR system. Gene-specific primers were designed
using online primer designing tools primer-blast (http://www.ncbi.nlm.nih.gov/tools/primer-blast/). The
primer sequences are listed in Table
I. The lengths of amplifications are between 100 and 250 bp.
Standard deviations were calculated from three PCR replicates. The
specificity of amplification was assessed by dissociation curve
analysis and the relative abundance of genes was determined using
the comparative ΔΔCt method.
 | Table IThe miRNA, lncRNA and mRNA primers
for qRT-PCR. |
Table I
The miRNA, lncRNA and mRNA primers
for qRT-PCR.
| Primers | | Sequences
(5′-3′) |
|---|
| miR-21 | RT: |
CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGATCAACATG |
| F: |
CTCAACTGGTGTCGTGGAGT |
| R: |
ACACTCCAGCTGGGTAGCTTATCAGACTG |
| miR-27a | RT: |
CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGCGGAACTT |
| F: |
CTCAACTGGTGTCGTGGAGT |
| R: |
ACACTCCAGCTGGGTTCACAGTGGCTAAG |
| miR-210 | RT: |
CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGATCAGCCGC |
| F: |
CTCAACTGGTGTCGTGGAGT |
| R: |
CACTCCAGCTGGGCTGTGCGTGTGACAG |
| miR-23a | RT: |
CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGGAAATCCC |
| F: |
CTCAACTGGTGTCGTGGAGT |
| R: |
CACTCCAGCTGGGATCACATTGCCAGGG |
| miR-155 | RT: |
CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAACCCCTATC |
| F: |
CTCAACTGGTGTCGTGGAGT |
| R: |
ACACTCCAGCTGGGTTAATGCTAATCGTG |
| miR-139 | RT: |
CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAACTGGAGA |
| F: |
CTCAACTGGTGTCGTGGAGT |
| R: |
CACTCCAGCTGGGTCTACAGTGCACGTG |
| hsa-miR-338 | RT: |
CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGACAACAAAAT |
| F: |
CTCAACTGGTGTCGTGGAGT |
| R: |
ACACTCCAGCTGGGTATTGCACTCGTCC |
| miR-137 | RT: |
CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGACTACGCGTA |
| F: |
CTCAACTGGTGTCGTGGAGT |
| R: |
ACACTCCAGCTGGGTTATTGCTTAAGAAT |
| miR-7 | RT: |
CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAACAACAAA |
| F: |
CTCAACTGGTGTCGTGGAGT |
| R: |
CACTCCAGCTGGGTGGAAGACTAGTGAT |
| miR-124a | RT: |
TCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGGCATTCAC |
| F: |
CTCAACTGGTGTCGTGGAGT |
| R: |
CACTCCAGCTGGGTAAGGCACGCGGTGA |
| miR-15a | RT: |
CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGACACAAACCA |
| F: |
CTCAACTGGTGTCGTGGAGT |
| R: |
CACTCCAGCTGGGTAGCAGCACATAATG |
| miR-29b | RT: |
CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAAACACTGAT |
| F: |
CTCAACTGGTGTCGTGGAGT |
| R: |
CACTCCAGCTGGGTAGCACCATTTGAAA |
| miR-29c | RT: |
CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGATAACCGATT |
| F: |
CTCAACTGGTGTCGTGGAGT |
| R: |
CACTCCAGCTGGGTAGCACCATTTGAAA |
|
ENST00000520186 | F: |
GTTGGACCTTACTGAGGCCG |
| R: |
GGAGACACCATGGCTGGAAC |
|
ENST00000559981 | F: |
AGAGTGAAATTTTGTATAAGCACCA |
| R: |
GCCTGGAGACATACTGAGATGG |
|
ENST00000547415 | F: |
TGCCATCTGCAGAGTGAAACT |
| R: |
GGCTTTCCAGTCTAGGGCAG |
|
ENST00000518554 | F: |
TGGCATTTTGTCAGTTTTCCCG |
| R: |
GCAAATGCACACACCACTCC |
| GAPDH | F: |
GCACCGTCAAGGCTGAGAAC |
| R: |
TGGTGAAGACGCCAGTGGA |
| RNU6 | F: |
CTCGCTTCGGCAGCACA |
| R: |
AACGCTTCACGAATTTGCGT |
| AKT3 | F: |
TTGCTTTCAGGGCTCTTGAT |
| R: |
CATAATTTCTTTGCATCATCTGG |
| PPP3CA | F: |
TGTGATATCCTGTGGTCAGA |
| R: |
CTGACTGTGTTGTGAGTGAA |
| LAMC1 | F: |
TGGGCATTCTTCTGTCTGTACAA |
| R: |
GCCACCCATCCTCATCAATC |
| TNFRSF1A | F: |
TGCCTACCCCAGATTGAGAA |
| R: |
ATTTCCCACAAACAATGGAGTAG |
Results
GBM demonstrates significantly altered
miRNA, lncRNA and mRNA expression patterns comparing with that of
the normal brain
In terms of the Sanger miRBase database, 866 human
and 89 human viral miRNAs were authenticated on the Agilent Human
MicroRNA array 8×15K. Based on the NetAffx annotation of the probe
sets, the Ensemble, NOCODE3.0, and UCSC annotations of lncRNAs, and
the RefSeq, Ensemble and GenBank annotations of mRNAs, we
identified 33,125 lncRNAs (corresponding to 44,482 probe sets) and
17,453 mRNAs (corresponding to 22,011 probe sets) represented on
the Affymetrix Human Exon 1.0 array (data not shown).
The expression patterns of miRNAs, lncRNAs and mRNAs
were detected in 8 GBMs and 8 normal brain samples. We identified
41 miRNAs, 398 lncRNAs and 1,995 mRNAs that had significant
differential expression in the GBM group comparing with the normal
brain group (fold change ≥2.0 or ≤0.5 and P-value <0.05, data
not shown).
The hierarchical clustering analysis showed that
with the differential expression of these miRNAs, lncRNAs and
mRNAs, samples were non-random partitioned, they were divided into
2 groups, the first group containing 8 normal brain samples and the
second group containing 8 GBM samples (Fig. 1). Thus, the miRNA, lncRNA and mRNA
expression signatures identified here were likely to be
representative.
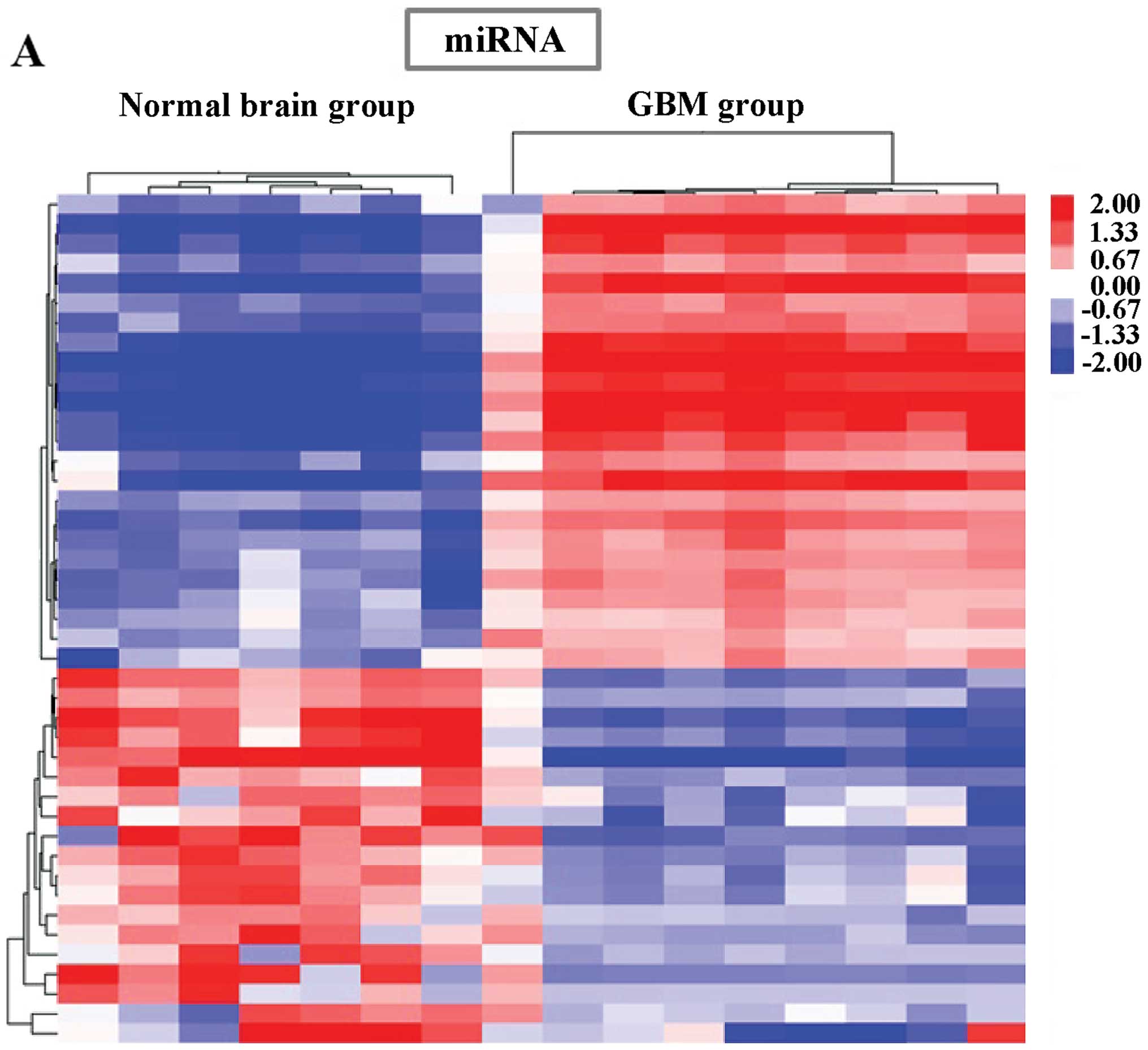 | Figure 1Hierarchical clustering analysis of
miRNA, lncRNA and mRNA expression levels change between two groups
(normal brain and GBM). (A) Forty-one miRNAs and (B) 398 lncRNAs.
(C) mRNAs (1,995) are differentially expressed in GBM tissue
(≥2-fold or ≤0.5-fold change; P<0.05 and FDR<0.05). Columns
represent samples and rows, respectively, represent miRNA, lncRNA
or mRNA probe sets. Red, represents high expression; green,
represents low expression, indicating expression above and below
the median expression value across all of the samples, respectively
(log scale, 2; from −1.80 to +1.80). miRNA, microRNA; lncRNA, long
non-coding RNA; GBM, glioblastoma multiforme; FDR, false discovery
rate. |
Construction of miRNA-lncRNA-mRNA
interaction network and identification lncRNAs acting as
ceRNAs
The miRNA-lncRNA-mRNA network was constructed
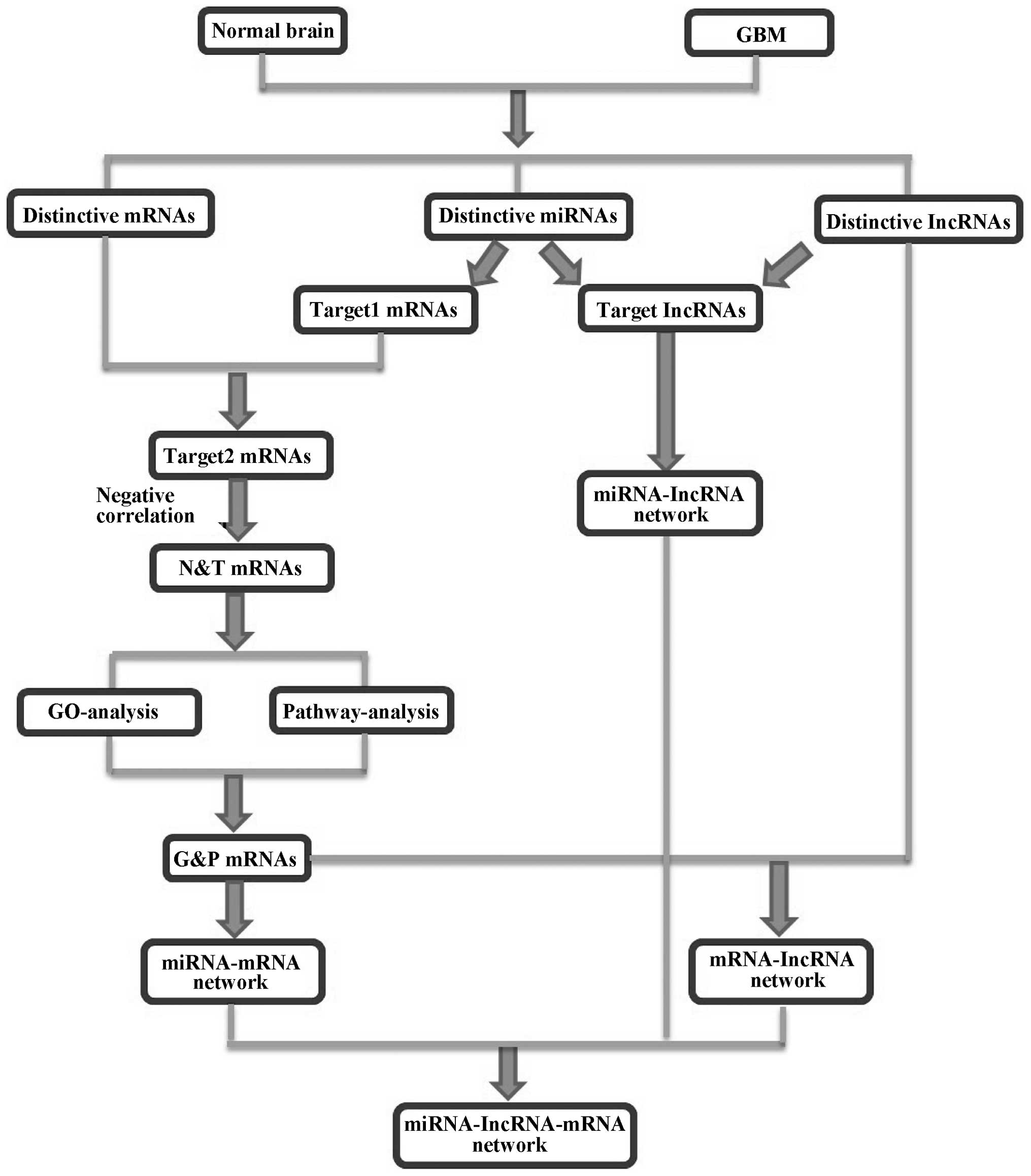
according to the study flow summarized in Fig. 2.
First, the target mRNAs of the differentially
expressed miRNA were analyzed by TargetScan and miRanda method,
termed as target 1 mRNAs (6,737 mRNAs, data not shown). The
intersection of the target 1 mRNAs and differentially expressed
mRNAs in GBM was picked and obtained target 2 mRNAs (1,034 mRNAs,
data not shown). Of the target 2 mRNAs, the mRNAs were selected
with expression levels negatively correlated with miRNA expression,
and were termed the N&T mRNAs (749 mRNAs, data not shown).
Then, GO and pathway analysis were applied to
analyze the significant function and pathway of the N&T mRNAs.
GO analysis results showed that upregulated and downregulated mRNAs
respectively were involved in 156 and 240 items with significant
functions (P-value <0.01, data not shown). The pathway analysis
revealed that there were 65 and 24 significant pathways
corresponding to the up and downregulated mRNAs respectively
(P-value <0.01, data not shown).
The third step, the mRNAs that contained both the
significant function and pathway were termed G&P mRNAs (248
mRNAs, data not shown). The G&P mRNAs and differently expressed
lncRNAs were used to build the lncRNA-mRNA co-expression network,
respectively, in the normal and GBM group (data not shown).
The TargetScan method was used to analysis the
target lncRNAs of differentially expressed miRNA and obtained the
55 miRNA targeted lncRNAs. These 55 lncRNAs were identified ceRNAs.
Based on the interaction network of miRNA-mRNA, miRNA-lncRNA and
lncRNA-mRNA, we obtained 224 feed-forward loop networks and
constructed general miRNA-lncRNA-mRNA feed-forward loop network
(data not shown). All of miRNAs, lncRNAs and mRNAs and their
relations in this network are listed in Table II.
 | Table IIThe 224 feed-forward loops including
miRNAs, lncRNAs and mRNAs. |
Table II
The 224 feed-forward loops including
miRNAs, lncRNAs and mRNAs.
| No. | miRNA | lncRNA | mRNA |
|---|
| 1 | miR-15a | n339339 | PAK7 |
| 2 | miR-15a | n339339 | CACNA1E |
| 3 | miR-15a |
ENST00000520186 | PAK7 |
| 4 | miR-15a |
ENST00000533229 | PAK7 |
| 5 | miR-15a |
ENST00000566630 | PAK7 |
| 6 | miR-15a | n341995 | PAK7 |
| 7 | miR-15a | n346032 | CACNA1E |
| 8 | miR-15a | n346032 | NMNAT2 |
| 9 | miR-15a |
ENST00000520186 | MAPK9 |
| 10 | miR-15a |
ENST00000520186 | AKT3 |
| 11 | miR-15a |
ENST00000559981 | VAMP1 |
| 12 | miR-15a |
ENST00000559981 | SYNJ1 |
| 13 | miR-15a |
ENST00000596580 | VAMP1 |
| 14 | miR-15a |
ENST00000596580 | AKT3 |
| 15 | miR-15a |
ENST00000596580 | SYNJ1 |
| 16 | miR-15a |
ENST00000596580 | SLC9A6 |
| 17 | miR-15a |
ENST00000569946 | VAMP1 |
| 18 | miR-15a |
ENST00000532691 | CACNA1E |
| 19 | miR-15a |
ENST00000566942 | HTR2A |
| 20 | miR-15a | n338128 | PTPRR |
| 21 | miR-15a |
ENST00000566630 | CACNA1E |
| 22 | miR-15a |
ENST00000524501 | CACNA1E |
| 23 | miR-15a |
ENST00000578746 | CACNA1E |
| 24 | miR-15a | n341995 | CACNA1E |
| 25 | miR-15a | n410578 | SYNJ1 |
| 26 | miR-15a |
ENST00000524501 | NMNAT2 |
| 27 | miR-15a |
ENST00000578746 | NMNAT2 |
| 28 | miR-15a | n411142 | AKT3 |
| 29 | miR-15a |
ENST00000492667 | AKT3 |
| 30 | miR-15a |
ENST00000434383 | SGCD |
| 31 | miR-15a | n411142 | SYNJ1 |
| 32 | miR-15a |
ENST00000492667 | SYNJ1 |
| 33 | miR-15b | n339339 | PAK7 |
| 34 | miR-15b | n339339 | CACNA1E |
| 35 | miR-15b |
ENST00000520186 | PAK7 |
| 36 | miR-15b |
ENST00000533229 | PAK7 |
| 37 | miR-15b |
ENST00000566630 | PAK7 |
| 38 | miR-15b | n341995 | PAK7 |
| 39 | miR-15b | n406352 | PAK7 |
| 40 | miR-15b | n346032 | CACNA1E |
| 41 | miR-15b | n346032 | NMNAT2 |
| 42 | miR-15b |
ENST00000520186 | MAPK9 |
| 43 | miR-15b |
ENST00000520186 | AKT3 |
| 44 | miR-15b |
ENST00000559981 | VAMP1 |
| 45 | miR-15b |
ENST00000559981 | SYNJ1 |
| 46 | miR-15b |
ENST00000596580 | VAMP1 |
| 47 | miR-15b |
ENST00000596580 | AKT3 |
| 48 | miR-15b |
ENST00000596580 | SYNJ1 |
| 49 | miR-15b |
ENST00000596580 | SLC9A6 |
| 50 | miR-15b |
ENST00000569946 | VAMP1 |
| 51 | miR-15b |
ENST00000532691 | CACNA1E |
| 52 | miR-15b |
ENST00000566942 | HTR2A |
| 53 | miR-15b | n338128 | PTPRR |
| 54 | miR-15b |
ENST00000566630 | CACNA1E |
| 55 | miR-15b |
ENST00000524501 | CACNA1E |
| 56 | miR-15b | n341995 | CACNA1E |
| 57 | miR-15b | n406352 | CACNA1E |
| 58 | miR-15b | n410578 | SYNJ1 |
| 59 | miR-15b |
ENST00000524501 | NMNAT2 |
| 60 | miR-15b | n411142 | AKT3 |
| 61 | miR-15b |
ENST00000492667 | AKT3 |
| 62 | miR-15b |
ENST00000434383 | SGCD |
| 63 | miR-15b | n411142 | SYNJ1 |
| 64 | miR-15b |
ENST00000492667 | SYNJ1 |
| 65 | miR-23a | n339339 | SLIT1 |
| 66 | miR-23a | n339339 | RXRG |
| 67 | miR-23a | n339339 | SLC1A1 |
| 68 | miR-23a |
ENST00000562191 | GABRB2 |
| 69 | miR-23a |
ENST00000562191 | NEFL |
| 70 | miR-23a |
ENST00000562191 | MEF2C |
| 71 | miR-23a | n346032 | GABRB3 |
| 72 | miR-23a | n346032 | PCLO |
| 73 | miR-23a |
ENST00000522102 | SRGAP3 |
| 74 | miR-23a | n385835 | GABRA4 |
| 75 | miR-23a | n383510 | FUT9 |
| 76 | miR-23a | n383510 | CADM3 |
| 77 | miR-23a |
ENST00000559981 | ATP6V1C1 |
| 78 | miR-23a |
ENST00000559981 | SYNJ1 |
| 79 | miR-23a |
ENST00000559981 | TLN2 |
| 80 | miR-23a |
ENST00000555811 | GABRB3 |
| 81 | miR-23a |
ENST00000566630 | GABRB3 |
| 82 | miR-23a |
ENST00000524501 | GABRB3 |
| 83 | miR-23a |
ENST00000555811 | FUT9 |
| 84 | miR-23a |
ENST00000555811 | PCLO |
| 85 | miR-23a | n411142 | ATP6V1C1 |
| 86 | miR-23a | n373066 | FUT9 |
| 87 | miR-23a |
ENST00000524501 | PCLO |
| 88 | miR-23a |
ENST00000524501 | NRXN3 |
| 89 | miR-23a |
ENST00000434383 | RXRG |
| 90 | miR-23a |
ENST00000434383 | SGCD |
| 91 | miR-23a |
ENST00000502752 | RAB11FIP2 |
| 92 | miR-23a | n384012 | TLN2 |
| 93 | miR-23a | n411142 | SYNJ1 |
| 94 | miR-30a | n346032 | GRM5 |
| 95 | miR-30a |
ENST00000522102 | SRGAP3 |
| 96 | miR-30a | n373066 | CAMK4 |
| 97 | miR-30a |
ENST00000520186 | PSD3 |
| 98 | miR-30a |
ENST00000473866 | GNAO1 |
| 99 | miR-30a |
ENST00000532691 | GRM5 |
| 100 | miR-30a |
ENST00000569946 | GRM3 |
| 101 | miR-30a |
ENST00000569946 | CACNA1C |
| 102 | miR-30a | n339481 | CACNA1C |
| 103 | miR-30a |
ENST00000532691 | B4GALT6 |
| 104 | miR-30a |
ENST00000569946 | NEFL |
| 105 | miR-30a |
ENST00000566630 | B4GALT6 |
| 106 | miR-30a | n374560 | GDA |
| 107 | miR-30a |
ENST00000471736 | GRIA2 |
| 108 | miR-30a |
ENST00000569946 | NEFM |
| 109 | miR-30a |
ENST00000530447 | B4GALT6 |
| 110 | miR-27a | n339339 | IQSEC1 |
| 111 | miR-27a | n339339 | PDE3B |
| 112 | miR-27a |
ENST00000564076 | CACNB2 |
| 113 | miR-27a |
ENST00000562191 | SNAP25 |
| 114 | miR-27a | n346032 | GABRB3 |
| 115 | miR-27a | n346032 | SYT1 |
| 116 | miR-27a | n346032 | PCLO |
| 117 | miR-27a |
ENST00000522102 | SRGAP3 |
| 118 | miR-27a |
ENST00000566630 | GABRB3 |
| 119 | miR-27a | n406352 | GABRB3 |
| 120 | miR-27a | n371475 | CDS1 |
| 121 | miR-27a | n338494 | CACNB2 |
| 122 | miR-27a | n345100 | SYT1 |
| 123 | miR-27a | n345100 | SNAP25 |
| 124 | miR-27a | n373066 | FUT9 |
| 125 | miR-27a | n339978 | SYT1 |
| 126 | miR-27a | n406352 | PDE3B |
| 127 | miR-27a | n406352 | ATP6V1A |
| 128 | miR-27a |
ENST00000492667 | ATP2B1 |
| 129 | miR-34a | n339339 | CACNA1E |
| 130 | miR-34a | n346032 | CACNA1E |
| 131 | miR-34a | n346032 | GABRA3 |
| 132 | miR-34a | n346032 | SYT1 |
| 133 | miR-34a | n383510 | FUT9 |
| 134 | miR-34a |
ENST00000549205 | PSD3 |
| 135 | miR-34a |
ENST00000549205 | PRKCE |
| 136 | miR-34a |
ENST00000596580 | SYNJ1 |
| 137 | miR-34a |
ENST00000555811 | CACNA1E |
| 138 | miR-34a |
ENST00000555811 | FUT9 |
| 139 | miR-34a |
ENST00000555811 | PCLO |
| 140 | miR-34a | n384244 | CNTN2 |
| 141 | miR-34a |
ENST00000566630 | CACNA1E |
| 142 | miR-34a | n406352 | CACNA1E |
| 143 | miR-34a | n410578 | SYNJ1 |
| 144 | miR-34a |
ENST00000569946 | CPLX2 |
| 145 | miR-34a | n339978 | SYT1 |
| 146 | miR-34a | n406352 | WASF1 |
| 147 | miR-34a | n406352 | PRKCE |
| 148 | miR-34a |
ENST00000492667 | SYNJ1 |
| 149 | miR-106b |
ENST00000440363 | PDE3B |
| 150 | miR-106b |
ENST00000522102 | SRGAP3 |
| 151 | miR-106b |
ENST00000596580 | PIP4K2A |
| 152 | miR-106b |
ENST00000596580 | AKT3 |
| 153 | miR-106b | n345100 | MAPK9 |
| 154 | miR-106b | n337874 | PIP4K2A |
| 155 | miR-106b | n337874 | GABBR1 |
| 156 | miR-106b |
ENST00000555811 | RIMS2 |
| 157 | miR-106b |
ENST00000555811 | B4GALT6 |
| 158 | miR-106b | n345100 | RIMS2 |
| 159 | miR-106b | n337874 | B4GALT6 |
| 160 | miR-106b | n345100 | SCN1A |
| 161 | miR-106b | n411142 | AKT3 |
| 162 | miR-106b |
ENST00000434383 | SGCD |
| 163 | miR-25 |
ENST00000493303 | PRKCE |
| 164 | miR-25 |
ENST00000549205 | MAP2K4 |
| 165 | miR-25 |
ENST00000555811 | ST6GAL2 |
| 166 | miR-25 | n337874 | ST6GAL2 |
| 167 | miR-25 |
ENST00000566630 | ST6GAL2 |
| 168 | miR-25 |
ENST00000530447 | ST6GAL2 |
| 169 | miR-25 |
ENST00000549205 | PRKCE |
| 170 | miR-25 | n345100 | CACNA1C |
| 171 | miR-25 | n345100 | NEFL |
| 172 | miR-25 |
ENST00000555811 | RIMS2 |
| 173 | miR-25 | n345100 | RIMS2 |
| 174 | miR-25 | n345100 | SYT1 |
| 175 | miR-2 | n410578 | SYNJ1 |
| 176 | miR-25 | n406352 | PRKCE |
| 177 | miR-25 | n411142 | SYNJ1 |
| 178 | miR-25 |
ENST00000492667 | SYNJ1 |
| 179 | miR-223 |
ENST00000493303 | PRKCE |
| 180 | miR-223 | n371741 | ATP2B1 |
| 181 | miR-223 | n406352 | PRKCE |
| 182 | miR-223 |
ENST00000492667 | ATP2B1 |
| 183 | miR-155 | n345100 | GABRA1 |
| 184 | miR-155 | n346032 | DYNC1I1 |
| 185 | miR-155 | n341006 | RAB11FIP2 |
| 186 | miR-155 |
ENST00000596580 | ATP6V1C1 |
| 187 | miR-155 | n345100 | CACNA1C |
| 188 | miR-155 |
ENST00000535764 | ATP6V1C1 |
| 189 | miR-155 | n411142 | ATP6V1C1 |
| 190 | miR-155 | n345100 | SCN1A |
| 191 | miR-155 | n345100 | DYNC1I1 |
| 192 | miR-155 |
ENST00000578746 | DYNC1I1 |
| 193 | miR-2 |
ENST00000559981 | PPP3CA |
| 194 | miR-21 | n411142 | PPP3CA |
| 195 | miR-21 |
ENST00000492667 | PPP3CA |
| 196 | miR-92b |
ENST00000555811 | ST6GAL2 |
| 197 | miR-92b | n337874 | ST6GAL2 |
| 198 | miR-92b |
ENST00000530447 | ST6GAL2 |
| 199 | miR-92b |
ENST00000596580 | SYNJ1 |
| 200 | miR-92b | n345100 | CACNA1C |
| 201 | miR-92b | n345100 | NEFL |
| 202 | miR-92b |
ENST00000555811 | RIMS2 |
| 203 | miR-92b | n345100 | RIMS2 |
| 204 | miR-92b | n345100 | SYT1 |
| 205 | miR-92b | n410578 | SYNJ1 |
| 206 | miR-92b | n411142 | SYNJ1 |
| 207 | miR-92b |
ENST00000492667 | SYNJ1 |
| 208 | miR-339 | n383510 | CADM3 |
| 209 | miR-339 |
ENST00000555811 | DAAM2 |
| 210 | miR-339 |
ENST00000555811 | PCLO |
| 211 | miR-339 |
ENST00000523571 | DAAM2 |
| 212 | miR-339 |
ENST00000481854 | SGCD |
| 213 | miR-10b |
ENST00000221169 | TIAM1 |
| 214 | miR-10b |
ENST00000555811 | RIMS2 |
| 215 | miR-10b |
ENST00000524501 | RIMS2 |
| 216 | miR-29b |
ENST00000481203 | TNFRSF1A |
| 217 | miR-29b |
ENST00000518554 | TNFRSF1A |
| 218 | miR-29b |
ENST00000547415 | LAMC1 |
| 219 | miR-29b |
ENST00000559148 | MYBL2 |
| 220 | miR-377 | n338229 | LAMC1 |
| 221 | miR-124 | n376998 | EDNRB |
| 222 | miR-29c |
ENST00000547415 | LAMC1 |
| 223 | miR-29c |
ENST00000559148 | MYBL2 |
| 224 | miR-29c |
ENST00000518554 | TNFRSF1A |
Biological prediction of lncRNA function
as ceRNAs in the GBM
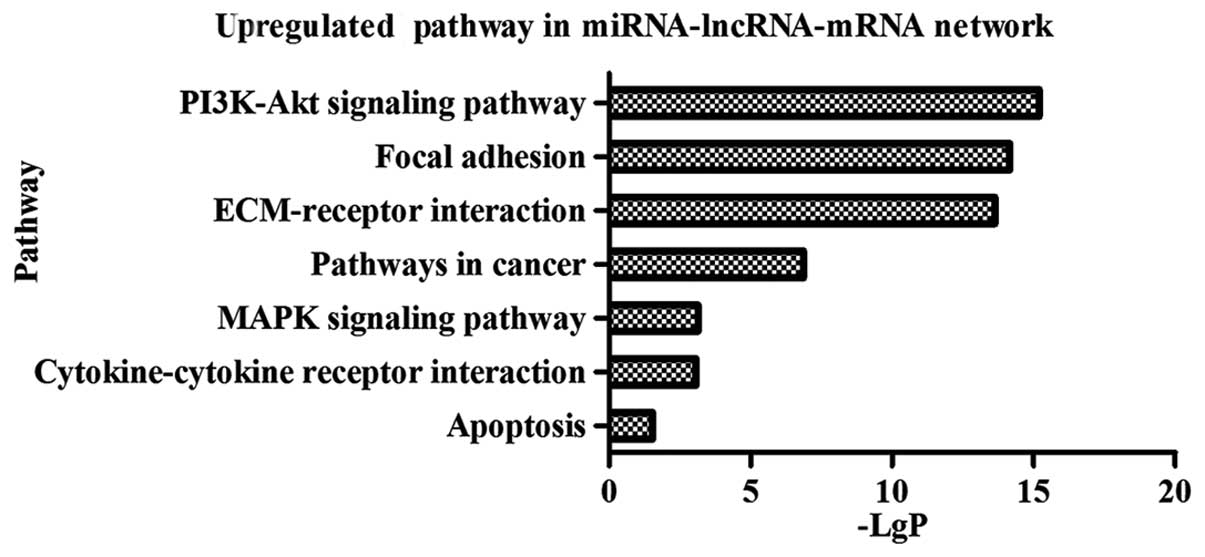
The functions of 55 lncRNAs acting as ceRNAs were
predicted through pathway analysis of 67 mRNAs in the
miRNA-lncRNA-mRNA interaction network. The results indicated that
30 mRNAs participated in 7 upregulated and 16 downregulated
pathways which involved in diverse biological processes of cancer,
including proliferation, cell apoptosis, adhesion, angiogenesis and
metastasis (Fig. 3A and B). As a
consequence, we predicted the important roles of the 39 ceRNAs in
GBM pathogenesis. The miRNAs, lncRNAs, mRNAs, and their
participated pathways are listed in Table III.
 | Table IIIFunctional prediction of the lncRNA
ceRNAs based on pathway analysis of mRNAs that location together in
the miRNA-lncRNA-mRNA feed-forward loop in GBM. |
Table III
Functional prediction of the lncRNA
ceRNAs based on pathway analysis of mRNAs that location together in
the miRNA-lncRNA-mRNA feed-forward loop in GBM.
| miRNA | lncRNA | mRNA | Pathway |
|---|
| hsa-miR-15a-5p | n339339 | PAK7 | ErbB signaling |
|
ENST00000520186 | | |
|
ENST00000533229 | | |
|
ENST00000566630 | | |
| n341995 | | |
| hsa-miR-15a-5p | n339339 | CACNA1E | Calcium, MAPK
signaling |
| n346032 | | |
|
ENST00000532691 | | |
|
ENST00000566630 | | |
|
ENST00000524501 | | |
|
ENST00000578746 | | |
| n341995 | | |
| hsa-miR-15a-5p |
ENST00000520186 | MAPK9 | MAPK, ErbB, Wnt
signaling, focal adhesion, pathways in cancer |
| hsa-miR-15a-5p |
ENST00000520186 | AKT3 | MAPK, ErbB,
PI3K-Akt, VEGF, mTOR signaling, glioma, apoptosis, focal adhesion,
pathways in cancer |
|
ENST00000596580 |
| n411142 |
|
ENST00000492667 |
| hsa-miR-15a-5p |
ENST00000559981 | SYNJ1 |
Phosphatidylinositol signaling |
|
ENST00000596580 | | |
| n410578 | | |
| n411142 | | |
|
ENST00000492667 | | |
| hsa-miR-15a-5p |
ENST00000566942 | HTR2A | Calcium
signaling |
| hsa-miR-15a-5p | n338128 | PTPRR | MAPK signaling |
| hsa-miR-15b-5p | n339339 | PAK7 | ErbB signaling |
|
ENST00000520186 | | |
|
ENST00000533229 | | |
|
ENST00000566630 | | |
| n341995 | | |
| n406352 | | |
| hsa-miR-15b-5p | n339339 | CACNA1E | Calcium, MAPK
signaling |
| n346032 | | |
|
ENST00000532691 | | |
|
ENST00000566630 | | |
|
ENST00000524501 | | |
| n341995 | | |
| n406352 | | |
| hsa-miR-15b-5p |
ENST00000520186 | AKT3 | MAPK, ErbB,
PI3K-Akt, VEGF, mTOR signaling, glioma, apoptosis, focal adhesion,
pathways in cancer |
|
ENST00000596580 |
| n411142 |
|
ENST00000492667 |
| hsa-miR-15b-5p |
ENST00000559981 | SYNJ1 |
Phosphatidylinositol signaling |
|
ENST00000596580 | | |
| n410578 | | |
| n411142 | | |
|
ENST00000492667 | | |
| hsa-miR-15b-5p |
ENST00000520186 | MAPK9 | MAPK, ErbB, Wnt
signaling, focal adhesion, pathways in cancer |
| hsa-miR-15b-5p |
ENST00000566942 | HTR2A | Calcium
signaling |
| hsa-miR-15b-5p | n338128 | PTPRR | MAPK signaling |
| hsa-miR-23a-3p | n339339 | SLIT1 |
Phosphatidylinositol signaling |
| hsa-miR-23a-3p | n339339 | RXRG | Pathways in
cancer |
|
ENST00000434383 | | |
| hsa-miR-23a-3p |
ENST00000559981 | SYNJ1 |
Phosphatidylinositol signaling |
| n411142 | | |
| hsa-miR-23a-3p |
ENST00000562191 | MEF2C | MAPK signaling |
| hsa-miR-23a-3p | n383510 | CADM3 | Cell adhesion
molecules |
| hsa-miR-23a-3p |
ENST00000559981 | TLN2 | Focal adhesion |
| hsa-miR-23a-3p |
ENST00000524501 | NRXN3 | Cell adhesion
molecules |
| hsa-miR-30a-5p | n346032 | GRM5 | Calcium
signaling |
|
ENST00000532691 | | |
| hsa-miR-30a-5p |
ENST00000569946 | CACNA1C | Calcium signaling,
MAPK signaling |
| n339481 | | |
| hsa-miR-30a-5p | n373066 | CAMK4 | Calcium
signaling |
| hsa-miR-27a-3p | n339339 | PDE3B | Proteoglycans in
cancer |
| hsa-miR-27a-3p |
ENST00000564076 | CACNB2 | MAPK signaling |
| n338494 | | |
| hsa-miR-27a-3p | n371475 | CDS1 |
Phosphatidylinositol signaling |
| hsa-miR-27a-3p |
ENST00000492667 | ATP2B1 | Calcium
signaling |
| hsa-miR-34a-5p | n339339 | CACNA1E | Calcium, MAPK
signaling |
| n346032 | | |
|
ENST00000555811 | | |
|
ENST00000566630 | | |
| n406352 | | |
| hsa-miR-34a-5p |
ENST00000596580 | SYNJ1 |
Phosphatidylinositol signaling |
| n410578 | | |
|
ENST00000492667 | | |
| hsa-miR-34a-5p |
ENST00000549205 | PRKCE | Tight junction |
| n406352 | | |
| hsa-miR-34a-5p | n384244 | CNTN2 | Cell adhesion
molecules |
| hsa-miR-34a-5p | n406352 | WASF1 | Regulation of actin
cytoskeleton, |
| | | Adherens
junction |
|
hsa-miR-106b-5p |
ENST00000596580 | PIP4K2A |
Phosphatidylinositol signaling, regulation
of actin cytoskeleton |
| n337874 |
|
hsa-miR-106b-5p |
ENST00000596580 | AKT3 | MAPK, ErbB,
PI3K-Akt, VEGF, |
| n411142 | | mTOR signaling,
glioma, apoptosis, focal adhesion, pathways in cancer |
|
hsa-miR-106b-5p | n345100 | MAPK9 | MAPK, ErbB, Wnt
signaling, focal adhesion, pathways in cancer |
| hsa-miR-25-3p |
ENST00000493303 | PRKCE | Tight junction |
|
ENST00000549205 | | |
| n406352 | | |
| hsa-miR-25-3p | n410578 | SYNJ1 |
Phosphatidylinositol signaling |
| n411142 | | |
|
ENST00000492667 | | |
| hsa-miR-25-3p |
ENST00000549205 | MAP2K4 | MAPK, ErbB
signaling |
| hsa-miR-223-3p |
ENST00000493303 | PRKCE | Tight junction |
| n406352 | | |
| hsa-miR-223-3p | n371741 | ATP2B1 | Calcium
signaling |
|
ENST00000492667 | | |
| hsa-miR-21-5p |
ENST00000559981 | PPP3CA | Calcium, MAPK, Wnt,
VEGF signaling |
| n411142 | | |
|
ENST00000492667 | | |
| hsa-miR-92b-3p |
ENST00000596580 | SYNJ1 |
Phosphatidylinositol signaling |
| n410578 | | |
| n411142 | | |
|
ENST00000492667 | | |
| hsa-miR-339-5p |
ENST00000555811 | DAAM2 | Wnt signaling |
|
ENST00000523571 | | |
| hsa-miR-339-5p | n383510 | CADM3 | Cell adhesion
molecules |
| hsa-miR-10b-5p |
ENST00000221169 | TIAM1 | Regulation of actin
cytoskeleton, proteoglycans in cancer |
| hsa-miR-29b-3p |
ENST00000481203 | TNFRSF1A | MAPK signaling,
cytokine-cytokine receptor interaction, apoptosis |
|
ENST00000518554 |
| hsa-miR-29b-3p |
ENST00000547415 | LAMC1 | PI3K-Akt signaling,
focal adhesion, ECM-receptor interaction, pathways in cancer |
| hsa-miR-377-3p | n338229 | LAMC1 | PI3K-Akt signaling,
focal adhesion, ECM-receptor interaction, pathways in cancer |
| hsa-miR-29c-3p |
ENST00000547415 | LAMC1 | PI3K-Akt signaling,
focal adhesion, ECM-receptor interaction, pathways in cancer |
| hsa-miR-29c-3p |
ENST00000518554 | TNFRSF1A | MAPK signaling,
cytokine-cytokine receptor interaction, apoptosis |
Quantitative real-time RT-PCR analysis of
the distinctive expression of lncRNAs, miRNAs and mRNAs in GBM
samples
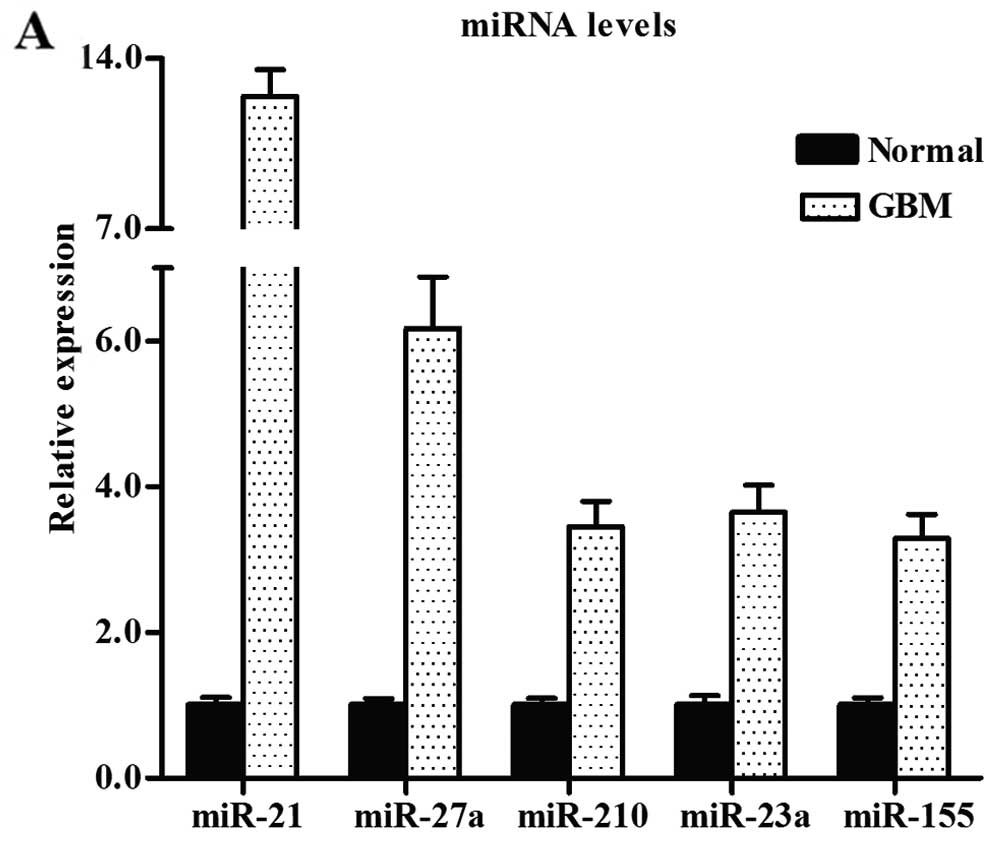
To validate the conclusions of microarray analysis,
we selected 10 miRNAs with larger fold change from the microarray
results and analyzed their expression levels by qRT-PCR in 20
normal brain and 30 GBM samples. Our results confirmed the findings
of the miRNA microarray dataset (Fig.
4A and B).
Based on the analysis of 224 miRNA-lncRNA-mRNA
feed-forward loops in Table II, we
evaluated the expression levels of 4 miRNA, 4 lncRNA and 4 mRNA
that, respectively, located in 4 feed-forward loops. The average
expression levels of miR-15a and miR-21 were significantly
increased, while miR-29b and miR-29c were reduced in GBM compared
with normal brain tissues. Analysis showed relatively high
expression of miRNA and low expression of lncRNA and mRNA, and low
expression of miRNA and high expression of lncRNA and mRNA
(Fig. 4C and D). The 4 feed-forward
loops detection by qRT-PCR are presented in Fig. 4E.
Discussion
In recent years, the emerging significance of ceRNAs
in cancers has drawn attention of researchers. ceRNA activity is
determined by factors such as miRNA/ceRNA abundance, ceRNA binding
affinity to miRNAs and RNA-binding proteins. The alteration of any
of these factors may lead to ceRNA network imbalance and thus
contribute to cancer (28). ceRNA
study processes generally include: ceRNA prediction, ceRNA
validation and ceRNA functional investigation.
Recently, several studies have confirmed the
dysregulation of lncRNAs by acting as ceRNAs have profound
implications for tumor initiation, maintenance or progression.
lncRNAs acting as ceRNAs are involvd in the pathogenesis of several
common cancers such as thyroid cancer, gastric cancer and
hepatocellular cancer (29–33). The ceRNA activity of lncRNAs has
also been shown to have an oncogenic effect: The lncRNA HOTAIR was
shown to display ceRNA activity in gastric cancer cells, in which
it was found to specifically bind the tumor suppressor miR-331-3p,
modulating HER2 derepression (31).
The other example of lncRNA-mediated ceRNA regulation involves the
tumor suppressor gene BARD1. The lncRNA BARD1 9′L is transcribed by
an alternative intronic promoter of the BARD1 gene and share both
miR-203 and miR-101 MREs with BARD1 mRNA in their homologous
3′UTRs. BARD1 mRNAs were downregulated by miR-203 and miR-101, and
BARD1 9′L counteracted the effect of these miRNAs. These data
support a role for BARD1 9′L as a tumor suppressor transcript
through its ceRNA activity (33).
These findings provide important clues for understanding the key
roles of lncRNA-miRNA functional network in cancers. Exploring the
interplay of lncRNA function as a ceRNA in cancer provides new
insight into cancer pathogenesis and opportunities for therapy
exploration.
Understanding the novel miRNA-lncRNA-mRNA crosstalk
will lead to significant insight into gene regulatory networks in
cancers. In this study, we investigated the miRNA, lncRNA and mRNA
expression signatures in GBM, constructed the miRNA-lncRNA-mRNA
regulation network, on this basis, identified the lncRNA acting as
ceRNAs and predicted the possible biology functions of these
ceRNAs.
We re-annotated the Affymetrix Human Exon 1.0 probe
sets and identified the lncRNAs and mRNAs on this array. The sample
matched miRNA expression profiling of the Agilent Human MicroRNA
array 8×15K was analyzed to determine differently expressed miRNAs
in GBM. We identified a set of 41 miRNAs, 398 lncRNAs and 1,995
mRNAs with differentiated expression between GBM and normal brain
tissues. Such differentiation signified their potential roles in
tumorigenesis.
The complexity and diversity of potential ceRNA
interactions have been described with the identification of
abundant lncRNAs. We discussed the effect of miRNA competition on
the regulation of both lncRNAs and mRNAs, as well as the
implications of lncRNA function as ceRNA for the development of
GBM. To our knowledge, this is the first study to show the roles of
lncRNA acting as ceRNAs in GBM. Understanding the key roles of
'miRNA-lncRNA' module will lead to the identification of new
therapeutic targets for treating GBM.
Our qRT-PCR expression analysis confirmed there are
a series of miRNAs, lncRNAs and mRNAs aberrantly expressed in GBM
tissues, which indicated that the differently expressed non-coding
and coding RNAs may be one of characters of GBM. The aberrant
miR-21, miR-27a, miR-210, miR-23a, miR-155, miR-139, miR-338,
miR-137, miR-7, miR-124a, miR-15a, miR-29b and miR-29c expression
levels in GBM were detected, our results were in concordance with
the previous findings, and these deregulated miRNAs have been
reported to be aberrantly expressed in GBM (34–42).
In our expression profiling analysis, the lncRNA ENST00000520186,
ENST00000559981, ENST00000547415 and ENST00000518554 were
separately considered as the ceRNA of miR-15a, miR-21, miR-29b and
miR-29c in GBM. So far, these ceRNAs have not been reported
implicated in GBM. Four mRNAs may be regulated by these miRNAs and
lncRNAs, the PPP3CA have been reported to be aberrantly expressed
in other tumors, but have not been studied in GBM; in addition,
AKT3, TNFRSF1A and LAMC1 have been studied to different expression
in GBM (36,43,44).
Overall, our study identified and analyzed lncRNA
function as ceRNA in GBM and showed they may play crucial
biological roles during GBM formation and development, and provide
important theory and experimental foundations for future study of
drug target and treatment for GBM.
Acknowledgments
This study was supported by the Natural Science Foun
dation of China (nos. 81572474 and 81303268), the Natural Science
Foundation of Beijing City (no. 7152098), the Excellence Talents
Training Projects of Beijing City (no. 2013D009008000006) and the
Science and Technology Development Fund Project of Traditional
Chinese Medicine of Beijing (JJ2015-14). The authors would like to
thank the Genminix Company (Shanghai, China) for assistance with
bioinformatics analysis.
References
|
1
|
Zamore PD and Haley B: Ribo-gnome: the big
world of small RNAs. Science. 309:1519–1524. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Guttman M, Amit I, Garber M, French C, Lin
MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al:
Chromatin signature reveals over a thousand highly conserved large
non-coding RNAs in mammals. Nature. 458:223–227. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Rinn JL and Chang HY: Genome regulation by
long noncoding RNAs. Annu Rev Biochem. 81:145–166. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Mercer TR, Dinger ME and Mattick JS: Long
non-coding RNAs: insights into functions. Nat Rev Genet.
10:155–159. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
5
|
Guttman M and Rinn JL: Modular regulatory
principles of large non-coding RNAs. Nature. 482:339–346. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bhan A and Mandal SS: Long noncoding RNAs:
emerging stars in gene regulation, epigenetics and human disease.
ChemMedChem. 9:1932–1956. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Spizzo R, Almeida MI, Colombatti A and
Calin GA: Long non-coding RNAs and cancer: a new frontier of
translational research? Oncogene. 31:4577–4587. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Gibb EA, Brown CJ and Lam WL: The
functional role of long non-coding RNA in human carcinomas. Mol
Cancer. 10:38–55. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bartel DP: MicroRNAs: target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lu M, Zhang Q, Deng M, Miao J, Guo Y, Gao
W and Cui Q: An analysis of human microRNA and disease
associations. PLoS One. 3:e34202008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nat Rev Cancer. 6:857–866. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Plaisier CL, Pan M and Baliga NS: A
miRNA-regulatory network explains how dysregulated miRNAs perturb
oncogenic processes across diverse cancers. Genome Res.
22:2302–2314. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Salmena L, Poliseno L, Tay Y, Kats L and
Pandolfi PP: A ceRNA hypothesis: the Rosetta stone of a hidden RNA
language? Cell. 146:353–358. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Cesana M, Cacchiarelli D, Legnini I,
Santini T, Sthandier O, Chinappi M, Tramontano A and Bozzoni I: A
long noncoding RNA controls muscle differentiation by functioning
as a competing endogenous RNA. Cell. 147:358–369. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Jeggari A, Marks DS and Larsson E:
miRcode: a map of putative microRNA target sites in the long
non-coding transcriptome. Bioinformatics. 28:2062–2063. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Louis DN, Ohgaki H, Wiestler OD, Cavenee
WK, Burger PC, Jouvet A, Scheithauer BW and Kleihues P: The 2007
WHO classification of tumours of the central nervous system. Acta
Neuropathol. 114:97–109. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Omuro A and DeAngelis LM: Glioblastoma and
other malignant gliomas: a clinical review. JAMA. 310:1842–1850.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Taylor LP: Diagnosis, treatment, and
prognosis of glioma: five new things. Neurology. 75(Suppl 1):
S28–S32. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhang X, Sun S, Pu JK, Tsang AC, Lee D,
Man VO, Lui WM, Wong ST and Leung GK: Long non-coding RNA
expression profiles predict clinical phenotypes in glioma.
Neurobiol Dis. 48:1–8. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Han L, Zhang K, Shi Z, Zhang J, Zhu J, Zhu
S, Zhang A, Jia Z, Wang G, Yu S, et al: LncRNA profile of
glioblastoma reveals the potential role of lncRNAs in contributing
to glioblastoma pathogenesis. Int J Oncol. 40:2004–2012.
2012.PubMed/NCBI
|
|
21
|
Yan Y, Zhang L, Jiang Y, Xu T, Mei Q, Wang
H, Qin R, Zou Y, Hu G, Chen J, et al: LncRNA and mRNA interaction
study based on transcriptome profiles reveals potential core genes
in the pathogenesis of human glioblastoma multiforme. J Cancer Res
Clin Oncol. 141:827–838. 2015. View Article : Google Scholar
|
|
22
|
Kim TM, Huang W, Park R, Park PJ and
Johnson MD: A developmental taxonomy of glioblastoma defined and
maintained by MicroRNAs. Cancer Res. 71:3387–3399. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Eisen MB, Spellman PT, Brown PO and
Botstein D: Cluster analysis and display of genome-wide expression
patterns. Proc Natl Acad Sci USA. 95:14863–14868. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ashburner M, Ball CA, Blake JA, Botstein
D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT,
et al The Gene Ontology Consortium: Gene ontology: tool for the
unification of biology. Nat Genet. 25:25–29. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kanehisa M, Goto S, Kawashima S, Okuno Y
and Hattori M: The KEGG resource for deciphering the genome.
Nucleic Acids Res. 32:D277–D280. 2004. View Article : Google Scholar :
|
|
26
|
Prieto C, Risueño A, Fontanillo C and De
las Rivas J: Human gene coexpression landscape: confident network
derived from tissue transcriptomic profiles. PLoS One. 3:e39112008.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Barabási AL and Oltvai ZN: Network
biology: understanding the cell's functional organization. Nat Rev
Genet. 5:101–113. 2004. View
Article : Google Scholar : PubMed/NCBI
|
|
28
|
Tay Y, Kats L, Salmena L, Weiss D, Tan SM,
Ala U, Karreth F, Poliseno L, Provero P, Di Cunto F, et al:
Coding-independent regulation of the tumor suppressor PTEN by
competing endogenous mRNAs. Cell. 147:344–357. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wang J, Liu X, Wu H, Ni P, Gu Z, Qiao Y,
Chen N, Sun F and Fan Q: CREB up-regulates long non-coding RNA,
HULC expression through interaction with microRNA-372 in liver
cancer. Nucleic Acids Res. 38:5366–5383. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Fan M, Li X, Jiang W, Huang Y, Li J and
Wang Z: A long non-coding RNA, PTCSC3, as a tumor suppressor and a
target of miRNAs in thyroid cancer cells. Exp Ther Med.
5:1143–1146. 2013.PubMed/NCBI
|
|
31
|
Liu XH, Sun M, Nie FQ, Ge YB, Zhang EB,
Yin DD, Kong R, Xia R, Lu KH, Li JH, et al: Lnc RNA HOTAIR
functions as a competing endogenous RNA to regulate HER2 expression
by sponging miR-331-3p in gastric cancer. Mol Cancer. 13:922014.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Braconi C, Kogure T, Valeri N, Huang N,
Nuovo G, Costinean S, Negrini M, Miotto E, Croce CM and Patel T:
microRNA-29 can regulate expression of the long non-coding RNA gene
MEG3 in hepatocellular cancer. Oncogene. 30:4750–4756. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Pilyugin M and Irminger-Finger I: Long
non-coding RNA and microRNAs might act in regulating the expression
of BARD1 mRNAs. Int J Biochem Cell Biol. 54:356–367. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Fan YC, Mei PJ, Chen C, Miao FA, Zhang H
and Li ZL: MiR-29c inhibits glioma cell proliferation, migration,
invasion and angiogenesis. J Neurooncol. 115:179–188. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Chung HJ, Choi YE, Kim ES, Han YH, Park MJ
and Bae IH: miR-29b attenuates tumorigenicity and stemness
maintenance in human glioblastoma multiforme by directly targeting
BCL2L2. Oncotarget. 6:18429–18444. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Fowler A, Thomson D, Giles K, Maleki S,
Mreich E, Wheeler H, Leedman P, Biggs M, Cook R, Little N, et al:
miR-124a is frequently down-regulated in glioblastoma and is
involved in migration and invasion. Eur J Cancer. 47:953–963. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Besse A, Sana J, Lakomy R, Kren L, Fadrus
P, Smrcka M, Hermanova M, Jancalek R, Reguli S, Lipina R, et al:
MiR-338-5p sensitizes glioblastoma cells to radiation through
regulation of genes involved in DNA damage response. Tumour Biol.
37:7719–7727. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Yue S, Wang L, Zhang H, Min Y, Lou Y, Sun
H, Jiang Y, Zhang W, Liang A, Guo Y, et al: miR-139-5p suppresses
cancer cell migration and invasion through targeting ZEB1 and ZEB2
in GBM. Tumour Biol. 36:6741–6749. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Qiu S, Lin S, Hu D, Feng Y, Tan Y and Peng
Y: Interactions of miR-323/miR-326/miR-329 and
miR-130a/miR-155/miR-210 as prognostic indicators for clinical
outcome of glioblastoma patients. J Transl Med. 11:10–21. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Rivera-Díaz M, Miranda-Román MA, Soto D,
Quintero-Aguilo M, Ortiz-Zuazaga H, Marcos-Martinez MJ and
Vivas-Mejía PE: MicroRNA-27a distinguishes glioblastoma multiforme
from diffuse and anaplastic astrocytomas and has prognostic value.
Am J Cancer Res. 5:201–218. 2014.
|
|
41
|
Malzkorn B, Wolter M, Liesenberg F,
Grzendowski M, Stühler K, Meyer HE and Reifenberger G:
Identification and functional characterization of microRNAs
involved in the malignant progression of gliomas. Brain Pathol.
20:539–550. 2010. View Article : Google Scholar
|
|
42
|
Koshkin PA, Chistiakov DA, Nikitin AG,
Konovalov AN, Potapov AA, Usachev DY, Pitskhelauri DI, Kobyakov GL,
Shishkina LV and Chekhonin VP: Analysis of expression of microRNAs
and genes involved in the control of key signaling mechanisms that
support or inhibit development of brain tumors of different grades.
Clin Chim Acta. 430:55–62. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Turner KM, Sun Y, Ji P, Granberg KJ,
Bernard B, Hu L, Cogdell DE, Zhou X, Yli-Harja O, Nykter M, et al:
Genomically amplified Akt3 activates DNA repair pathway and
promotes glioma progression. Proc Natl Acad Sci USA. 112:3421–3426.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Jain R, Poisson L, Narang J, Scarpace L,
Rosenblum ML, Rempel S and Mikkelsen T: Correlation of perfusion
parameters with genes related to angiogenesis regulation in
glioblastoma: a feasibility study. AJNR Am J Neuroradiol.
33:1343–1348. 2012. View Article : Google Scholar : PubMed/NCBI
|