Introduction
Glioblastoma multiforme (GBM) is the most common
form of human primary malignant brain tumors and it accounts for
>60% of all primary brain tumors in adults (1,2).
Because of resistance to conventional therapies, the prognosis of
GBM remains dismal with median survival of ~14 months and 5-year
survival only ~3% (3).
Understanding molecular mechanisms underpinning resistance of
conventional therapies of glioblastoma will offer novel targets for
effective therapies.
MicroRNAs (miRNAs) are small, non-coding RNAs that
post-transcriptionally regulate gene expression (4) and play significant roles in
maintaining normal cellular functions (5). Deregulation of miRNA expression leads
to diverse disease types, including cancers (6) as exemplified by their differential
expression in carcinomas (7),
sarcomas (8,9), and hematologic tumors (10). Let-7g-5p is significantly
downregulated in the serum of GBM patients and it has been proposed
as a tumor suppressive gene (11,12).
Glioma stem cells (GSCs) or glioma initiating cells
(GICs) have been identified and shown to constitute a primitive
cell population capable of self-renewal and differentiation that
has the unique capacity to give rise to new tumors upon serial
transplantation (13–16). Cancer stem/initiating cells are
believed to play an essential role in tumor recurrence after
therapeutic intervention (17), and
their high chemo-resistance and radiation resistance (18) require the identification of
alternative therapeutic strategies that could effectively lead to
their functional or physical eradication. Although a few signaling
pathways, including Sonic-Hedgehog (19), and the bone morphogenic proteins
BMP4 and BMPR1B (20,21) have been shown to be implicated in
GSCs maintenance, the mechanisms underlying GSCs generation, and
propagation have yet to be elucidated.
Epithelial to mesenchymal transition (EMT) is an
essential process for driving plasticity during development, but is
also an unintentional behavior of cells during progression of
malignant tumor (22–24). EMT confers mesenchymal properties on
epithelial cells and has been closely associated with the
acquisition of aggressive traits by carcinoma cells (25). Disturbance of a controlled
epithelial balance is triggered by altering several layers of
regulation, including the transcriptional and translational
machinery, expression of non-coding RNAs, alternative splicing and
protein stability (26–28).
In this study, we found that VSIG4 protein is
upregulated in glioblastoma. Overexpressing VSIG4 induced
epithelial-mesenchymal transition (EMT) and significantly promoted
invasion and migration in glioblastoma U-87MG cells. Moreover, we
showed that its overexpression promoted formation of glioma stem
cell phenotypes in U-87MG cells. We found that let-7g-5p can
downregulate VSIG4 protein expression, but it cannot degrade VSIG4
mRNA in U-87MG cells. Contrary to VSIG4, we demonstrated that
overexpressing let-7g-5p promoted mesenchymal-epithelial transition
(MET) and significantly inhibited invasion and migration consistent
with the reduction of glioblastoma stem cell phenotypes in U-87MG
cells.
Materials and methods
Glioblastoma tissues
Glioblastoma tissues and adjacent normal tissues
were obtained from the Department of Neurosurgery, The Affiliated
Hospital of Taishan Medical University, Shandong, China. All
tissues were examined histologically, and pathologists confirmed
the diagnosis. Medical ethics committee approved the experiments.
The use of human's tissue samples follows internationally
recognized guidelines as well as local and national regulations.
Informed consent was obtained from each individual.
Glioblastoma U-87MG cell line, VSIG4
expressing plasmids/empty vectors, pre-let-7g-5p/control miR and
transfection
Human glioblastoma cell line U-87MG was obtained
from American Type Culture Collection. Briefly, cells were
maintained in RPMI-1640 medium supplemented with 10% fetal bovine
serum (FBS) (Gibco, Grand Island, NY, USA) and
penicillin/streptomycin at 37°C in a humidified atmosphere with 5%
CO2. VSIG4 expressing plasmids/empty vectors (pcDNA3.1)
were purchased from Tiangen (Beijing, China). Pre-let-7g and
control miR were purchased from Ambion, Inc. (Ambion, Austin, TX,
USA). For transfection experiments, the cells were cultured in
serum-free medium without antibiotics at 60% confluence for 24 h,
and then transfected with transfection reagent (Lipofectamine 2000,
Invitrogen, Carlsbad, CA, USA) according to the manufacturer's
instructions. After incubation for 6 h, the medium was removed and
replaced with normal culture medium for 48 h, unless otherwise
specified.
Western blot analysis
Western blot analysis was performed as described
before (29). Briefly, after
incubation with primary antibody anti-VSIG4 (1:500; Abcam,
Cambridge, MA, USA), anti-CD133 (1:500; Abcam), anti-EZH2 (1:500;
Abcam), anti-c-Met (1:500; Abcam), anti-P4HB (1:500; Abcam),
anti-VAMP8 (1:500; Abcam), anti-CX43 (1:500; Abcam),
anti-E-cadherin (1:500; Abcam), anti-TGFB1 (1:500; Abcam),
anti-vimentin (1:500; Abcam), anti-SNAIL (1:500; Abcam),
anti-Notch1 (1:500; Abcam), anti-TLR9 (1:500; Abcam), anti-EphA2
(1:500; Abcam), anti-MLK4 (1:500; Abcam) and anti-β-actin (1:500;
Abcam) overnight at 4°C, IRDye™-800 conjugated anti-rabbit
secondary antibodies (LI-COR, Biosciences, Lincoln, NE, USA) were
used for 30 min at room temperature. The specific proteins were
visualized by Odyssey™ Infrared Imaging System (Gene Co., Lincoln,
NE, USA).
Sphere growth
Cells (103/ml) in serum-free RPMI-1640/1
mM Na-pyruvate were seeded on 0.5% agar precoated 6-well plates.
After 1 week, half the medium was changed every third day. Single
spheres were picked and counted.
Immunofluorescence analyses
For U-87MG cell immunofluorescence analyses, U-87MG
cells were plated on glass coverslips in 6-well plates and
transfected as indicated. At 48 h after transfection, coverslips
were stained with CD44 (1:500; Abcam) or antibody anti-VSIG4
(1:500; Abcam). Alexa Fluor 488 goat anti-rabbit IgG antibody was
used as secondary antibody (Invitrogen). Coverslips were
counterstained with DAPI (Invitrogen-Molecular Probes, Eugene, OR,
USA) for visualization of the nuclei. Microscopic analysis was
performed with a confocal laser-scanning microscope (Leica
Microsystems, Bensheim, Germany). Fluorescence intensities were
measured in a few viewing areas for 300 cells per coverslip and
analyzed using ImageJ 1.37v software (http://rsb.info.nih.gov/ij/index.html).
Wound healing assay
Wound healing assay was performed as described
before (30).
Migration and invasion assay
Migration and invasion assay was performed as
described before (29).
Methods of bioinformatics
The analysis of potential microRNA target site using
the commonly used prediction algorithms - miRanda (http://www.microrna.org/).
Real-time PCR for microRNAs
Total RNA from cultured cells, with efficient
recovery of small RNAs, was isolated using the mirVana miRNA
Isolation kit (Ambion). Detection of the mature form of miRNAs was
performed using the mirVana qRT-PCR miRNA Detection kit and qRT-PCR
Primer Sets, according to the manufacturer's instructions (Ambion).
The U6 small nuclear RNA was used as an internal control.
Reverse transcription-polymerase chain
reaction
It was performed as described before (31). Primers for VSIG4: forward,
5′-GTGTCCAGTTTGGCTAGTGCC-3′; reverse, 5′-GACTGGAGAACAGAAGCAGGC-3′.
Primers for GAPDH: forward, 5′-CGGAGTCAACGGATTTGGTCG TAT-3′;
reverse, 5′-AGCCTTCTCCATGGTGGTGAAGAC-3′.
Northern blot analysis
Northern blot analysis for miRNAs were performed as
described previously (32). Probes
were labeled with [γ-32P]-ATP complementary to let-7g-5p
and U6 snRNA.
Statistical analysis
Data are presented as mean ± SEM. Student's t-test
(two-tailed) was used to compare two groups (P<0.05 was
considered significant), unless otherwise indicated (χ2
test).
Results
VSIG4 promotes formation of stem
cell-like population in glioblastoma U-87MG cells
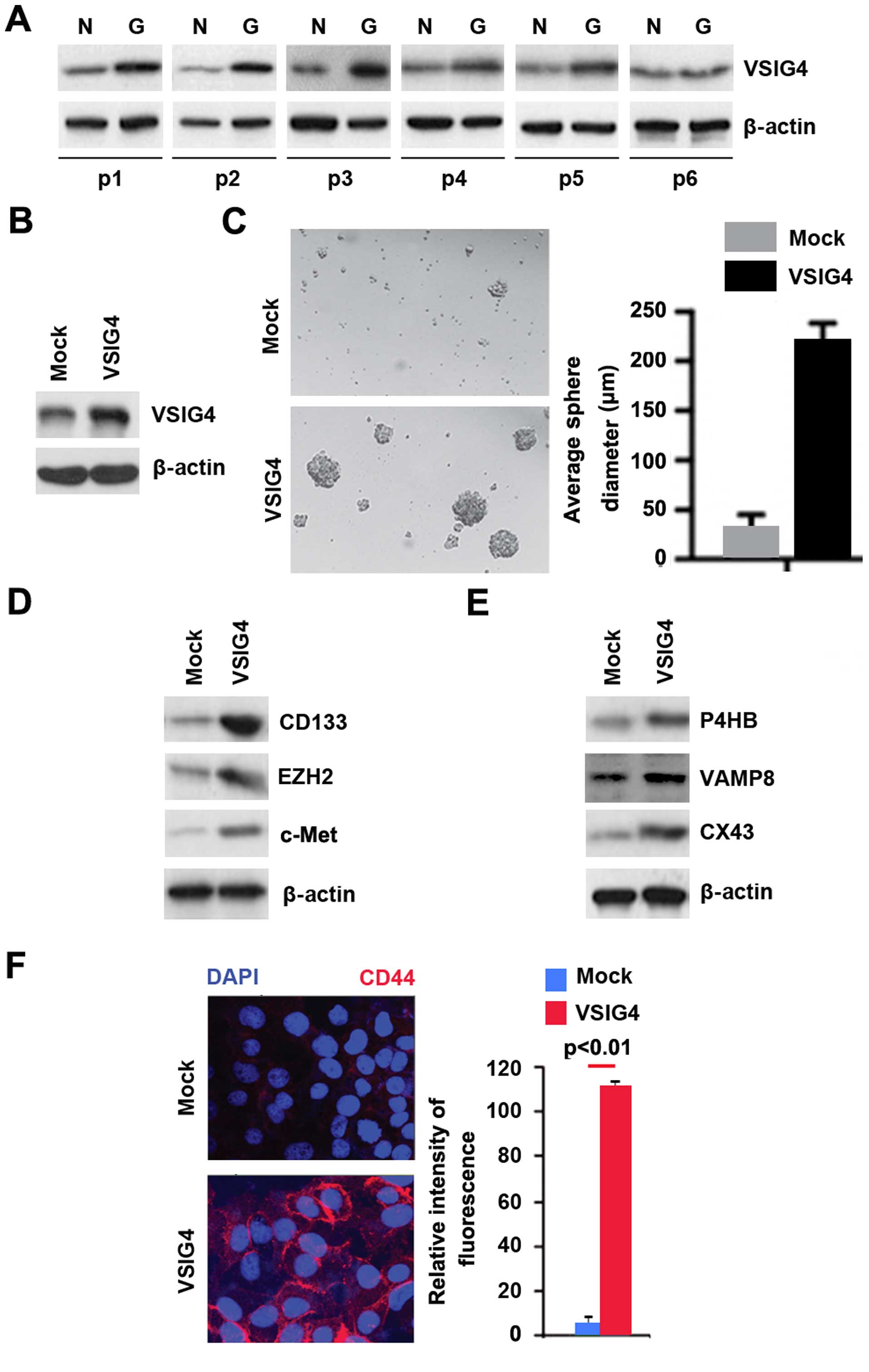
In an attempt to identify VSIG4 protein expression
between glioblastoma tissues and adjacent normal tissues, we
performed western blotting in tumor tissues versus normal tissues.
Protein was isolated from 6 pairs of glioblastoma tissues and
normal tissues (patient nos. 1–6). We found that VSIG4 protein was
significantly increased in cancer tissues, compared with adjacent
normal tissues (Fig. 1A). It
implied that VSIG4 could be an oncogene in glioblastoma.
In order to assess the role of VSIG4 in
glioblastoma, we transfected U-87MG cells with VSIG4 expressing
plasmids and then western blotting was performed. We found that
VSIG4 protein was significantly increased in the cells transfected
with VSIG4 expressing plasmids (Fig.
1B). To determine whether VSIG4 can affect GSCs, we performed
sphere forming assay to assess formation of stem cell-like
population. We found that formations of spheres were increased by
VSIG4 in U-87MG cells (Fig. 1C).
CD133, EZH2, c-Met and CD44 are robust markers and are of
functional importance for GSC for tumor initiation (33–36).
In order to detect whether CD133, EZH2, c-Met and CD44 protein
expression can be affected by VSIG4, we performed western blotting
and immunofluorescence. The results showed that CD133, EZH2, c-Met
(Fig. 1D) and CD44 protein were
upregulated by VSIG4 (Fig. 1F).
P4HB, VAMP8 and Connexin 43 (CX43) can promote
temozolomide (TMZ) resistance in human glioma cells (37–39).
To identify whether VSIG4 could have potential to affect
temozolomide (TMZ) resistance, we performed western blotting to
detect P4HB, VAMP8 and Connexin 43 (CX43) protein. The results
showed that P4HB, VAMP8 and Connexin 43 (CX43) protein were
upregulated by VSIG4 in U-87MG cells (Fig. 1E).
Overexpressing VSIG4 promotes EMT in
glioblastoma U-87MG cells
EMT has been shown to result in cancer cells with
stem cell-like characteristics that have a propensity to invade
surrounding tissue and display resistance to certain therapeutic
interventions (40). In order to
assess the role of VSIG4 in EMT of U-87MG, we transfected U-87MG
cells with VSIG4 expressing plasmids and then we found that its
overexpression caused significant changes in the cell morphology
(EMT, phenotype from a cobblestone-like to a spindle-like
morphology) (Fig. 2A). To further
verify that the changes in cell morphology are caused by EMT, we
performed western blotting to detect expression of epithelial and
mesenchymal markers in U-87MG cells transfected with VSIG4
expressing plasmids and the cells transfected with empty vectors.
The results revealed that epithelial marker (E-cadherin) was
inhibited and the mesenchymal markers (TGFB1, Vimentin, SNAIL, ZEB1
and Notch1) were induced by VSIG4 in U-87 MG cells (Fig. 1B).
EMT can result in increased cell invasion and
migration (41–43). Thus, we reasoned that VSIG4 could
also affect invasion and migration in U-87 MG cells. To identify
this reason, we performed would healing, invasion, and migration
assays. We found that overexpressing VSIG4 resulted in enhanced
migration (Fig. 1C and D) and
invasion (Fig. 1E) in the
cells.
Let-7g-5p inhibits VSIG4 in glioblastoma
U-87MG cells
Having demonstrated that overexpressing VSIG4
promoted formation of stem cell-like population and EMT, next we
studied the mechanisms regulating VSIG4 expression in the disease.
MicroRNAs (miRs) are a class of small non-coding RNAs (~22
nucleotides) and negatively regulate protein-coding gene expression
by targeting mRNA degradation or translation inhibition (44–46).
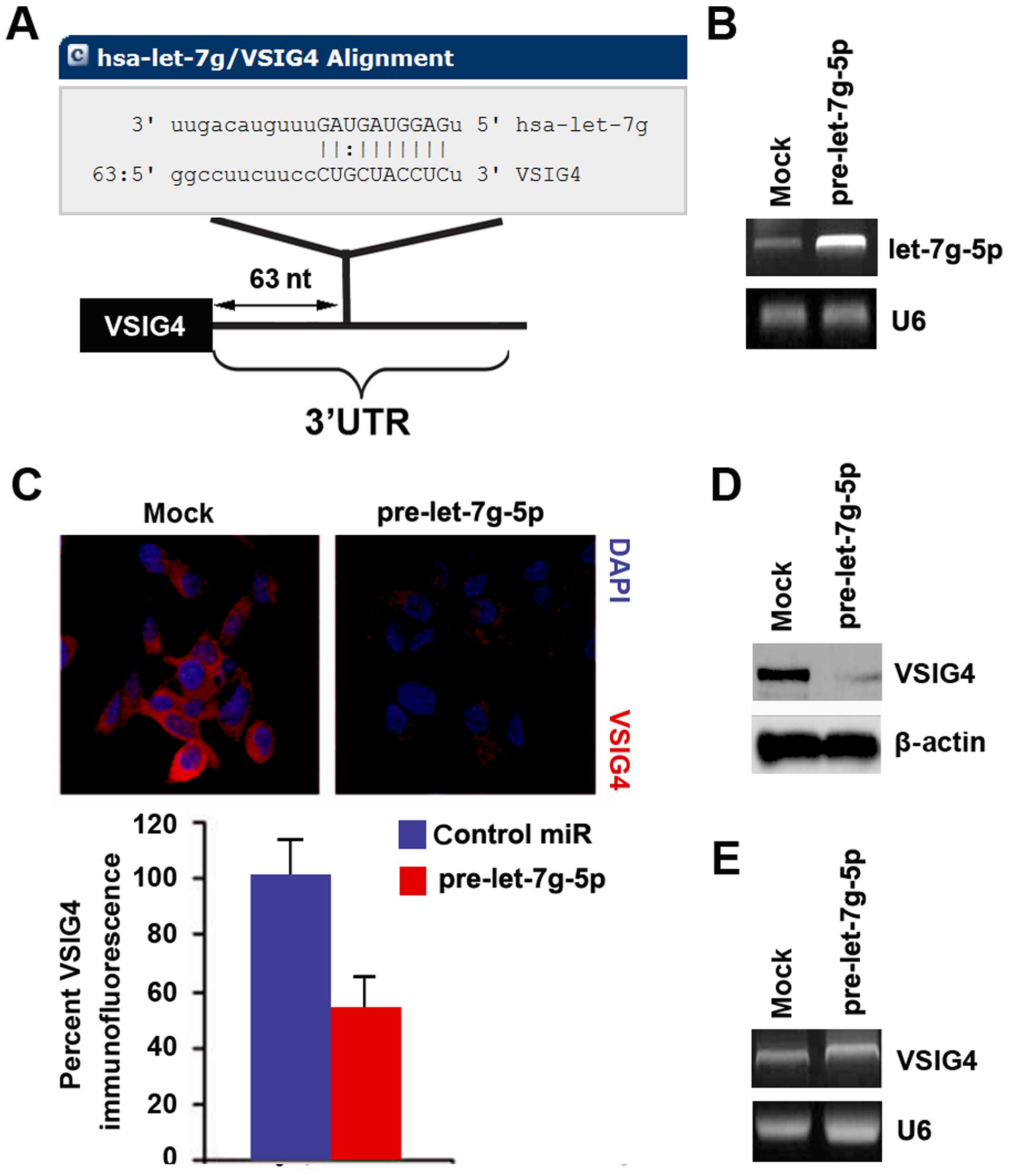
To further confirm whether VSIG4 could be regulated by microRNA, we
used the commonly used prediction algorithm - miRanda (http://www.microrna.org/microrna/home.do) to analyze
3′UTR of VSIG4. A dozen of microRNAs were found by the algorithm.
Nonetheless, we are interested in let-7g-5p, because let-7g-5p has
been proposed as a tumor suppressive gene (47,48).
However, its role still keeps emerging in glioblastoma.
Target sites on 3′UTR of VSIG4 are shown in Fig. 3A. We reasoned that let-7g-5p could
downregulate VSIG4 expression by targeting its 3′UTR in
glioblastoma. Downregulation of let-7g-5p can contribute to
upregulation of VSIG4 in glioblastoma. In an attempt to identify
the role of let-7g-5p in regulating VSIG4 expression in
glioblastoma, we transfected U-87MG cells with pre-let-7g-5p and
control miR. After transfection, let-7g-5p expression was detected
by real-time PCR and the results showed that let-7g-5p was
significantly increased by pre-let-7g-5p in the cells (Fig. 3B).
To confirm the reason, we performed
immunofluorescence analyses in U-87MG cells transfected with
pre-let-7g-5p and control miR. The results showed that VSIG4
protein was evidently inhibited in the cells transfected with
pre-let-7g-5p (Fig. 3C). We next
performed western blotting and RT-PCR to detect VSIG4 expression in
U-87MG cells transfected with pre-let-7g-5p and control miR. The
results showed that VSIG4 protein (Fig.
3D) was significantly downregulated in the cells transfected
with pre-let-7g-5p. However, we found that let-7g-5p did not
degrade VSIG4 mRNA (Fig. 3E).
Let-7g-5p inhibits formation of stem
cell-like population in glioblastoma U-87MG cells
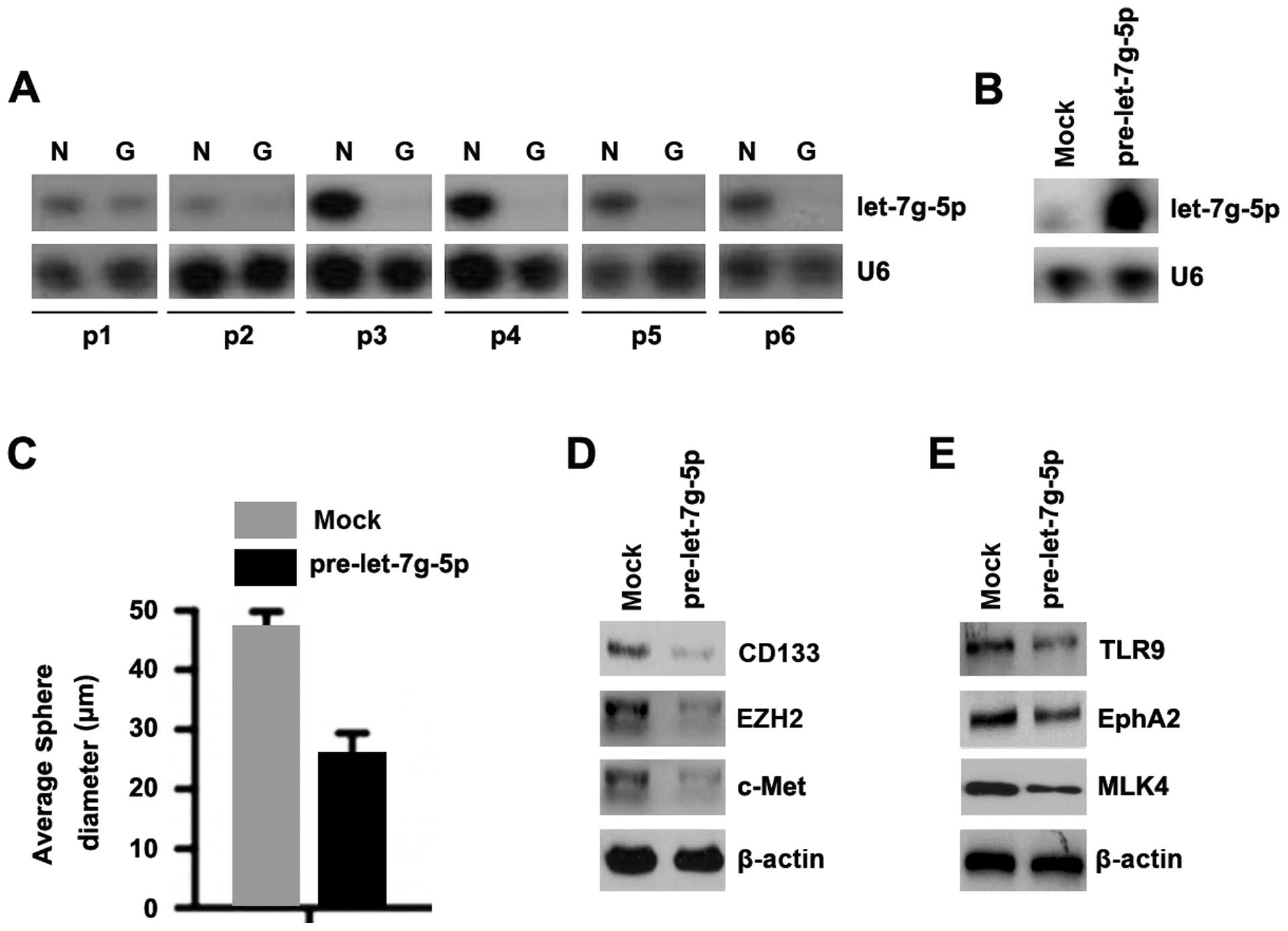
In an attempt to identify let-7g-5p expression
between glioblastoma tissues and adjacent normal tissues, we
performed northern blotting in tumor tissues versus normal tissues.
Protein was isolated from 6 pairs of glioblastoma tissues and
normal tissues (patient nos. 1–6). We found that let-7g-5p was
significantly decreased in glioblastoma tissues, compared with
adjacent normal tissues (Fig. 4A).
It indicated that let-7g-5p could be a tumor suppressive gene in
glioblastoma. In order to assess the role of let-7g-5p in
glioblastoma, we transfected U-87MG cells with pre-let-7g-5p and
then northern blot analyses were performed. We found that let-7g-5p
was significantly increased in the cells transfected with
pre-let-7g-5p (Fig. 4B).
To determine whether let-7g-5p could affect
stem-like cell characteristics, we performed sphere forming assay
to assess the capacity of CSC or CSC-like cell self-renewal in this
study. We found that formations of spheres were decreased by
let-7g-5p in U-87MG cells (Fig.
4C). We also performed western blotting to detect whether GSCs
markers, CD133, EZH2, c-MET, TLR9, EphA2 and MLK4 can be affected
by let-7g-5p in the cells. The results showed that CD133, EZH2,
c-MET, TLR9, EphA2 and MLK4 protein was significantly decreased by
let-7g-5p in U-87MG cells (Fig. 5D and
E).
Overexpressing let-7g-5p promotes MET in
glioblastoma U-87MG cells
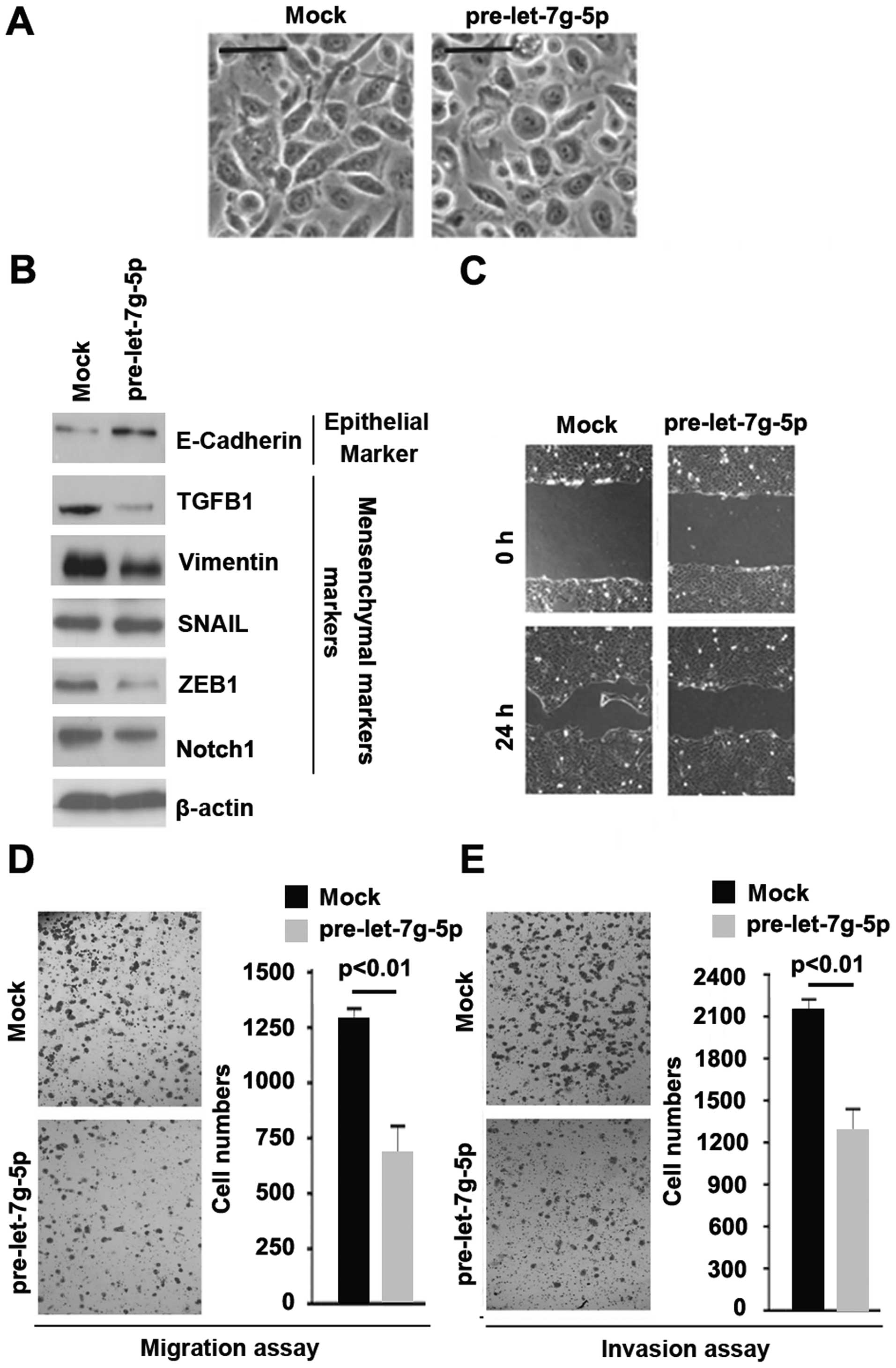
To assess the role of let-7g-5p in U-87MG cells, we
transfected U-87MG cells with pre-let-7g-5p and control miR. We
found that its overexpression caused slight changes in the cell
morphology (MET, phenotype from a spindle-like morphology to a
cobblestone-like) (Fig. 5A). To
further verify that the changes in cell morphology are caused by
MET, we performed western blotting to detect expression levels of
epithelial and mesenchymal markers in U-87MG cells transfected with
pre-let-7g-5p and the cells transfected with control miR. The
results revealed that epithelial marker (E-cadherin) was induced
and the mesenchymal markers (TGFB1, Vimentin, SNAIL, ZEB1 and
Notch1) were inhibited by let-7g-5p in U-87 MG cells (Fig. 5B).
To identify whether let-7g-5p could inhibit
migration and invasion, we performed wound-healing, migration and
invasion assays. We found that overexpressing let-7g-5p resulted in
decreased migration (Fig. 5C and D)
and invasion (Fig. 5E) in the
cells.
Discussion
Recently, it was reported that VSIG4 is highly
expressed in glioblastoma and correlated with poor prognosis of
high-grade glioma patients (49).
However, its role has not been reported in glioblastoma cells.
Consistent with the previous report, we found that VSIG4 protein is
upregulated in glioblastoma. Ionizing radiation represents the most
effective therapy for glioblastoma (50), but radiotherapy remains only
palliative (51) because of
radio-resistance. Glioma stem cells can promote radio-resistance
(18). We showed that
overexpressing VSIG4 promoted glioma stem cell phenotypes in U87MG
cells, implying that VSIG4 might play an important role in
radio-resistance. The emerging role of VSIG4 in glioblastoma
response to radiotherapy urges further investigation. Notch1 and
Notch2 can promote radio-resistance of GSCs in glioma (52). We found that VSIG4 can evidently
promote Notch1 protein expression. The results further indicated
that VSIG4 is a potential candidate to prevent radiotherapy
resistance. Moreover, VAMP8 can promote temozolomide resistance in
human glioma cells (37). Our
results also found that VSIG4 can upregulate VAMP8 protein
expression in glioblastoma cells, indicating its upregulation might
be a cause for temozolomide resistance.
Although the cell origin of cancer stem cells (CSCs)
remains to be fully elucidated, mounting evidence has demonstrated
that epithelial-to-mesenchymal transition, induced by different
factors, is associated with tumor aggressiveness and metastasis and
these cells share molecular characteristics with CSCs (53). We found that VSIG4 induced
epithelial-to-mesenchymal transition consistent with glioma stem
cell phenotypes in glioblastoma cells.
Let-7g-5p is significantly downregulated in the
serum of GBM patients and it has been proposed as a tumor
suppressive gene in glioblastoma (11,12).
Our results showed that its overexpression inhibited VSIG4 protein
in glioblastoma cells. Contrary to VSIG4, overexpressing let-7g-5p
promoted mesenchymal-epithelial transition and significantly
inhibited invasion and migration consistent with the reduction of
glioblastoma stem cells phenotypes in U87MG cells.
Elucidating the mechanism that let-7g-5p inhibits
epithelial-mesenchymal transition consistent with the reduction of
glioma stem cell phenotypes by targeting VSIG4 in glioblastoma will
help us to better understand the molecular mechanism of
epithelial-mesenchymal transition and glioma stem cells in
glioblastoma. Thus, restoration of let-7g-5p may represent a
promising therapeutic way to inhibit VSIG4-mediated EMT and GSCs
regulation. However, the roles of let-7g-5p/VSIG4 need to be
further confirmed in vivo.
References
|
1
|
Jemal A, Siegel R, Xu J and Ward E: Cancer
statistics, 2010. CA Cancer J Clin. 60:277–300. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Furnari FB, fenton T, Bachoo RM, Mukasa A,
Stommel JM, Stegh A, Hahn WC, Ligon KL, Louis DN, Brennan C, et al:
Malignant astrocytic glioma: Genetics, biology, and paths to
treatment. Genes Dev. 21:2683–2710. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Stupp R, Hegi ME, Mason WP, van den Bent
MJ, Taphoorn MJ, Janzer RC, Ludwin SK, Allgeier A, Fisher B,
Belanger K, et al European Organisation for Research and Treatment
of Cancer Brain Tumour and Radiation Oncology Groups; National
Cancer Institute of Canada Clinical Trials Group: Effects of
radiotherapy with concomitant and adjuvant temozolomide versus
radiotherapy alone on survival in glioblastoma in a randomised
phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet
Oncol. 10:459–466. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kim VN: Small RNAs: Classification,
biogenesis, and function. Mol Cells. 19:1–15. 2005.PubMed/NCBI
|
|
6
|
Mendell JT: miRiad roles for the miR-17-92
cluster in development and disease. Cell. 133:217–222. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lu J, Getz G, Miska EA, Alvarez-Saavedra
E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA,
et al: MicroRNA expression profiles classify human cancers. Nature.
435:834–838. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Subramanian S, Lui WO, Lee CH, Espinosa I,
Nielsen TO, Heinrich MC, Corless CL, Fire AZ and van de Rijn M:
MicroRNA expression signature of human sarcomas. Oncogene.
27:2015–2026. 2008. View Article : Google Scholar
|
|
9
|
Sarver AL, Phalak R, Thayanithy V and
Subramanian S: S-MED: Sarcoma microRNA expression database. Lab
Invest. 90:753–761. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ward A, Balwierz A, Zhang JD, Küblbeck M,
Pawitan Y, Hielscher T, Wiemann S and Sahin Ö: Re-expression of
microRNA-375 reverses both tamoxifen resistance and accompanying
EMT-like properties in breast cancer. Oncogene. 32:1173–1182. 2013.
View Article : Google Scholar
|
|
11
|
Dong L, Li Y, Han C, Wang X, She L and
Zhang H: miRNA microarray reveals specific expression in the
peripheral blood of glioblastoma patients. Int J Oncol. 45:746–756.
2014.PubMed/NCBI
|
|
12
|
Mao XG, Hütt-Cabezas M, Orr BA, Weingart
M, Taylor I, Rajan AK, Odia Y, Kahlert U, Maciaczyk J, Nikkhah G,
et al: LIN28A facilitates the transformation of human neural stem
cells and promotes glioblastoma tumorigenesis through a proinvasive
genetic program. Oncotarget. 4:1050–1064. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Huang Z, Cheng L, Guryanova OA, Wu Q and
Bao S: Cancer stem cells in glioblastoma - molecular signaling and
therapeutic targeting. Protein Cell. 1:638–655. 2010. View Article : Google Scholar
|
|
14
|
Galli R, Binda E, Orfanelli U, Cipelletti
B, Gritti A, De Vitis S, Fiocco R, Foroni C, Dimeco F and Vescovi
A: Isolation and characterization of tumorigenic, stem-like neural
precursors from human glioblastoma. Cancer Res. 64:7011–7021. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Singh SK, Clarke ID, Terasaki M, Bonn VE,
Hawkins C, Squire J and Dirks PB: Identification of a cancer stem
cell in human brain tumors. Cancer Res. 63:5821–5828.
2003.PubMed/NCBI
|
|
16
|
Singh SK, Hawkins C, Clarke ID, Squire JA,
Bayani J, Hide T, Henkelman RM, Cusimano MD and Dirks PB:
Identification of human brain tumour initiating cells. Nature.
432:396–401. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Dick JE: Stem cell concepts renew cancer
research. Blood. 112:4793–4807. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Bao S, Wu Q, McLendon RE, Hao Y, Shi Q,
Hjelmeland AB, Dewhirst MW, Bigner DD and Rich JN: Glioma stem
cells promote radioresistance by preferential activation of the DNA
damage response. Nature. 444:756–760. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Clement V, Sanchez P, de Tribolet N,
Radovanovic I and Ruiz i Altaba A: HEDGEHOG-GLI1 signaling
regulates human glioma growth, cancer stem cell self-renewal, and
tumorigenicity. Curr Biol. 17:165–172. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lee J, Son MJ, Woolard K, Donin NM, Li A,
Cheng CH, Kotliarova S, Kotliarov Y, Walling J, Ahn S, et al:
Epigenetic-mediated dysfunction of the bone morphogenetic protein
pathway inhibits differentiation of glioblastoma-initiating cells.
Cancer Cell. 13:69–80. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Piccirillo SG, Reynolds BA, Zanetti N,
Lamorte G, Binda E, Broggi G, Brem H, Olivi A, Dimeco F and Vescovi
AL: Bone morphogenetic proteins inhibit the tumorigenic potential
of human brain tumour-initiating cells. Nature. 444:761–765. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Nieto MA: The ins and outs of the
epithelial to mesenchymal transition in health and disease. Annu
Rev Cell Dev Biol. 27:347–376. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Savagner P, Yamada KM and Thiery JP: The
zinc-finger protein slug causes desmosome dissociation, an initial
and necessary step for growth factor-induced epithelial-mesenchymal
transition. J Cell Biol. 137:1403–1419. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Thiery JP: Epithelial-mesenchymal
transitions in tumour progression. Nat Rev Cancer. 2:442–454. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yang J and Weinberg RA:
Epithelial-mesenchymal transition: At the crossroads of development
and tumor metastasis. Dev Cell. 14:818–829. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chang CJ, Chao CH, Xia W, Yang JY, Xiong
Y, Li CW, Yu WH, Rehman SK, Hsu JL, Lee HH, et al: p53 regulates
epithelial-mesenchymal transition and stem cell properties through
modulating miRNAs. Nat Cell Biol. 13:317–323. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Gravdal K, Halvorsen OJ, Haukaas SA and
Akslen LA: A switch from E-cadherin to N-cadherin expression
indicates epithelial to mesenchymal transition and is of strong and
independent importance for the progress of prostate cancer. Clin
Cancer Res. 13:7003–7011. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hader C, Marlier A and Cantley L:
Mesenchymal-epithelial transition in epithelial response to injury:
The role of foxc2. Oncogene. 29:1031–1040. 2010. View Article : Google Scholar :
|
|
29
|
Lu Y, Chopp M, Zheng X, Katakowski M,
Buller B and Jiang F: MiR-145 reduces ADAM17 expression and
inhibits in vitro migration and invasion of glioma cells. Oncol
Rep. 29:67–72. 2013.
|
|
30
|
Zhang BG, Li JF, Yu BQ, Zhu ZG, Liu BY and
Yan M: microRNA-21 promotes tumor proliferation and invasion in
gastric cancer by targeting PTEN. Oncol Rep. 27:1019–1026.
2012.PubMed/NCBI
|
|
31
|
Zhang HY, Li JH, Li G and Wang SR:
Activation of ARK5/miR-1181/HOXA10 axis promotes
epithelial-mesenchymal transition in ovarian cancer. Oncol Rep.
34:1193–1202. 2015.PubMed/NCBI
|
|
32
|
Yu J, Ryan DG, Getsios S,
Oliveira-Fernandes M, Fatima A and Lavker RM: MicroRNA-184
antagonizes microRNA-205 to maintain SHIP2 levels in epithelia.
Proc Natl Acad Sci USA. 105:19300–19305. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zeppernick F, Ahmadi R, Campos B, Dictus
C, Helmke BM, Becker N, Lichter P, Unterberg A, Radlwimmer B and
Herold-Mende CC: Stem cell marker CD133 affects clinical outcome in
glioma patients. Clin Cancer Res. 14:123–129. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Suvà ML, Riggi N, Janiszewska M,
Radovanovic I, Provero P, Stehle JC, Baumer K, Le Bitoux MA, Marino
D, Cironi L, et al: EZH2 is essential for glioblastoma cancer stem
cell maintenance. Cancer Res. 69:9211–9218. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Li Y, Li A, Glas M, Lal B, Ying M, Sang Y,
Xia S, Trageser D, Guerrero-Câzares H, Eberhart CG, et al: c-Met
signaling induces a reprogramming network and supports the
glioblastoma stem-like phenotype. Proc Natl Acad Sci USA.
108:9951–9956. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Pietras A, Katz AM, Ekström EJ, Wee B,
Halliday JJ, Pitter KL, Werbeck JL, Amankulor NM, Huse JT and
Holland EC: Osteopontin-CD44 signaling in the glioma perivascular
niche enhances cancer stem cell phenotypes and promotes aggressive
tumor growth. Cell Stem Cell. 14:357–369. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Chen Y, Meng D, Wang H, Sun R, Wang D,
Wang S, Fan J, Zhao Y, Wang J, Yang S, et al: VAMP8 facilitates
cellular proliferation and temozolomide resistance in human glioma
cells. Neuro Oncol. 17:407–418. 2015.
|
|
38
|
Gielen PR, Aftab Q, Ma N, Chen VC, Hong X,
Lozinsky S, Naus CC and Sin WC: Connexin43 confers Temozolomide
resistance in human glioma cells by modulating the mitochondrial
apoptosis pathway. Neuropharmacology. 75:539–548. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Sun S, Lee D, Ho AS, Pu JK, Zhang XQ, Lee
NP, Day PJ, Lui WM, Fung CF and Leung GK: Inhibition of prolyl
4-hydroxylase, beta polypeptide (P4HB) attenuates temozolomide
resistance in malignant glioma via the endoplasmic reticulum stress
response (ERSR) pathways. Neuro-oncol. 15:562–577. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Polyak K and Weinberg RA: Transitions
between epithelial and mesenchymal states: Acquisition of malignant
and stem cell traits. Nat Rev Cancer. 9:265–273. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Zuo JH, Zhu W, Li MY, Li XH, Yi H, Zeng
GQ, Wan XX, He QY, Li JH, Qu JQ, et al: Activation of EGFR promotes
squamous carcinoma SCC10A cell migration and invasion via inducing
EMT-like phenotype change and MMP-9-mediated degradation of
E-cadherin. J Cell Biochem. 112:2508–2517. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Jung H, Lee KP, Park SJ, Park JH, Jang YS,
Choi SY, Jung JG, Jo K, Park DY, Yoon JH, et al: TMPRSS4 promotes
invasion, migration and metastasis of human tumor cells by
facilitating an epithelial-mesenchymal transition. Oncogene.
27:2635–2647. 2008. View Article : Google Scholar
|
|
43
|
Christiansen JJ and Rajasekaran AK:
Reassessing epithelial to mesenchymal transition as a prerequisite
for carcinoma invasion and metastasis. Cancer Res. 66:8319–8326.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Lee RC, Feinbaum RL and Ambros V: The C.
elegans heterochronic gene lin-4 encodes small RNAs with antisense
complementarity to lin-14. Cell. 75:843–854. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Pasquinelli AE, Reinhart BJ, Slack F,
Martindale MQ, Kuroda MI, Maller B, Hayward DC, Ball EE, Degnan B,
Müller P, et al: Conservation of the sequence and temporal
expression of let-7 heterochronic regulatory RNA. Nature.
408:86–89. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Reinhart BJ, Slack FJ, Basson M,
Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR and Ruvkun G:
The 21-nucleotide let-7 RNA regulates developmental timing in
Caenorhabditis elegans. Nature. 403:901–906. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Nakajima G, Hayashi K, Xi Y, Kudo K,
Uchida K, Takasaki K, Yamamoto M and Ju J: Non-coding microRNAs
hsa-let-7g and hsa-miR-181b are associated with chemoresponse to
S-1 in colon cancer. Cancer Genomics Proteomics. 3:317–324.
2006.
|
|
48
|
Ji JF, Zhao L, Budhu AL, et al: Let-7g
targets collagen type I α2 and inhibits cell migration in
hepatocellular carcinoma. Exp Mol Med. 43:298–304. 2011.
|
|
49
|
Xu T, Jiang Y, Yan Y, Wang H, Lu C, Xu H,
Li W, Fu D, Lu Y and Chen J: VSIG4 is highly expressed and
correlated with poor prognosis of high-grade glioma patients. Am J
Transl Res. 7:1172–1180. 2015.PubMed/NCBI
|
|
50
|
Legler JM, Ries LA, Smith MA, Warren JL,
Heineman EF, Kaplan RS and Linet MS: Brain and other central
nervous system cancers: Recent trends in incidence and mortality. J
Natl Cancer Inst. 91:1382–1390. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Garden AS, Maor MH, Yung WK, Bruner JM,
Woo SY, Moser RP and Lee YY: Outcome and patterns of failure
following limited-volume irradiation for malignant astrocytomas.
Radiother Oncol. 20:99–110. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Wang J, Wakeman TP, Lathia JD, Hjelmeland
AB, Wang XF, White RR, Rich JN and Sullenger BA: Notch promotes
radioresistance of glioma stem cells. Stem Cells. 28:17–28.
2010.
|
|
53
|
Kong D, Li Y, Wang Z and Sarkar FH: Cancer
stem cells and epithelial-to-mesenchymal transition
(EMT)-phenotypic cells: Are they cousins or twins? Cancers (Basel).
3:716–729. 2011. View Article : Google Scholar
|