Introduction
Breast cancer is one of the most common malignancies
worldwide. In the US, it is estimated that 234,190 patients were
diagnosed with breast cancer and 40,730 succumbed to the disease in
2015 (1). Breast cancer is a
heterogeneous disease. In addition, this heterogeneity occurs both
between tumors (inter-tumor heterogeneity) and within tumors
(intra-tumor heterogeneity). This high diversity determines the
danger of disease progression and challenges the effectiveness of
treatment strategies (2,3). Therefore, in-depth understanding of
the heterogeneity of the disease may elucidate strategies by which
to conquer the disease.
Triple-negative breast cancer (TNBC) is a subtype of
invasive breast cancer with an estrogen receptor-negative
(ER−), progesterone receptor-negative (PR−),
and HER-2-negative (HER2−) phenotype. It is a
significant and noticeable subtype of breast cancer due to its
relationship with basal-like breast cancer (4). Although TNBC is highly proliferative
and sensitive to systemic chemotherapies, patient outcomes are poor
compared with other subtypes of breast cancer (HER2+ or
ER+) (5–7). TNBC is the only major type of breast
cancer for which no specific FDA-approved targeted therapy is
available to improve patient outcomes; it is resistant to targeted
therapies such as hormonal and HER2-targeting therapies (6).
The epidermal growth factor receptor (EGFR), a
cell-surface receptor, which is a member of the ErbB family of
tyrosine kinases, plays a critical role in the regulation of cell
proliferation, survival and differentiation (8). Dysregulation of EGFR has been reported
in a wide variety of carcinomas, including head and neck, breast,
bladder, ovarian, renal, colon and lung cancer (8–10). In
breast cancer, abnormal activation of EGFR is associated with
aggressive phenotypes, such as large tumor size, poor
differentiation and poor clinical outcomes (11–13).
Moreover, overexpression and activation of EGFR have been
frequently reported in TNBC and provide a potential target for TNBC
(14,15). Therefore, a full understanding of
the EGFR pathway in TNBC is a precondition for conquering the
disease.
G-protein-coupled receptor family C group 5 member A
(GPRC5A) is a member of the type 3-G protein-coupling receptor
family, characterized by the signature 7-transmem-brane domain
motif. Its role in cancer was first unveiled in lung cancer and it
has been recognized as a growth-promoting gene and a novel P53
transcriptional target (16).
Further study showed that GPRC5A was downregulated in 61% of lung
tumors compared with adjacent normal tissues (17). Results from Gprc5a-knockout
mice suggest a tumor-suppressive role in lung adenocarcinoma
(18). Molecular and pathway
analysis indicates interaction between EGFR and GPRC5A and negative
regulation of EGFR by GPRC5A (19,20).
In breast cancer, a high prevalence of GPRC5A germline
mutations was found in BRCA1-mutant breast cancer patients
(21). However, the exact
biological functions of GPRC5A and its correlation with EGFR in
breast cancer remain obscure. In the present study, we explored the
functional role of GPRC5A and the related molecular mechanisms in
breast cancer cells in an attempt to discover useful information
and clues for developing a novel treatment strategy for TNBC.
Materials and methods
Chemicals, cell culture and plasmid
transfection
Breast cancer cell lines MDA-MB-231 and MCF7 were
obtained from the Type Culture Collection of the Chinese Academy of
Sciences and were maintained in Dulbecco's modified Eagle's medium
(DMEM) with 10% fetal bovine serum (FBS) in a 5%
CO2-humidified, 95% air incubator at 37°C. A plasmid
expressing EGFR (plasmid #11011) was obtained from Addgene
(Cambridge, MA, USA). Transient transfection was performed using
the Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA, USA)
following the manufacturer's instructions. MEK inhibitor, PD98059,
was obtained from Cell Signaling Technology (Danvers, MA, USA).
Lentiviral infection
GPRC5A-shRNA and the control lentivirus were
obtained from GenePharma (Shanghai, China). The GPRC5A-shRNA1
target sequence was: 5′-GCTTATGTTA GTCCCGAGTTT-3′; and the
GPRC5A-shRNA2 target sequence was: 5′-CCTGACCATGAATAGGACCAA-3′. The
virus-containing supernatant was incubated on target cells for 12 h
with 8 μg/ml Polybrene, following the manufacturer's
instructions. Infected cells were selected in puromycin, as
optimized for each cell line.
Colony formation assay
MDA-MB-231 and MCF7 cells infected with
GPRC5A-shRNA1, GPRC5A-shRNA2 or the scramble were plated into a
6-well plate (1,000 cells/well). Medium was replaced every 2 days,
and the cells were cultured for 2 weeks in a 5% CO2
environment at 37°C. Then, the colonies were stained with 0.01%
(w/v) crystal violet. Colonies containing >50 cells were
counted.
Apoptosis and BrdU incorporation
assay
Apoptosis rates were measured using the PE Annexin V
apoptosis detection kit (BD Pharmingen, San Diego, CA, USA)
following the manufacturer's instructions. Briefly, MCF7 and
MDA-MB-231 cells were incubated with cisplatin (40 μM) for
24 h, and then trypsinized and resuspended in 100 μl binding
buffer. Annexin V and PI (5 μl) were added to each tube.
Then, 400 μl binding buffer was added to each reaction tube
and the cells were analyzed by flow cytometry. Cell proliferation
rates were determined with the BrdU incorporation assay using cell
proliferation ELISA kit (Roche). The measurements were performed
following the manufacturer's instructions as previously described
(22).
RNA extraction and qRT-PCR
Total RNA was isolated using TRIzol reagent
(Invitrogen) following the manufacturer's instructions. Total RNA
(2 μg) was used for the synthesis of first-strand cDNA using
a High Capacity cDNA reverse transcription kit (Invitrogen,
Beijing, China). Quantitative real-time PCR was performed using
SYBR-Green Mix (Applied Biosystems, Foster City, CA, USA). The
reactions were performed using a 7500 Fast Real-Time PCR system
(Applied Biosystems). The data are displayed as 2−ΔΔCt
values and are representative of at least 3 independent
experiments. Primers used in the present study were: GPRC5a,
ATGGCTA CAACAGTCCCTGAT and CCACCGTTTCTAGGACGA TGC; EGFR,
AGGCACGAGTAACAAGCTCAC and ATGA GGACATAACCAGCCACC.
CCK-8 assay
MCF7 (8,000) and MBA-MD-231 (5,000) cells were
placed into 96-well plates. The cells were continually cultured for
24–72 h. At 24, 48 and 72 h, 10 μl of Cell Counting Kit-8
(CCK-8) reagent (Dojindo, Shanghai, China) was added to each well.
Then, the cells were incubated at 37°C for another 2 h. After
shaking for 20 min, the absorbance was detected at 450 nm on a
microplate spectrophotometer (Bio-Tek Instruments, Winooski, VT,
USA).
Migration and invasion assays
For the Transwell migration assay, the breast cancer
cells were trypsinized and placed in the upper chamber of each
insert (Corning, Cambridge, MA, USA, USA) containing the non-coated
membrane. For the invasion assay, the cells were placed on the
upper chamber of each insert coated with 40 μl of Matrigel
(BD Biosciences, Franklin Lakes, NJ, USA), which was diluted to 4
μg/μl with serum-free medium. Then, the medium
supplemented with 20% FBS (600 μl) was added to the lower
chambers. After 24 h of incubation at 37°C, the upper surface of
the membrane was wiped with a cotton tip, and the cells that had
attached to the lower surface were stained for 10 min with crystal
violet. Cells in 5 random fields of view at a magnification of ×100
were counted and values are expressed as the average number of
cells/field of view. All of the assays were performed in
triplicate.
Western blot assay
Whole-cell protein extracts were obtained using RIPA
buffer (Beyotime Institute of Biotechnology, Nanjing, China). Then,
the extracts were separated on 10% SDS denatured polyacrylamide gel
electrophoresis (PAGE) gels, transferred to nitrocellulose
membranes and blocked in phosphate-buffered saline/Tween-20
containing 5% non-fat milk. The membranes were incubated with the
antibodies overnight at 4°C. The membranes were then incubated with
the HRP-labeled corresponding IgG for 1 h. The protein expression
level was assessed by enhanced chemiluminescence and exposure to
film (Fujifilm, Tokyo, Japan). The anti-GPRC5A antibody (cat. no.
HPA007928) was obtained from Sigma-Aldrich (Shanghai, China). The
anti-E-cadherin (ab15148), anti-β-actin (ab8229), anti-EGFR
(ab2430) and anti-phospho-EGFR (ab40815) antibodies were obtained
from Abcam (Cambridge, MA, USA). The anti-Akt (9272),
anti-phospho-Akt (9271), anti-Stat3 (9132) and anti-phospho-Stat3
(9131) antibodies were purchased from Cell Signaling
Technology.
Statistical analysis
Experimental results are expressed as mean ±
standard deviation (SD). Statistically significant differences
between groups were determined using a two-tailed unpaired
Student's t-test. All statistical analyses were performed using
SPSS 16.0 software (SPSS, Inc., Chicago, IL, USA). P<0.05 was
considered to indicate a statistically significant result.
Results
GPRC5A knockdown promotes colony
formation in MDA-MB-231 cells but not in MCF7 cells
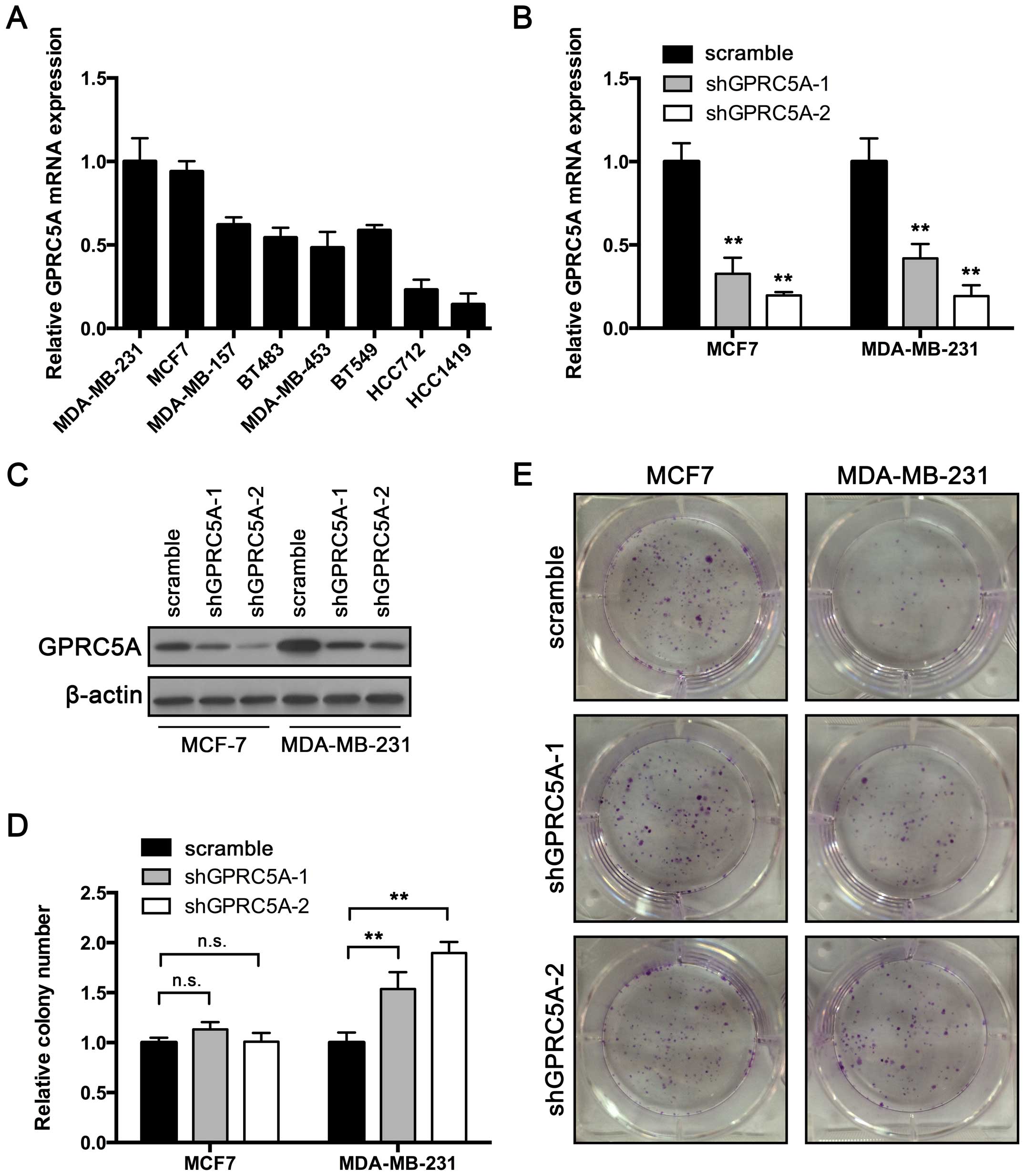
To investigate its role in breast cancer, we first
examined GPRC5A expression in established breast cancer cell
lines (Fig. 1A). We chose high
GPRC5A-expressing cell lines, MCF7 and MDA-MB-231, for
further investigation. Expression of GPRC5A in both cell
lines was suppressed using short hairpin RNAs (shRNAs), which
targeted 2 different sequences in RASAL2. RT-qPCR and western blot
assay were performed to confirm the knockdown efficiency. As shown
in Fig. 1B, the expression of
GPRC5A mRNA and protein were inhibited in cells transfected
with shRNA-GPRC5A-1 or shRNA-GPRC5A-2 compared with scramble shRNA.
Then a colony formation assay was performed. The results showed
that the colony number was unaffected by GPRC5A knockdown in the
MCF7 cells (Fig. 1D and E).
However, MDA-MB-231 cells transfected with GPRC5A shRNA produced
more colonies than the control cells in the colony formation assay
(Fig. 1D and E). These results
indicate that GPRC5A may play a suppressive role in breast cancer
cells.
GPRC5A knockdown enhances cell
proliferation in MDA-MB-231 cells but not in MCF7 cells
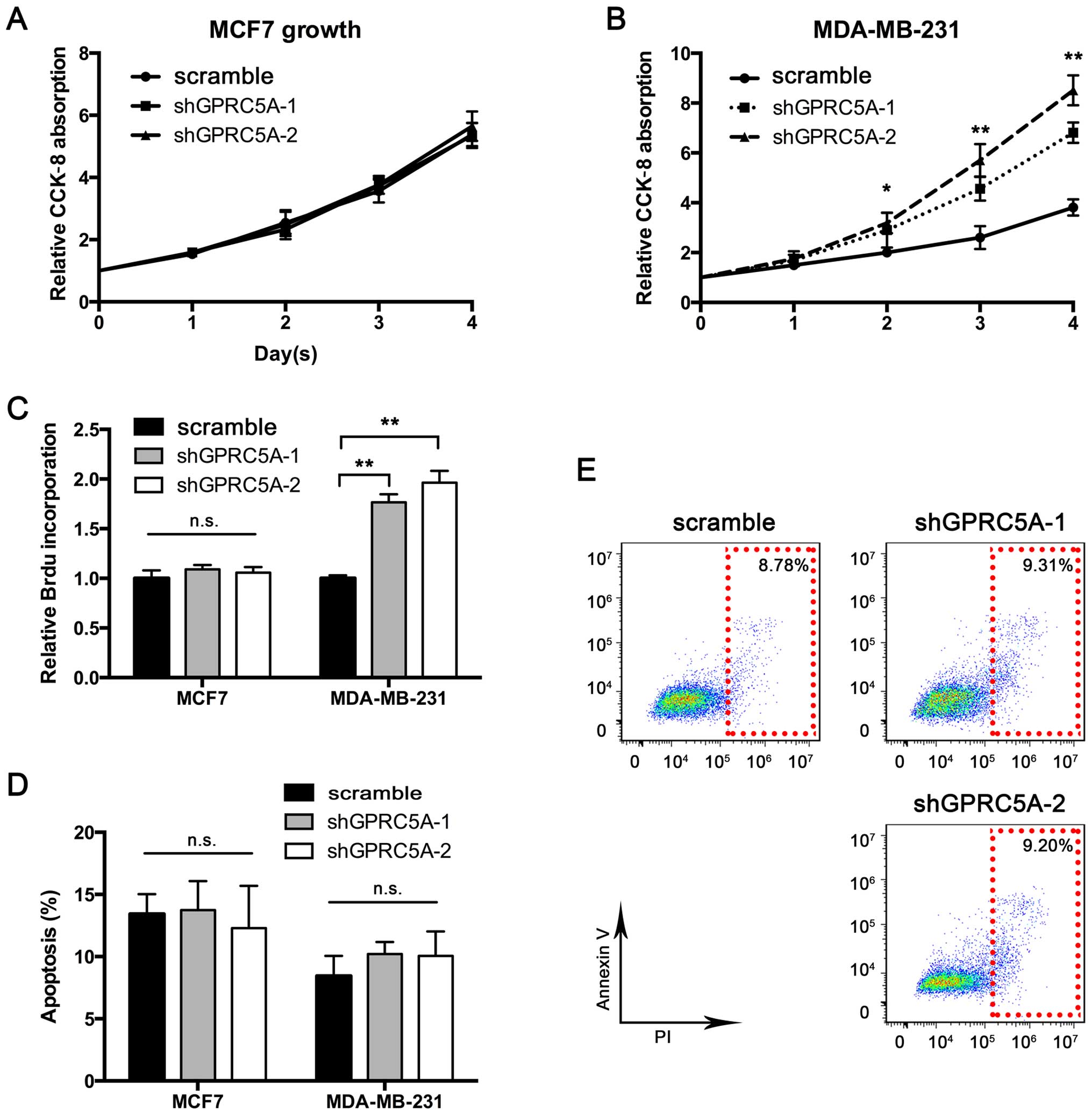
To further investigate the significance of GPRC5A in
breast cancer, we determined whether GPRC5A affects breast cancer
cell proliferation and apoptosis. We first performed the CCK-8
assay to construct a proliferation curve. As shown in Fig. 2A, although GPRC5A knockdown had no
impact on MCF7 proliferation, MDA-MB-231 cells exhibited
accelerated proliferation after GPRC5A knockdown (Fig. 2B). To further validate the role of
GPRC5A in proliferation, we performed BrdU incorporation assay.
Consistent with the previous results, GPRC5A knockdown aggravated
BrdU incorporation in the MDA-MB-231 cells, while BrdU uptake
remained unaffected in the MCF7 cells (Fig. 2C). Furthermore, we examined whether
GPRC5A knockdown controls cell apoptosis. The results showed that
GPRC5A knockdown did not regulate apoptosis in the MCF7 and
MDA-MB-231 cells (Fig. 2D and E).
Taken together, these results confirmed the suppressive role of
GPRC5A in MDA-MB-231 cells, particularly in regards to
proliferation.
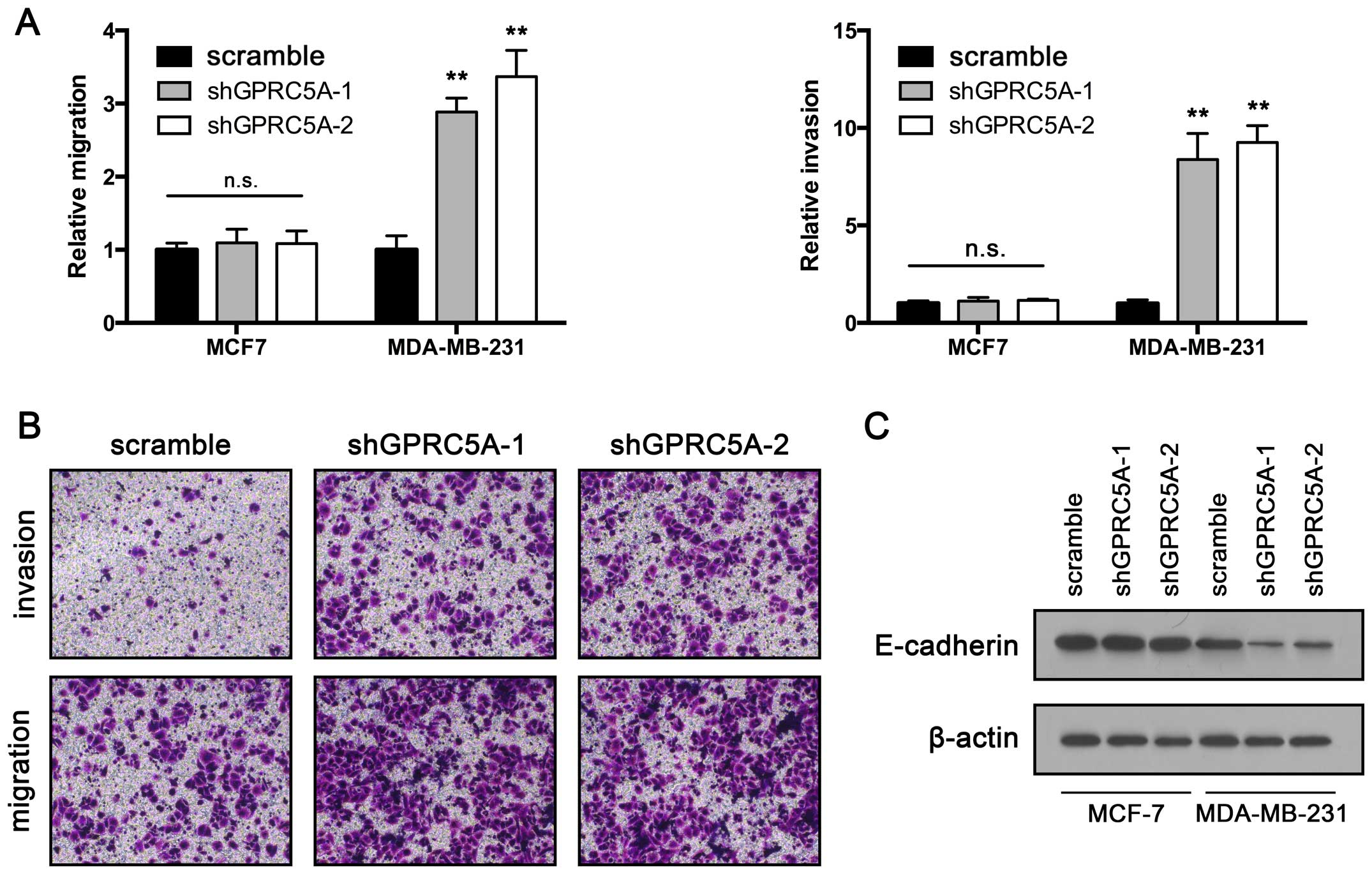
GPRC5A knockdown promotes migration and
invasion in MDA-MB-231 cells
Migration and invasion are critical steps in the
initial process of cancer metastasis (23). To further investigate the
significance of GPRC5A downregulation in breast cancer, we
determined whether inhibition of GPRC5A also promotes migration and
invasion. A Transwell migration assay was performed to assess
migratory ability. The results showed that inhibition of GPRC5A
significantly increased MDA-MB-231 cell migration. However, the
migratory ability of the MCF7 cells was unaltered after GPRC5A
knockdown (Fig. 3A and B). We
further confirmed these findings using a wound healing assay.
Consistent with the data obtained from the Transwell migration
assay, GPRC5A downregulation significantly promoted the migration
of the MDA-MB-231 cells, but had no impact on MCF7 cell migration.
In addition, we used a Matrigel-coated Transwell to assess cell
invasive ability. The MDA-MB-231 cells exhibited increased invasive
ability after GPRC5A knockdown. In agreement with the previous
results, MCF7 cells transfected with GPRC5A shRNA showed no
increased invasive ability when compared with that noted in the
control cells (Fig. 3A and B).
E-cadherin is a key player in the regulation of cancer cell EMT and
metastasis (24). We further
determined whether GPRC5A participates in the regulation of
E-cadherin. RT-qPCR and western blot assay were performed. The
results showed that GPRC5A downregulation suppressed GPRC5A
expression in the MDA-MB-231 cells but not in the MCF7 cells
(Fig. 3C). Taken together, our
results showed that GPRC5A may be a potential metastatic inhibitor
in breast cancer.
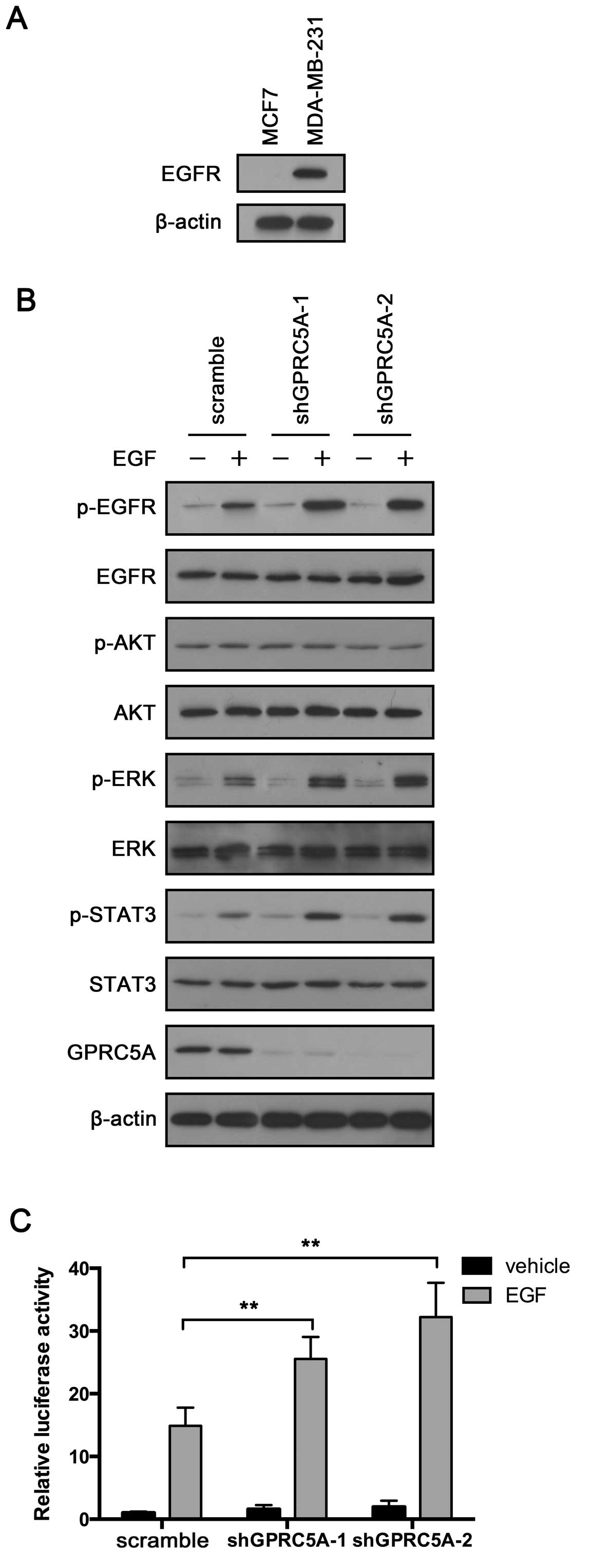
GPRC5A knockdown enhances EGFR activation
in the MDA-MB-231 cells but not in the MCF7 cells
The previous results confirmed the tumor-suppressive
role of GPRC5A in MDA-MB-231 cells. Notably, GPRC5A did not exhibit
any tumor-suppressive effects in the MCF7 cells. MDA-MB-231 cells
express EGFR, while MCF7 do not. We aimed to ascertain whether
GPRC5A exerts its tumor-suppressive effects through EGFR. First, we
confirmed its expression in the MCF7 and MDA-MB-231 cells. We did
not detect EGFR expression in the MCF7 cells, while the MDA-MB-231
cells exhibited relatively strong expression (Fig. 4A). Then, we treated MDA-MB-231 cells
infected with GPRC5A shRNA or scramble shRNA with EGF for 4 h and
examined EGFR activity. The results showed that GPRC5A knockdown
significantly enhanced EGF-induced EGFR activation (Fig. 4B). In addition, p-STAT3 was also
increased in the GPRC5A-knockdown cells compared with that noted in
the control cells (Fig. 4B). ERK is
one of the key downstream effectors of EGFR. Consistent with EGFR
and STAT3, total expression of ERK was not altered, while p-ERK was
significantly increased after EGF treatment in the GPRC5A-knockdown
cells (Fig. 4B). To further
validate the role of GPRC5A in EGF-induced EGFR activation, we
performed STAT3 luciferase reporter assay. MDA-MB-231 cells
infected with GPRC5A shRNA or scramble shRNA were transfected with
STAT3 luciferase and treated with EGF for 4 h. GPRC5A inhibition
enhanced STAT3 luciferase activity in the MDA-MB-231 cells
(Fig. 4C). Taken together, these
results indicate that GPRC5A may participate in the negative
regulation of EGFR in breast cancer cells.
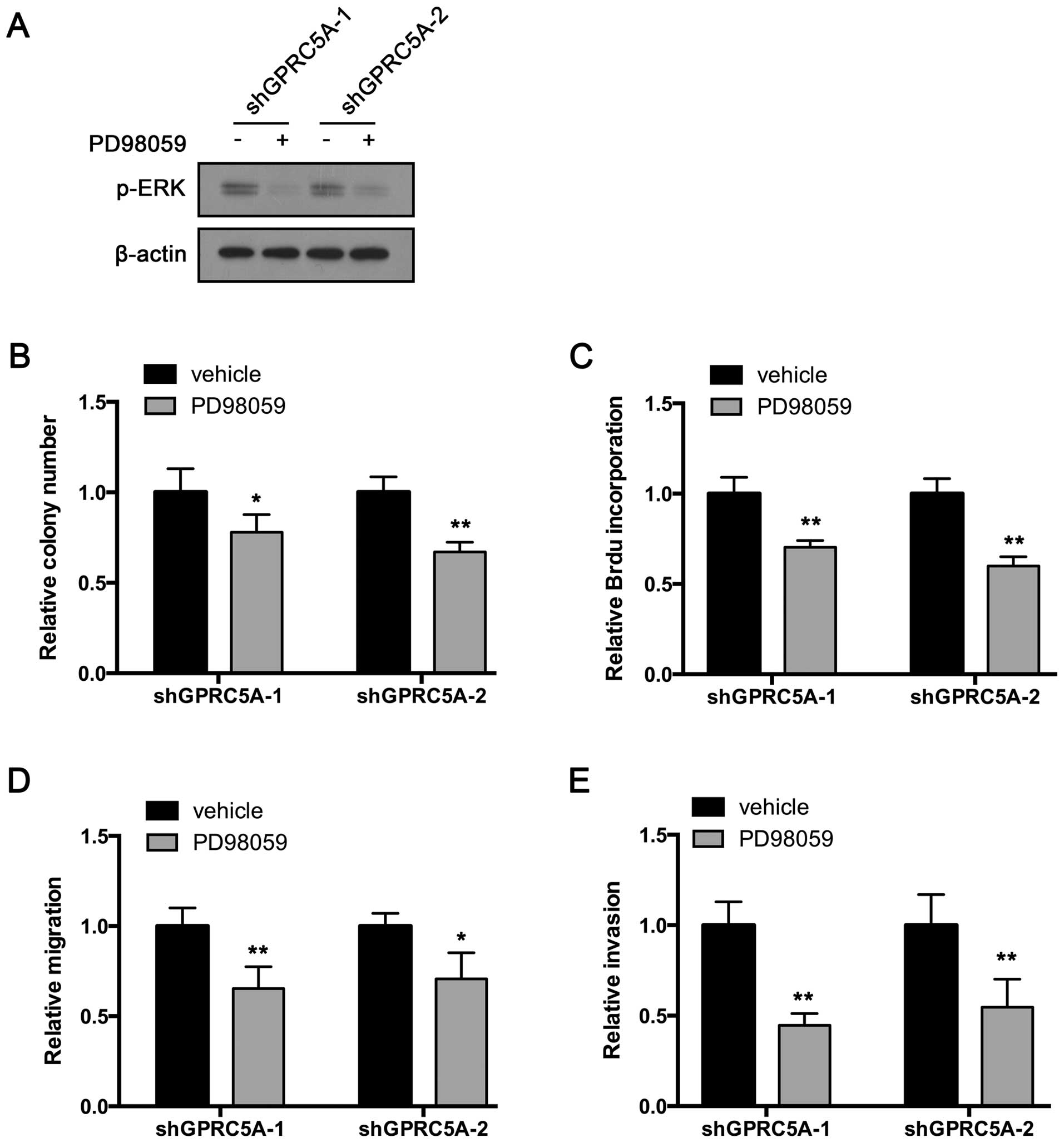
PD98059 inhibits the oncogenic effects of
GPRC5A knockdown in the MDA-MB-231 cells
ERK is the key downstream effector of EGFR. We aimed
to ascertain whether GPRC5A exerts its tumor-suppressive effects
through the EGFR-ERK pathway. Thus, we inhibited ERK activation by
treating the MDA-MB-231 cells with PD98059, an MEK inhibitor. The
inhibition was confirmed by western blot assay (Fig. 5A). Then, colony formation was
performed. As expected, PD98059 suppressed the colony formation of
the GPRC5A-knockdown cells (Fig.
5B). Furthermore, BrdU incorporation rate was also decreased
after PD98059 treatment (Fig. 5C).
Next, we examined whether PD98059 had an impact on the regulation
of migration and invasion by GPRC5A. Transwell migration assay
showed that PD98059 reversed GPRC5A knockdown-induced enhancement
of migration (Fig. 5D). In
addition, PD98059 also inhibited the enhanced invasive ability of
the GPRC5A-knockdown cells (Fig.
5E). Taken together, these results indicate that GPRC5A exerts
its tumor-suppressive effects by inhibiting, at least in part, the
EGFR-ERK pathway.
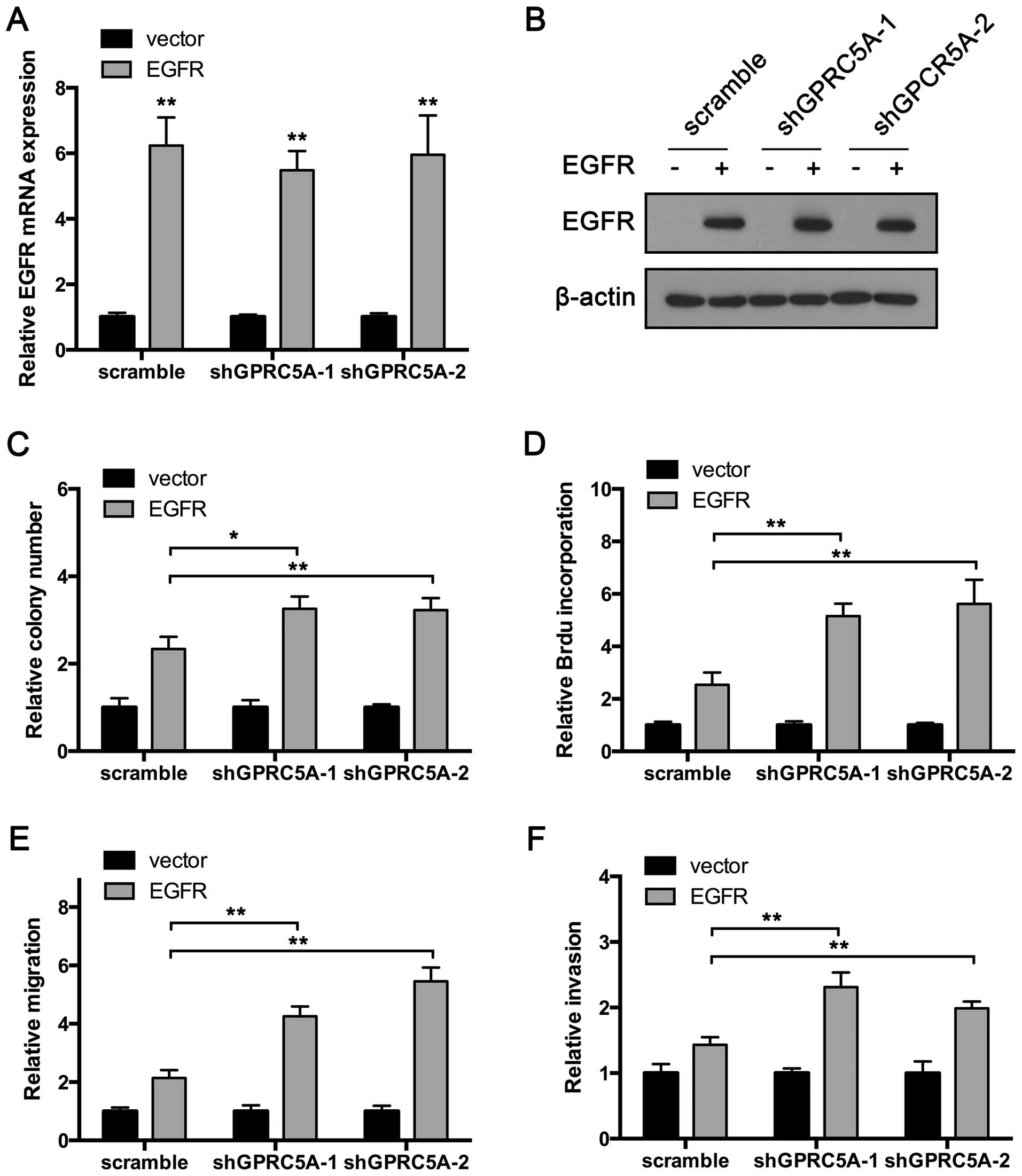
GPRC5A knockdown exhibits oncogenic
effects in the MCF7 cells transfected with EGFR
Previous results showed that GPRC5A inhibits EGFR
activation, and therefore suppresses breast cancer cell
proliferation, migration and invasion. To further confirm this
conclusion, we overexpressed EGFR in the MCF7 cells which do not
express EGFR. The overexpression was confirmed by RT-qPCR and
western blot assay (Fig. 6A and B).
Then, the colony formation assay was performed. As shown in
Fig. 6C, EGFR overexpression
increased the colonies produced by the MCF7 cells. Consistent with
the previous experiment, GPRC5A suppression in MCF7 cells
transfected with the vector plasmid produced the same number of
colonies compared with that noted in the control cells (Fig. 6C). Notably, GPRC5A knockdown
significantly increased the colonies produced by
EGFR-overexpressing MCF7 cells (Fig.
6C). Furthermore, EGFR overexpression enhanced BrdU
incorporation in the MCF7 cells. Consistent with the colony
formation assay, GPRC5A inhibition aggravated BrdU incorporation in
the MCF7 cells transfected with the EGFR-expressing plasmid, while
no impact on the MCF7 cells transfected with the vector plasmid was
observed (Fig. 6D).
In addition, we performed Transwell migration and
invasion assays. The results showed that EGFR overexpression
significantly promoted MCF7 cell migration and invasion. GPRC5A
inhibition suppressed the invasion and migration of the MCF7 cells
transfected with the EGFR-expressing plasmid (Fig. 6E and F). Taken together, these
results indicate that GPRC5A exerts its tumor-suppressive functions
by targeting EGFR.
Discussion
GPRC5A was initially discovered as a retinoic
acid-inducible gene strongly expressed in the lung (16,25,26).
Subsequent study provided evidence for a role of GPRC5A in lung
carcinogenesis. The level of GPRC5A mRNA is often lower in lung
tumors compared with normal tissues, and a somatic loss of GPRC5A
is associated with poor survival of lung cancer patients.
GPRC5A-knockout mice are prone to sporadic and carcinogen- or
inflammation-induced lung adenocarcinomas strongly suggesting its
role as a lung cancer tumor-suppressor gene. In contrast to lung
cancer, GPRC5A is frequently over-expressed in myelodysplastic
syndrome as well as in breast, colon and gastric cancers. This
indicates that GPRC5A may play a pro-survival role at least in some
circumstances.
In the present study, we knocked down GPRC5A
expression in the TNBC cell line, MDA-MB-231. The cells produced
more and larger colonies when compared with the control cells.
Consistent with this result, cell proliferation was also enhanced
after GPRC5A suppression. Furthermore, we observed enhanced in
vitro migration and invasion in the GPRC5A-downregulated cells.
These results implicate GPRC5A as a tumor-suppressor in MDA-MB-231
cells.
Signaling pathway analysis showed that GPRC5A
suppression significantly enhanced EGF-induced EGFR pathway
activation in the MDA-MB-231 cells. Inhibition of ERK activation
reversed the GPRC5A-induced oncogenic effects. However, the
oncogenic effects of GPRC5A knockdown were not reproducible in the
MCF7 cells. Notably, when the MCF7 cells were transfected with the
EGFR-expressing plasmid, GPRC5A suppression enhanced EGFR
activation and EGFR induced oncogenic effects. These results
indicate that GPRC5A is a downstream regulator of the EGFR pathway
in breast cancer.
TNBC refers to any breast cancer that does not
express the genes for estrogen receptor (ER), progesterone receptor
(PR) or Her2/neu. Thus, TNBC is more difficult to treat since most
chemotherapies target one of the three receptors. Consequently,
triple-negative cancers often require combinatorial therapies
(27). Exploring novel approaches
to treat this subtype is critical since <30% of women with
metastatic breast cancer survive 5 years and virtually all women
with metastatic TNBC ultimately die of their disease despite
systemic therapy (27). EGRF is
frequently overexpressed and has been proposed as a therapeutic
target for TNBC (2,5,8,27,28).
Our results provide a new regulatory mechanism of the EGFR pathway
in TNBC cells.
In summary, we found that GPRC5A functions as a
tumor suppressor in breast cancer cells. GPRC5A inhibits cell
proliferation, migration and invasion in vitro and exerts
its tumor-suppressive functions by inhibiting EGFR and its
downstream pathway. Furthermore, our results highlight the
potential of targeting EGFR and the ERK pathway and provide a new
therapeutic target for TNBC treatment.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2015. CA Cancer J Clin. 65:5–29. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Polyak K: Heterogeneity in breast cancer.
J Clin Invest. 121:3786–3788. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
Qin XJ and Ling BX: Proteomic studies in
breast cancer (Review). Oncol Lett. 3:735–743. 2012.PubMed/NCBI
|
|
4
|
Sun B, Zhang S, Zhang D, Li Y, Zhao X, Luo
Y and Guo Y: Identification of metastasis-related proteins and
their clinical relevance to triple-negative human breast cancer.
Clin Cancer Res. 14:7050–7059. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Teng YH, Tan WJ, Thike AA, Cheok PY, Tse
GM, Wong NS, Yip GW, Bay BH and Tan PH: Mutations in the epidermal
growth factor receptor (EGFR) gene in triple negative breast
cancer: Possible implications for targeted therapy. Breast Cancer
Res. 13:R352011. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ueno NT and Zhang D: Targeting EGFR in
triple negative breast cancer. J Cancer. 2:324–328. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Liu M, Mo QG, Wei CY, Qin QH, Huang Z and
He J: Platinum- based chemotherapy in triple-negative breast
cancer: A meta-analysis. Oncol Lett. 5:983–991. 2013.PubMed/NCBI
|
|
8
|
Yewale C, Baradia D, Vhora I, Patil S and
Misra A: Epidermal growth factor receptor targeting in cancer: A
review of trends and strategies. Biomaterials. 34:8690–8707. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Herbst RS: Review of epidermal growth
factor receptor biology. Int J Radiat Oncol Biol Phys. 59(Suppl 2):
21–26. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Tomas A, Futter CE and Eden ER: EGF
receptor trafficking: Consequences for signaling and cancer. Trends
Cell Biol. 24:26–34. 2014. View Article : Google Scholar :
|
|
11
|
Masuda H, Zhang D, Bartholomeusz C,
Doihara H, Hortobagyi GN and Ueno NT: Role of epidermal growth
factor receptor in breast cancer. Breast Cancer Res Treat.
136:331–345. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Bhargava R, Gerald WL, Li AR, Pan Q, Lal
P, Ladanyi M and Chen B: EGFR gene amplification in breast cancer:
Correlation with epidermal growth factor receptor mRNA and protein
expression and HER-2 status and absence of EGFR-activating
mutations. Mod Pathol. 18:1027–1033. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Masuda H, Zhang D, Bartholomeusz C,
Doihara H, Hortobagyi GN and Ueno NT: Role of epidermal growth
factor receptor in breast cancer. Breast Cancer Res Treat.
136:331–345. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Gluz O, Liedtke C, Gottschalk N, Pusztai
L, Nitz U and Harbeck N: Triple-negative breast cancer - current
status and future directions. Ann Oncol. 20:1913–1927. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Corkery B, Crown J, Clynes M and O'Donovan
N: Epidermal growth factor receptor as a potential therapeutic
target in triple-negative breast cancer. Ann Oncol. 20:862–867.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wu Q, Ding W, Mirza A, Van Arsdale T, Wei
I, Bishop WR, Basso A, McClanahan T, Luo L, Kirschmeier P, et al:
Integrative genomics revealed RAI3 is a cell growth-promoting gene
and a novel P53 transcriptional target. J Biol Chem.
280:12935–12943. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Tao Q, Fujimoto J, Men T, Ye X, Deng J,
Lacroix L, Clifford JL, Mao L, Van Pelt CS, Lee JJ, et al:
Identification of the retinoic acid-inducible Gprc5a as a new lung
tumor suppressor gene. J Natl Cancer Inst. 99:1668–1682. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kadara H, Fujimoto J, Men T, Ye X, Lotan
D, Lee JS and Lotan R: A Gprc5a tumor suppressor loss of expression
signature is conserved, prevalent, and associated with survival in
human lung adenocarcinomas. Neoplasia. 12:499–505. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lin X, Zhong S, Ye X, Liao Y, Yao F, Yang
X, Sun B, Zhang J, Li Q, Gao Y, et al: EGFR phosphorylates and
inhibits lung tumor suppressor GPRC5A in lung cancer. Mol Cancer.
13:2332014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhong S, Yin H, Liao Y, Yao F, Li Q, Zhang
J, Jiao H, Zhao Y, Xu D, Liu S, et al: Lung tumor suppressor GPRC5A
binds EGFR and restrains its effector signaling. Cancer Res.
75:1801–1814. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Sokolenko AP, Bulanova DR, Iyevleva AG,
Aleksakhina SN, Preobrazhenskaya EV, Ivantsov AO, Kuligina ES,
Mitiushkina NV, Suspitsin EN, Yanus GA, et al: High prevalence of
GPRC5A germline mutations in BRCA1-mutant breast cancer patients.
Int J Cancer. 134:2352–2358. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ma T, Yang L and Zhang J: MiRNA-542–3p
downregulation promotes trastuzumab resistance in breast cancer
cells via AKT activation. Oncol Rep. 33:1215–1220. 2015.PubMed/NCBI
|
|
23
|
Chaffer CL and Weinberg RA: A perspective
on cancer cell metastasis. Science. 331:1559–1564. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lamouille S, Xu J and Derynck R: Molecular
mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell
Biol. 15:178–196. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
25
|
Cheng Y and Lotan R: Molecular cloning and
characterization of a novel retinoic acid-inducible gene that
encodes a putative G protein-coupled receptor. J Biol Chem.
273:35008–35015. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Tao Q, Cheng Y, Clifford J and Lotan R:
Characterization of the murine orphan G-protein-coupled receptor
gene Rai3 and its regulation by retinoic acid. Genomics.
83:270–280. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Mayer IA, Abramson VG, Lehmann BD and
Pietenpol JA: New strategies for triple-negative breast
cancer--deciphering the heterogeneity. Clin Cancer Res. 20:782–790.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Taurin S, Allen KM, Scandlyn MJ and
Rosengren RJ: Raloxifene reduces triple-negative breast cancer
tumor growth and decreases EGFR expression. Int J Oncol.
43:785–792. 2013.PubMed/NCBI
|