Introduction
The incidence of thyroid cancer, one of the most
common endocrine malignancies, has increased rapidly in recent
years based on worldwide statistics (1). The annual number of newly diagnosed
thyroid cancer cases is 129/1,000,000, and the associated deaths
are 5/1,000,000 (2). Thyroid cancer
can be divided into well-differentiated carcinomas (such as
papillary or follicular carcinomas) and anaplastic thyroid cancer
(ATC) (3). The percentage of ATC is
low, ranging from 1 to 5% of all thyroid cancers, but accounts for
~14–50% of deaths (4). The poor
prognosis and the high rate of distant metastases of ATC lead to a
5-year survival rate of 7% (5).
Considering the complexity of the molecular
mechanisms, various studies have been carried out such as mRNA
(6) and miRNA expression profiling
(5) by microarray, gene mutation
whole exome sequencing (7),
cytogenetic analysis and comparative genomic hybridization (CGH)
microarray (8). Several de
novo mutations including TP53, β-catenin and
PIK3CA have been identified in ATC whereas in pre-existing
mutations in PTC (papillary thyroid cancer) mutations such as
RAS and BRAF have been found (4). mRNA and miRNA expression levels have
also been demonstrated to be critical in tumor progression. A study
by von Roemeling et al showed that stearoyl-CoA desaturase 1
(SCD1) associated with fatty acid metabolism is highly
expressed in ATC compared with that in normal samples (9). Gene expression analysis of ATC and PTC
demonstrated that most of the DEGs were common in both, but ATC
contained more genes associated with epithelial to mesenchymal
transition (EMT), dedifferentiation and glycolytic phenotypes
(6). Furthermore, miRNA expression
profile analysis of 11 ATC samples showed that 17 common miRNAs
were downregulated and one was upregulated (5).
Although previous research has been carried out for
ATC, the combination analysis of mRNA and miRNA expression profiles
has not yet been systematically explored. A substantial amount of
mRNA expression datasets have been submitted to the GEO (Gene
Expression Omnibus) database and bioinformatic methods have been
demonstrated to be valuable for molecular mechanism investigation
(10). In the present study, mRNA
expression datasets from different laboratories were analyzed and
several DEGs were identified by RT-PCR. Then common DEGs were
subjected to subsequent function analysis. Furthermore, miRNA and
mRNA expression levels were integrated for the elucidation of the
molecular mechanisms.
Materials and methods
Gene expression profiles
Two gene expression profiles GSE33630 and GSE65144
submitted by Dom et al (11)
and von Roemeling et al (9),
respectively, were downloaded from the Gene Expression Omnibus
(GEO, http://www.ncbi.nlm.nih.gov/geo/). For the GSE33630
dataset, 11 ATC samples and 11 paired normal samples were selected
for subsequent analysis. The GSE65144 dataset consisted of 12 ATC
samples and 13 normal samples (12 matched and 1 unmatched).
Experiments for the two datasets were carried out using GPL570
platform (HG-U133_Plus_2, Affymetrix Human Genome U133 Plus 2.0
array).
Data preprocessing and screening of
DEGs
The mRNA expression profiles were subjected to log2
transformation, background correction and normalization using the
GeneChip Robust Multi-array Analysis (GCRMA) method within the
Bioconductor package (http://www.bioconductor.org) (12). The uninformative probe sets such as
control probe sets, and genes with low expression variance were
filtered out. For genes with multiple probes, the average
expression value was calculated. Finally, the significant DEGs were
identified using the Limma (Linear Models for Microarray Analysis)
(13) package with criteria of
adjustment of p<0.01 and |log2 fold change (FC)| ≥2. DEGs with a
fold change >5 were used for downstream analysis. The heat map
was constructed using the heat map method in Bioconductor.
Quantitative RT-PCR analysis
The mRNA expression levels of three randomly
selected DEGs, VCAN, COL5A1 and KCNJ16, were
examined using RT-PCR. The total RNA was extracted from the 10 ATC
samples and the adjacent normal tissues using TRIzol reagent
(Thermo Fisher Scientific, Inc., USA) according to the
manufacturer's instructions. cDNA was obtained using M-MLV reverse
transcriptase based on the manufacturer's protocol (Promega,
Madison, WI, USA). The VCAN, COL5A1 and KCNJ16
mRNA expression levels were detected using 7500 Real-Time PCR
system (Thermo Fisher Scientific). Relative quantification was
normalized using GAPDH mRNA expression and calculated with
the 2−ΔCt method. The primer sequence list is as
follows: VCAN forward, 5′-CACAACCCGCATTTGAACTTG-3′ and
reverse, 5′-CGCACGCCTGGAGTTCTT-3′; COL5A1 forward,
5′-ACAACTTGCCTGATGGAA TAACAA-3′ and reverse,
5′-CCGGGCCTTTGGAAGATC-3′; KCNJ16 forward,
5′-TCAATGCGGACGCAAAATAC-3′ and reverse,
5′-AATCGTCTTCTTGCTCTTCTCTTCTC-3′; and GAPDH forward,
5′-TGACTTCAACAGCGACACCCA-3′ and reverse,
5′-CACCCTGTTGCTGTAGCCAAA-3′.
Functional and pathway enrichment
analysis
In order to explore the biological processes
involved in ATC, functional and pathway enrichment analyses for the
common DEGs were carried out using Database for Annotation,
Visualization and Integrated Discovery (DAVID) online tools
(14), which is based on the Gene
Ontology (GO) (15) and Kyoto
Encyclopedia of Genes and Genomes (KEGG) (16) databases. The criterion for
significantly enriched pathways was set as p≤0.05. GO terms,
consisting of biological processes (BP), cellular components (CC)
and molecular functions (MF), were screened with a p≤0.05.
Gene interaction and miRNA regulation
network analysis
The physical interaction and validated pathway
interaction network for the common DEGs was constructed using the
Gene Multiple Association Network Integration Algorithm (GeneMANIA,
http://www.genemania.org/) (17). Moreover, 17 deregulated miRNAs from
the study of Hebrant et al (5) were integrated for the exploration of
miRNA-mRNA interaction by using the CyTargetLinker plugin of
Cytoscape (18).
Results
Common DEGs
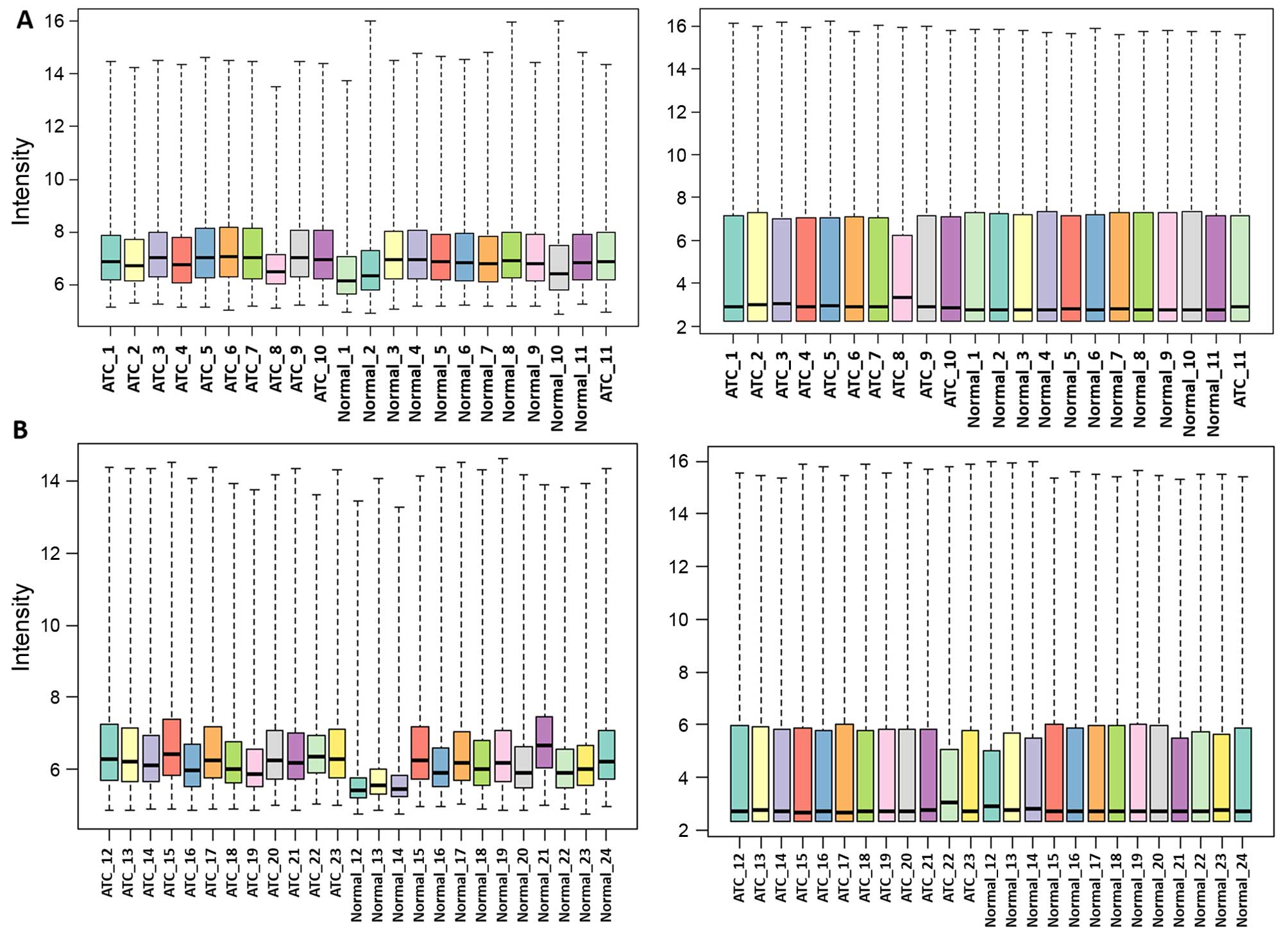
After background correction and normalization, the
medians of the gene expression values were almost at the same level
indicating that the data were suitable for subsequent analysis
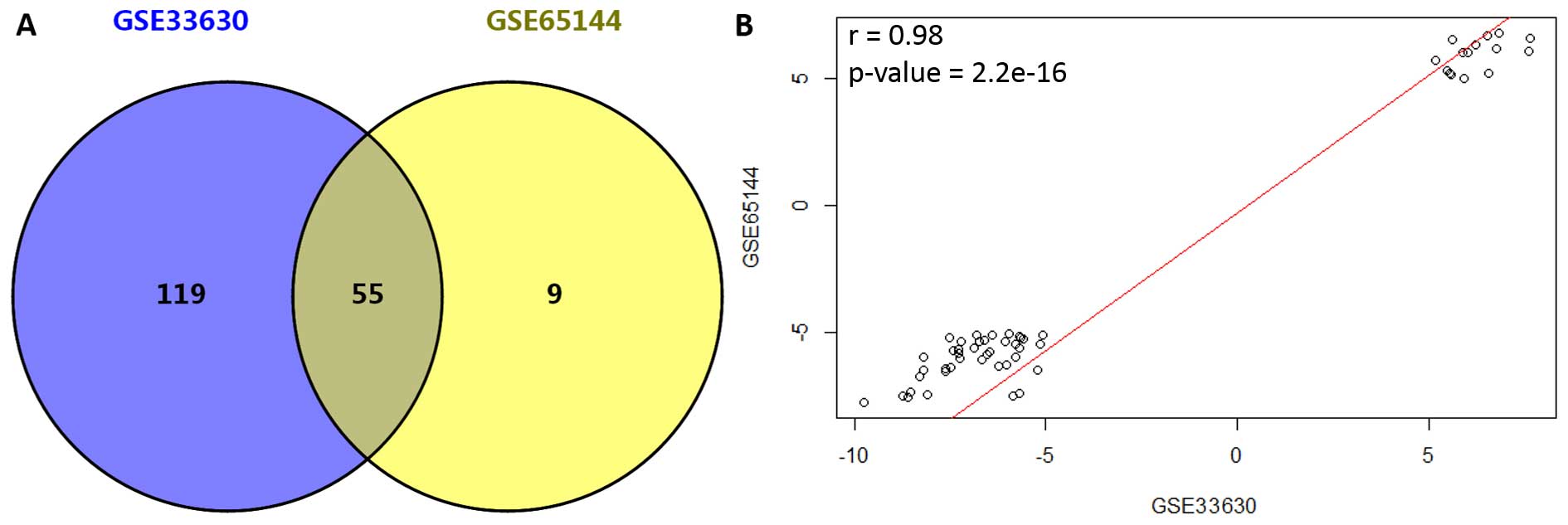
(Fig. 1). DEGs for the two datasets
were identified independently, and a total of 174 and 64 DEGs were
screened out for GSE33630 and GSE65144, respectively. Moreover, 55
DEGs (accounting for 32 and 85% for GSE33630 and GSE65144,
respectively) were found to be simultaneously differentially
expressed in these two datasets (Fig.
2A). Among the common DEGs, 15 genes were upregulated and 40
were downregulated (data not shown). In addition, correlation of
expression values for the 55 common DEGs was 0.98 (p<2.2e−16,
Fig. 2B).
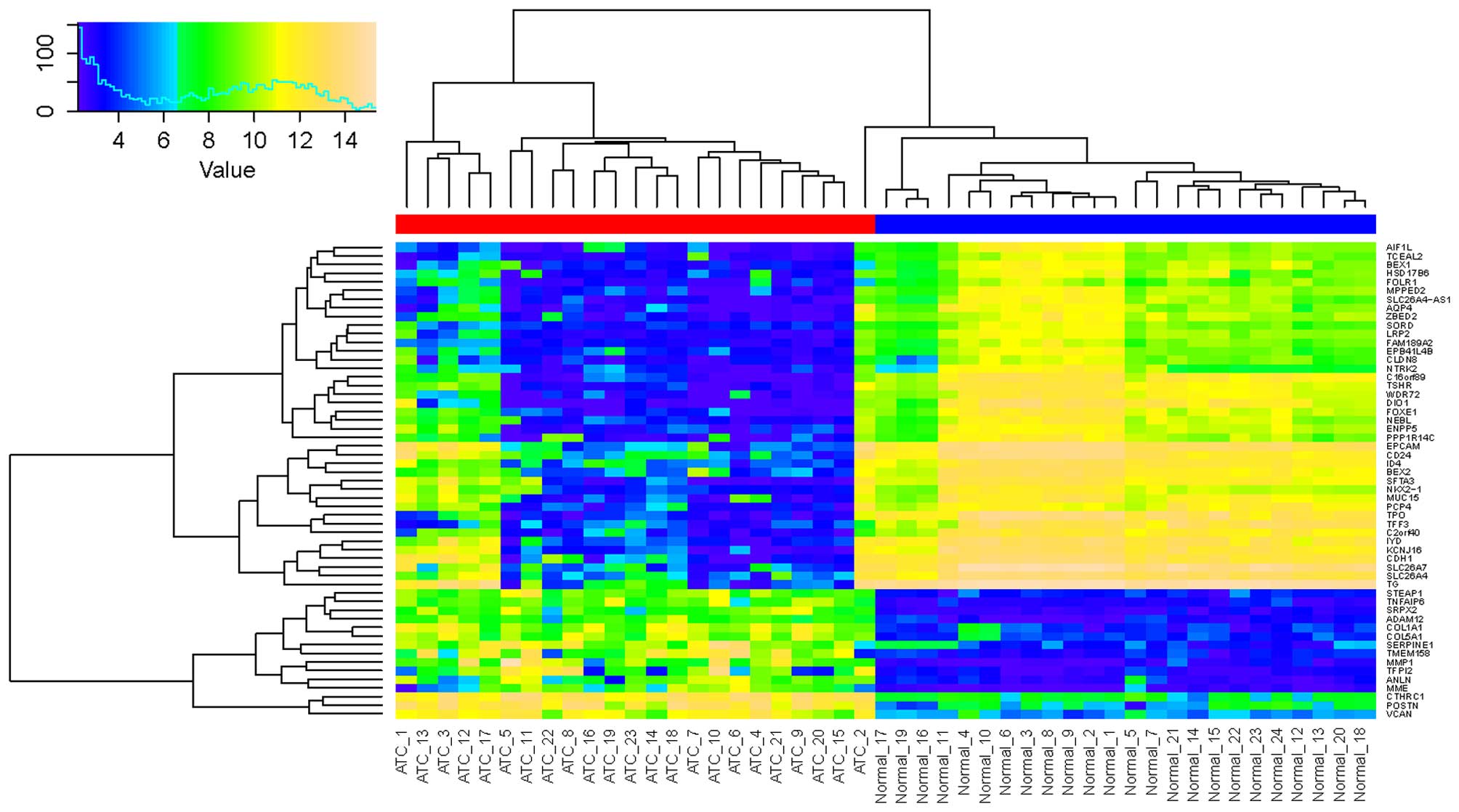
Furthermore, the 55 common DEGs were used for the
classification of ATC and normal samples. As indicated in Fig. 3, the ATC and normal samples were
clearly classified into two groups except for sample ATC_2, which
was possibly due to the smaller expression value variation, ranging
from 8–12.
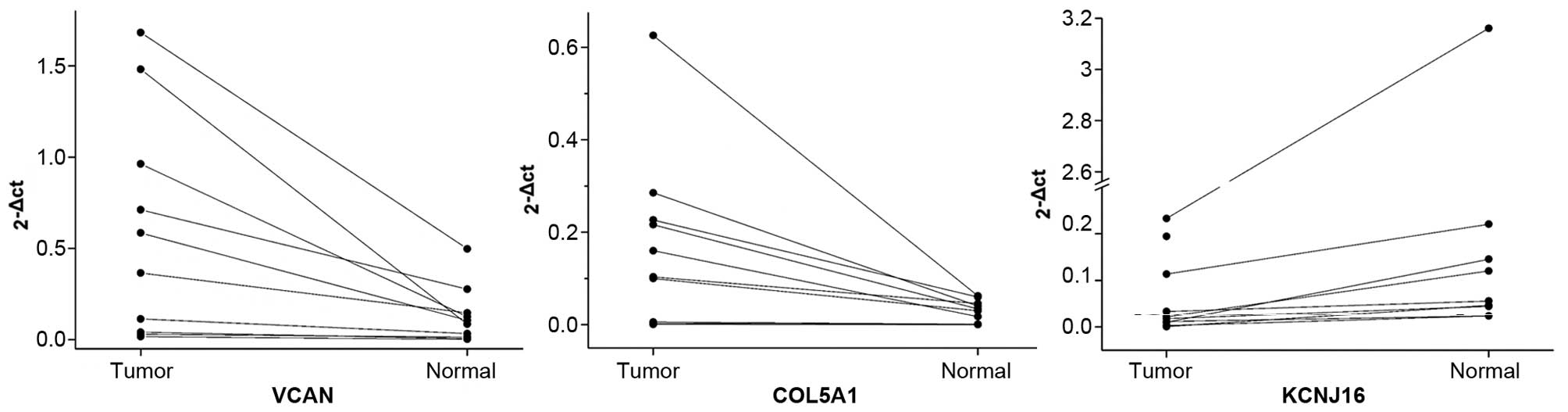
mRNA expression validation by RT-PCR
To verify mRNA expression levels of the identified
DEGs, RT-PCR experiments were carried out for three randomly
selected genes including VCAN, COL5A1 and
KCNJ16. The mRNA expression levels in the 10 ATC samples and
adjacent normal tissues were analyzed. Results showed that the
expression levels of VCAN and COL5A1 were higher in
the tumor tissues than the levels in the adjacent normal tissues;
whereas, the mRNA expression level of KCNJ16 was lower in
the tumor tissues (Table I and
Fig. 4). These results were nearly
consistent with those in the microarray analysis.
 | Table IRelative expression values of
VCAN, COL5A1 and KCNJ16 in 10 paired tumor and
adjacent normal tissues. |
Table I
Relative expression values of
VCAN, COL5A1 and KCNJ16 in 10 paired tumor and
adjacent normal tissues.
| Sample | VCAN
(2−ΔCt)
| COL5A1
(2−ΔCt)
| KCNJ16
(2−ΔCt)
|
|---|
| Tumor | Normal | Tumor | Normal | Tumor | Normal |
|---|
| 1 | 0.3647021 | 0.1466735 | 0.1601689 | 0.0177641 | 0.0101683 | 0.145485 |
| 2 | 0.0286243 | 0.0132945 | 0.216439 | 0.0340146 | 0.0031314 | 0.0238292 |
| 3 | 0.0162446 | 0.0018798 | 0.0059821 | 0.0012272 | 0.0218826 | 0.1202726 |
| 4 | 0.039647 | 0.0060125 | 0.2266865 | 0.0599975 | 0.1942628 | 0.3765939 |
| 5 | 0.5845333 | 0.1055037 | 0.2854041 | 0.0404317 | 0.0180416 | 0.0442845 |
| 6 | 1.6831185 | 0.4974194 | 0.6257863 | 0.0625064 | 0.2327167 | 3.1607958 |
| 7 | 0.962266 | 0.1256899 | 0.0009169 | 0.0003568 | 0.1135657 | 0.2204115 |
| 8 | 1.4817526 | 0.0848966 | 0.003156 | 0.0010707 | 0.0332327 | 0.0556471 |
| 9 | 0.1130402 | 0.0329317 | 0.1000809 | 0.0302228 | 0.0117418 | 0.0237591 |
| 10 | 0.7113506 | 0.2759241 | 0.1035169 | 0.0460322 | 0.0009712 | 0.0465934 |
| Mean | 0.5985279 | 0.1290226 | 0.1728138 | 0.0293624 | 0.0639715 | 0.4217672 |
| SD | 0.6117568 | 0.1542157 | 0.1882915 | 0.0236354 | 0.085654 | 0.9687421 |
Functional and pathway enrichment
analysis
In order to explore the functions of these common
DEGs, functional and pathway enrichment were carried out. The
results indicated that the two pathways were significantly enriched
for the common DEGs (Table II) and
that TG, TPO and TSHR participated in the
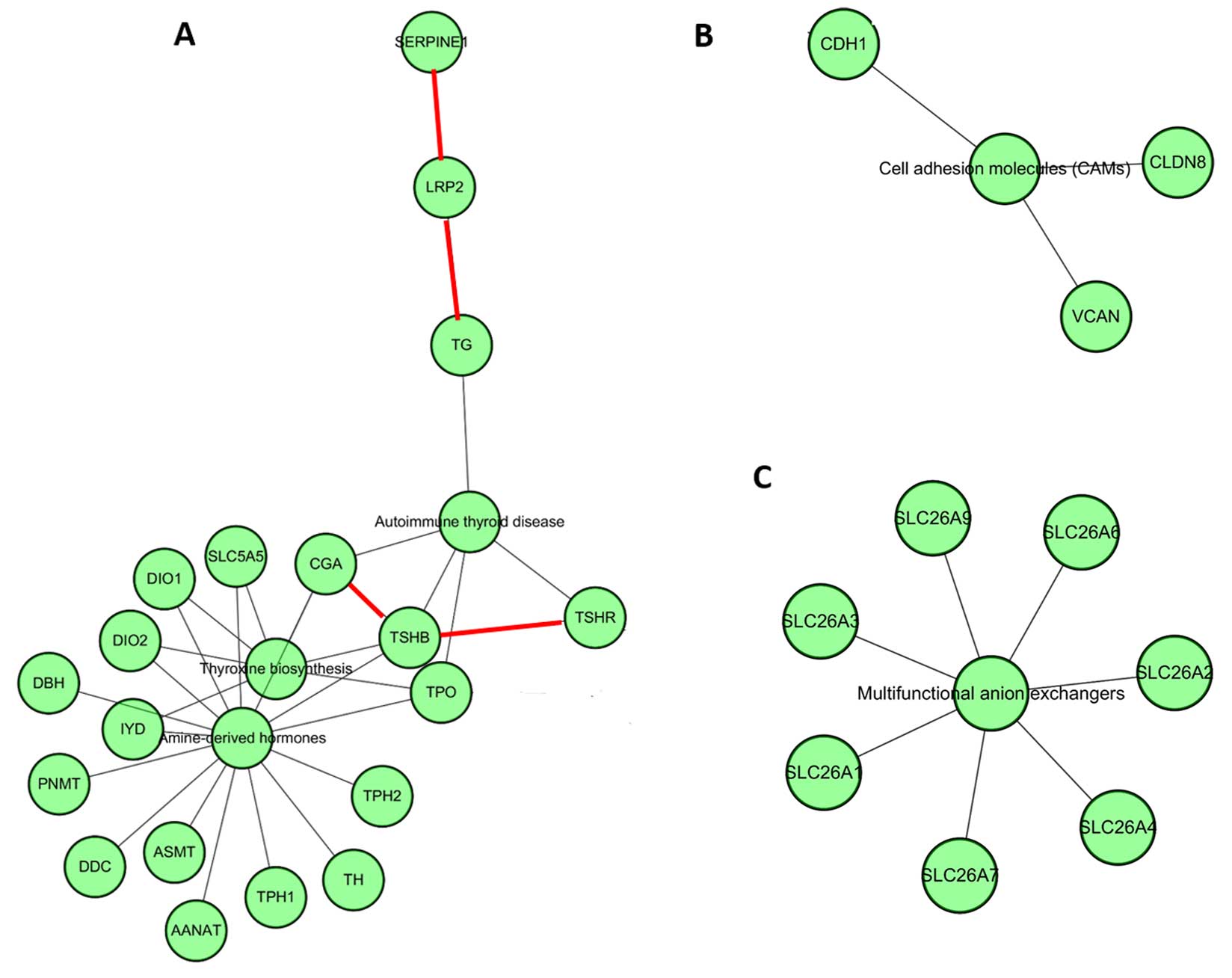
autoimmune thyroid disease pathway (p=0.0083), and CLDN8,
CDH1 and VCAN were significantly involved in the cell
adhesion molecule (CAM) pathway (p=0.049).
 | Table IIThe enriched pathways for the common
differentially expressed genes. |
Table II
The enriched pathways for the common
differentially expressed genes.
| Pathway ID | Pathway name | Count | Genes | P-value |
|---|
| hsa05320 | Autoimmune thyroid
disease | 3 | TG,
TPO, TSHR | 0.0083 |
| hsa04514 | Cell adhesion
molecules (CAMs) | 3 | CLDN8,
CDH1, VCAN | 0.049 |
Futhermore, the common DEGs were mainly related to
the cellular components of the extracellular region (p=2.5E−05) and
the proteinaceous extracellular matrix (p=7.3E−05). Some common
DEGs, such as TNFAIP6, VCAN, POSTN and
COL5A1, were significantly enriched in the carbohydrate
binding activity (p=0.002) and glycosaminoglycan binding function
(p=0.006). In addition, hormone regulation-related processes such
as regulation of hormone levels (p=4.6E−05), hormone biosynthetic
processes (p=7.3E−05) and thyroid hormone generation (p=1.0E−04)
biological processes were also enriched.
Gene interaction and miRNA regulation
analysis
Furthermore, the interaction between these common
DEGs was explored to elucidate the potential regulatory mechanism.
The results indicated that several important subnetworks were
formed based on different pathways. Fig. 5A shows that LRP2 can interact
with SERPINE and TG physically and TSHR can
interact with TSHB physically. Also these common DEGs
participated in the autoimmune thyroid disease pathway similar to
our results from the pathway enrichment analysis. In addition,
CDH1, CLDN8 and VCAN were found to
independently participate in cell adhesion molecules. The
SLC26A gene family was significantly involved in the
multifunctional anion exchanger pathway (Fig. 5B).
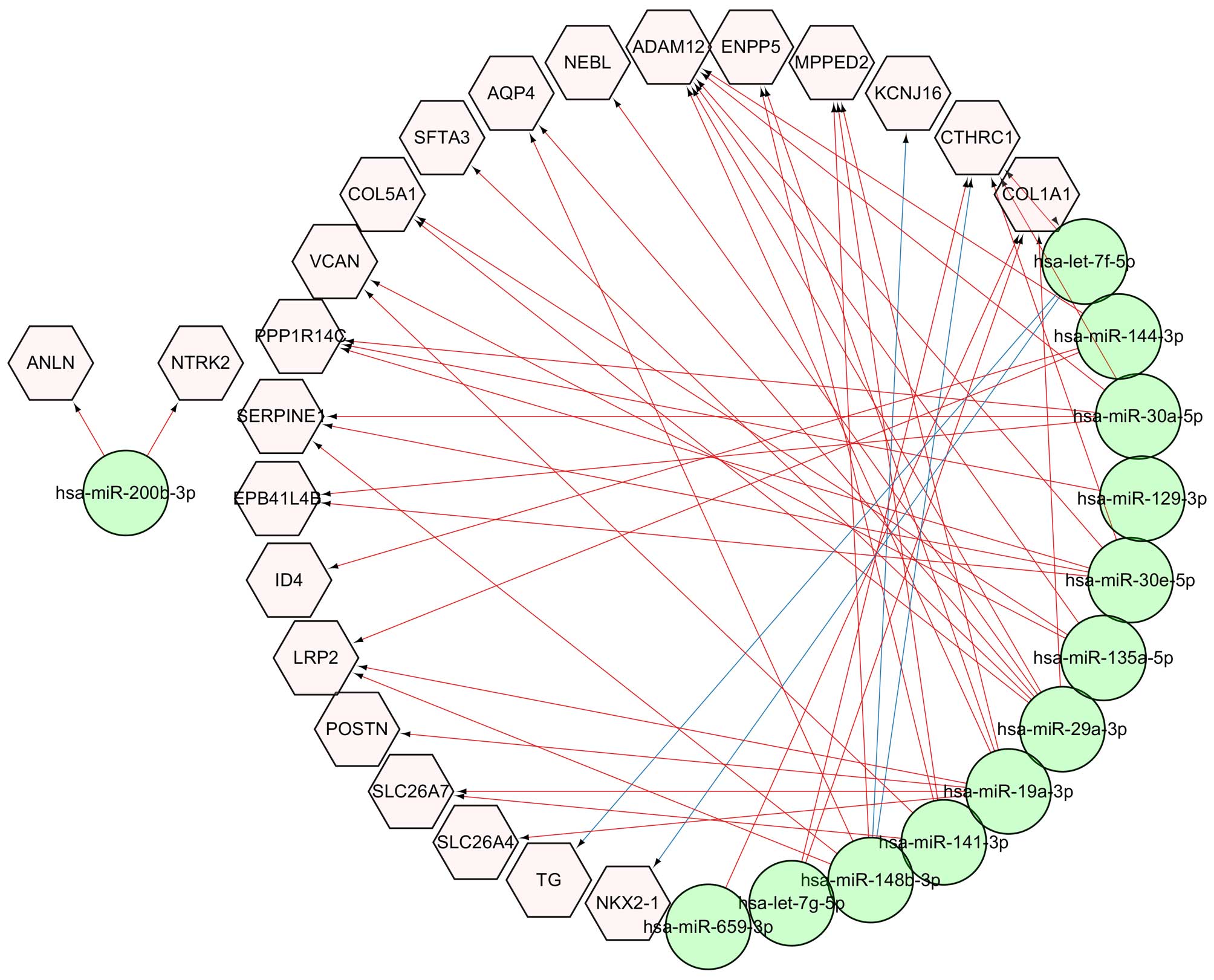
For the discovery of miRNAs that can regulate the
expression of these common DEGs, the potential targets of 17
reported miRNAs (5) in ATC were
predicted. The results showed that 870 and 13,841 target genes were
found in the miRTarBase (version 4.4) (19) and TargetScan (version 6.2)
(http://www.targetscan.org/) databases,
respectively. Among these target genes, 23 genes were identified
among the 55 common DEGs, and the miRNA and mRNA interaction
network between them is shown in Fig.
6. The network indicated that hsa-let-7f-5p miRNA regulates
TG and NKX2-1, and hsa-miR-148b-3p miRNA regulates
KCNJ16 and CTHRC1 which was validated by our
experiments. Other interactions were predicted based on the
TargetScan database.
Discussion
ATC is a type of thyroid cancer with poor prognosis
and has been widely studied in mRNA expression, miRNA expression or
genome mutational landscape levels. However, efforts trying to
elucidate the molecular mechanisms of ATC with the combination of
different omics data are rare. In this study, mRNA and miRNA
expression levels were combined to explore the potential molecular
mechanisms. The mRNA expression analysis showed that 55 common DEGs
were simultaneously differentially expressed in the tumor samples
of the GSE33630 and GSE65144 datasets. Based on the 15 upregulated
and 40 downregulated genes, the ATC and normal samples were clearly
classified into two groups. The error assignment of ATC_2 was
possibly due to sample quality or tumor heterogeneity. RT-PCR
analysis established that VCAN and COL5A1 were
significantly expressed in 10 tumor tissues, compared with that in
adjacent normal tissues and that the mRNA expression level of
KCNJ16 was lower in the 10 tumor tissues.
Notably, the pathway enrichment analysis revealed
that three downregulated DEGs including TG, TPO and
TSHR were significantly involved in the autoimmune thyroid
disease pathway. Although the relationship between thyroid cancer
and autoimmune thyroid disease is unclear, the co-existance of
these two clinical afflictions has been demonstrated by
retrospective cohort analysis. One study showed that thyroid cancer
is significantly associated with an elevated concentration of TgAb
(thyroglobulin antibodies, OR=1.57; CI=1.11–2.23) based on the
study of 253 patients with thyroid cancer (20). Moreover, one review systematically
summarized the reports concerning the link between thyroid
autoimmunity and differentiated thyroid cancer (DTC) (2). The low expression of TG,
TPO and TSHR in this study was possibly caused by the
lower expression of NKX2-1 which can bind to thyroglobulin
promoter and regulate thyroid functional gene expression (21). Research has shown that promoter
hypermethylation of TSHR is significantly related to
TSHR gene silencing (22).
With relatively rich CpG dinucleotides, TSHR can be methylated
(23) and suppress thyroid
iodide-metabolizing molecules. Hence thyroid tumor cells are unable
to concentrate iodine and are insensitive to radioiodine (24). These downregulated genes are
possibly the results of thyroid cancer (25,26).
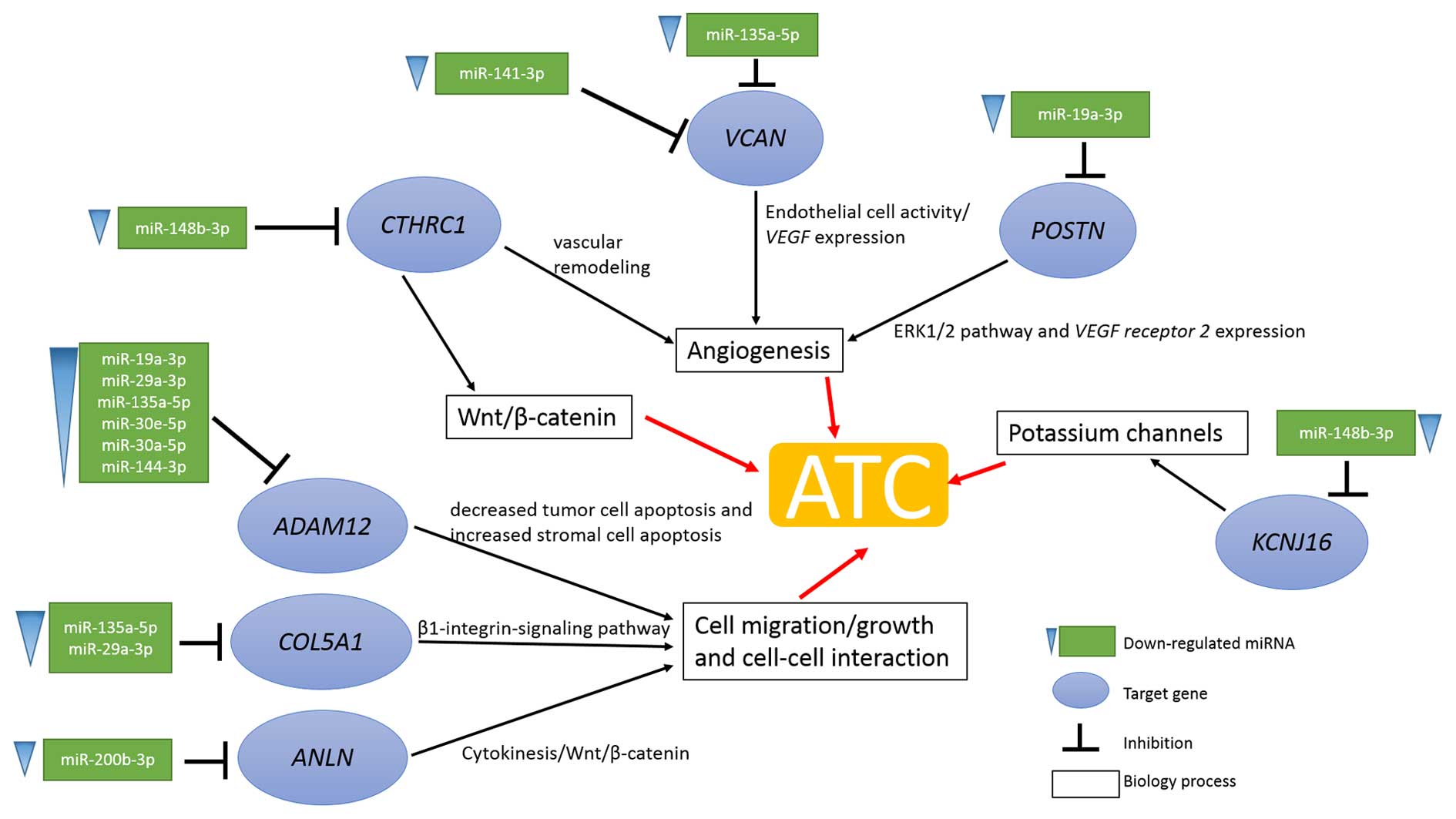
Considering the complexity of cancer, the molecular
mechanisms of ATC are far from clear. Based on the integration
analysis of mRNA and miRNA expression (5), the unknown mechanisms of ATC were
further revealed. Integrative analysis results showed that the
progression of ATC is considered to be a multipath process
including the angiogenesis process, the Wnt/β-catenin pathway, cell
migration or cell-cell interaction and potassium channel function
(Fig. 7).
Research has shown that angiogenesis is critical for
cancer cell proliferation and metastatic spread due to the
requirement of tumors for adequate oxygen and nutrient supply
(27). In the present study, the
angiogenesis process was activated by the upregulated expression of
CTHRC1, VCAN and POSTN and based on the miRNA
regulation analysis results. The upregulated expression of
CTHRC1, VCAN and POSTN was the consequence of
the downregulated expression of miR-148b-3p, miR-141-3p,
miR-135a-5p and miR-19a-3p, respectively (Fig. 7). Additionally, somatic mutation of
VCAN (D748G, missense) is possibly related to its functional
upregulation (7). CTHRC1, as
a novel oncogene, has been proved to be abnormally overexpressed in
malignant tumors such as melanoma, breast, pancreas, human
non-small cell lung and thyroid cancers (28–32).
Overexpression of CTHRC1 may contribute to vascular
remodeling and cell migration by suppressing collage matrix
deposition (33). Moreover, reports
have shown that CTHRC1 anchoring on the cell membrane may
stabilize the physical interaction between frizzled receptors and
Wnt ligands, and activate the non-canonical Wnt pathway regulating
cell motility (34). Moreover,
VCAN, as a member of the versican proteoglycan family, is
also valuable for angiogenesis. Yang and Yee reported that
VCAN-transfected tumor cells, exhibited enriched
vascularization and accumulation of red blood cells by H&E
staining compared with vector-transfected cells (35). In addition, the endothelial marker
of blood vessel formation CD34 was significantly overexpressed in
tumor sections (35).
Immunocytochemistry analysis also showed that
VCAN-transfected cells contained more and larger blood
vessels than the control cells (36). These angiogenesis-related processes
participated in the enhancement of endothelial cell activities and
fibronectin and vascular endothelial growth factor (VEGF)
expression (36). Furthermore,
POSTN, an adhesion molecule in osteoblasts, was identified
to be overexpressed and be related to the angiogenesis process.
Research has been carried out to explore the relationship between
POSTN and angiogenesis. In keloids, overexpression of
POSTN was found to promote angiogenesis by inducing ERK1/2
and focal adhesion kinase pathways and by upregulating expression
of VEGF and angiopoietin-1 (37). Additionally, the upregulation of
VEGF receptor 2 was identified in human breast cancer with
acquired POSTN expression (38), and POSTN promoted
angiogenesis via the paracrine pathway by interacting in a
αVβ3- and
αVβ5-dependent process in ovarian cancer
(39). All in all, these three
overexpressed genes possibly enhance ATC metastasis and progression
via the angiogenesis process.
Moreover, cell migration, cell growth and cell-cell
interactions are also critical in the development or metastasis of
tumors. In the present study, ADAM12, COL5A1 and
ANLN were identified to be overexpressed and their
upregulation was possibly caused by the low expression of
corresponding miRNAs such as miR-19a-3p, miR-29a-3p, miR-135a-5p,
miR-30e-5p, miR-30a-5p, miR-144-3p, miR-135a-5p, miR-29a-3p and
miR-200b-3p. Additionally somatic mutations of ANLN (R1095W,
missense) and COL5A1 (G1348A, missense) were found to
contribute to its upregulation (40). ADAM12, as one of the
disintegrins and metalloproteases, has been demonstrated to be
involved in several pathological processes. Microarray experiments
indicate that ADAM12 is upregulated in aggressive
fibromatosis. The mechanism of ADAM12 in breast tumor has
been systematically investigated and the results showed that
ADAM12 reduced tumor cell apoptosis and simultaneously
increased stromal cell apoptosis (41). In serous ovarian carcinoma, high
levels of ADAM12 mRNA were detected possibly caused by
TGFβ signaling (42).
Moreover, COL5A1 encoding an α-chain of fibrillar collagens
also regulated cell migration and motility (43,44).
COL5A1 binds to α2β1-integrin or β1-integrin receptor and
activates the corresponding signaling pathways. In pancreatic
ductal adenocarcinoma, high expression of COL5A1
significantly affected cell adhesion, migration and viability based
on results from β1-integrin inhibition, siRNA ablation of
COL5A1 expression and COL5A1 knockdown experiments
(45). In regards to colorectal
carcinogenesis, RT-PCR results showed that COL5A1 was
co-expressed with COL11A1 in tumor samples rather than in
normal samples (46). In addition,
the actin-binding protein ANLN is also critical for cell
growth, migration and cytokinesis (47). Zhou et al showed that
knockdown of ANLN markedly inhibited breast cancer cell line
proliferation and colony formation, and more cells were blocked at
the G2/M phase (47).
Alhough a detailed mechanism of ANLN in carcinoma
progression is still unclear, in silico pathway prediction
indicated that the Wnt/β-catenin signaling pathway is associated
with ANLN downstream regulation (48).
Moreover, ion channels especially potassium channels
have been demonstrated to play a crucial role in tumors (49). In the present study, KCNJ16
was downregulated which led to an ion concentration unbalance
between the extracellular and intracellular compartments. Rather
than a single mechanism, potassium channel regulation may influence
tumor progression via multiple paths such as cell adhesion or
migration, angiogenesis and apoptosis (49). The complex regulation mechanism of
KCNJ16 in ATC progression still remains to be explored.
In summary, the poor prognosis of ATC is possibly
induced by various processes. Firstly, upregulation of
CTHRC1, VCAN and POSTN promotes angiogenesis
and provides necessary nutrition for tumor cells. Then
ADAM12, COL5A1 and ANLN induce cell migration,
cell growth or cell-cell interaction leading to tumor distant
metastasis. Finally, KCNJ16 regulates intracellular and
extracellular ion concentrations and promotes ATC progression.
Acknowledgments
This study was funded by the National Natural
Science Foundation of Shanghai (12ZR1438700).
References
|
1
|
Nguyen QT, Lee EJ, Huang MG, Park YI,
Khullar A and Plodkowski RA: Diagnosis and treatment of patients
with thyroid cancer. Am Health Drug Benefits. 8:30–40.
2015.PubMed/NCBI
|
|
2
|
Feldt-Rasmussen U and Rasmussen AK:
Autoimmunity in differentiated thyroid cancer: Significance and
related clinical problems. Hormones (Athens). 9:109–117. 2010.
View Article : Google Scholar
|
|
3
|
Carling T and Udelsman R: Thyroid cancer.
Annu Rev Med. 65:125–137. 2014. View Article : Google Scholar
|
|
4
|
Smallridge RC, Marlow LA and Copland JA:
Anaplastic thyroid cancer: Molecular pathogenesis and emerging
therapies. Endocr Relat Cancer. 16:17–44. 2009. View Article : Google Scholar
|
|
5
|
Hébrant A, Floor S, Saiselet M, Antoniou
A, Desbuleux A, Snyers B, La C, de Saint Aubain N, Leteurtre E,
Andry G, et al: miRNA expression in anaplastic thyroid carcinomas.
PLoS One. 9:e1038712014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hébrant A, Dom G, Dewaele M, Andry G,
Trésallet C, Leteurtre E, Dumont JE and Maenhaut C: mRNA expression
in papillary and anaplastic thyroid carcinoma: Molecular anatomy of
a killing switch. PLoS One. 7:e378072012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kunstman JW, Juhlin CC, Goh G, Brown TC,
Stenman A, Healy JM, Rubinstein JC, Choi M, Kiss N, Nelson-Williams
C, et al: Characterization of the mutational landscape of
anaplastic thyroid cancer via whole-exome sequencing. Hum Mol
Genet. 24:2318–2329. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wilkens L, Benten D, Tchinda J, Brabant G,
Pötter E, Dralle H and von Wasielewski R: Aberrations of
chromosomes 5 and 8 as recurrent cytogenetic events in anaplastic
carcinoma of the thyroid as detected by fluorescence in situ
hybridisation and comparative genomic hybridisation. Virchows Arch.
436:312–318. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
von Roemeling CA, Marlow LA, Pinkerton AB,
Crist A, Miller J, Tun HW, Smallridge RC and Copland JA: Aberrant
lipid metabolism in anaplastic thyroid carcinoma reveals stearoyl
CoA desaturase 1 as a novel therapeutic target. J Clin Endocrinol
Metab. 100:E697–E709. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Berger B, Peng J and Singh M:
Computational solutions for omics data. Nat Rev Genet. 14:333–346.
2013. View
Article : Google Scholar : PubMed/NCBI
|
|
11
|
Dom G, Tarabichi M, Unger K, Thomas G,
Oczko-Wojciechowska M, Bogdanova T, Jarzab B, Dumont JE, Detours V
and Maenhaut C: A gene expression signature distinguishes normal
tissues of sporadic and radiation-induced papillary thyroid
carcinomas. Br J Cancer. 107:994–1000. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Gentleman RC, Carey VJ, Bates DM, Bolstad
B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al:
Bioconductor: Open software development for computational biology
and bioinformatics. Genome Biol. 5:R802004. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kerr MK: Linear models for microarray data
analysis: Hidden similarities and differences. J Comput Biol.
10:891–901. 2003. View Article : Google Scholar
|
|
14
|
Dennis G Jr, Sherman BT, Hosack DA, Yang
J, Gao W, Lane HC and Lempicki RA: DAVID: Database for Annotation,
Visualization, and Integrated Discovery. Genome Biol. 4:32003.
View Article : Google Scholar
|
|
15
|
Harris MA, Clark J, Ireland A, Lomax J,
Ashburner M, Foulger R, Eilbeck K, Lewis S, Marshall B, Mungall C,
et al: Gene Ontology Consortium: The Gene Ontology (GO) database
and informatics resource. Nucleic Acids Res. 32:D258–D261. 2004.
View Article : Google Scholar
|
|
16
|
Kanehisa M and Goto S: KEGG: Kyoto
encyclopedia of genes and genomes. Nucleic Acids Res. 28:27–30.
2000. View Article : Google Scholar
|
|
17
|
Warde-Farley D, Donaldson SL, Comes O,
Zuberi K, Badrawi R, Chao P, Franz M, Grouios C, Kazi F, Lopes CT,
et al: The GeneMANIA prediction server: Biological network
integration for gene prioritization and predicting gene function.
Nucleic Acids Res. 38:W214–W220. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kutmon M, Kelder T, Mandaviya P, Evelo CT
and Coort SL: CyTargetLinker: A cytoscape app to integrate
regulatory interactions in network analysis. PLoS One.
8:e821602013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hsu SD, Lin FM, Wu WY, Liang C, Huang WC,
Chan WL, Tsai WT, Chen GZ, Lee CJ, Chiu CM, et al: miRTarBase: A
database curates experimentally validated microRNA-target
interactions. Nucleic Acids Res. 39:D163–D169. 2011. View Article : Google Scholar
|
|
20
|
Azizi G and Malchoff CD: Autoimmune
thyroid disease: A risk factor for thyroid cancer. Endocr Pract.
17:201–209. 2011. View Article : Google Scholar
|
|
21
|
Ma R, Latif R and Davies TF: Thyroid
follicle formation and thyroglobulin expression in multipotent
endodermal stem cells. Thyroid. 23:385–391. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Khan MS, Pandith AA, Masoodi SR, Wani KA,
Ul Hussain M and Mudassar S: Epigenetic silencing of TSHR gene in
thyroid cancer patients in relation to their BRAF V600E mutation
status. Endocrine. 47:449–455. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Xing M, Usadel H, Cohen Y, Tokumaru Y, Guo
Z, Westra WB, Tong BC, Tallini G, Udelsman R, Califano JA, et al:
Methylation of the thyroid-stimulating hormone receptor gene in
epithelial thyroid tumors: A marker of malignancy and a cause of
gene silencing. Cancer Res. 63:2316–2321. 2003.PubMed/NCBI
|
|
24
|
Smith JA, Fan CY, Zou C, Bodenner D and
Kokoska MS: Methylation status of genes in papillary thyroid
carcinoma. Arch Otolaryngol Head Neck Surg. 133:1006–1011. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kowalska A, Pałyga I, Gąsior-Perczak D,
Walczyk A, Trybek T, Słuszniak A, Mężyk R and Góźdź S: The cut-off
level of recombinant human TSH-stimulated thyroglobulin in the
follow-up of patients with differentiated thyroid cancer. PLoS One.
10:e01338522015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Teama SH, Agwa SH, Fawzy A, Sayed MM,
Ibrahim WA and Eid YM: Molecular detection of circulating thyroid
specific transcripts (TSHR/Tg-mRNAs) in thyroid cancer patients:
Their diagnostic significance. Egypt J Med Hum Genet. 12:201–209.
2011. View Article : Google Scholar
|
|
27
|
Nishida N, Yano H, Nishida T, Kamura T and
Kojiro M: Angiogenesis in cancer. Vasc Health Risk Manag.
2:213–219. 2006. View Article : Google Scholar
|
|
28
|
Ip W, Wellman-Labadie O, Tang L, Su M, Yu
R, Dutz J, Wang Y, Huang S, Zhang X, Huang C, et al: Collagen
triple helix repeat containing 1 promotes melanoma cell adhesion
and survival. J Cutan Med Surg. 15:103–110. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
LeClair R and Lindner V: The role of
collagen triple helix repeat containing 1 in injured arteries,
collagen expression, and transforming growth factor beta signaling.
Trends Cardiovasc Med. 17:202–205. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Durmus T, LeClair RJ, Park KS, Terzic A,
Yoon JK and Lindner V: Expression analysis of the novel gene
collagen triple helix repeat containing-1 (Cthrc1). Gene Expr
Patterns. 6:935–940. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Tang L, Dai DL, Su M, Martinka M, Li G and
Zhou Y: Aberrant expression of collagen triple helix repeat
containing 1 in human solid cancers. Clin Cancer Res. 12:3716–3722.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Turashvili G, Bouchal J, Ehrmann J,
Fridman E, Skarda J and Kolar Z: Novel immunohistochemical markers
for the differentiation of lobular and ductal invasive breast
carcinomas. Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub.
151:59–64. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Pyagay P, Heroult M, Wang Q, Lehnert W,
Belden J, Liaw L, Friesel RE and Lindner V: Collagen triple helix
repeat containing 1, a novel secreted protein in injured and
diseased arteries, inhibits collagen expression and promotes cell
migration. Circ Res. 96:261–268. 2005. View Article : Google Scholar
|
|
34
|
Ke Z, He W, Lai Y, Guo X, Chen S, Li S,
Wang Y and Wang L: Overexpression of collagen triple helix repeat
containing 1 (CTHRC1) is associated with tumour aggressiveness and
poor prognosis in human non-small cell lung cancer. Oncotarget.
5:9410–9424. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Yang W and Yee AJ: Versican V2 isoform
enhances angiogenesis by regulating endothelial cell activities and
fibronectin expression. FEBS Lett. 587:185–192. 2013. View Article : Google Scholar
|
|
36
|
Zheng PS, Wen J, Ang LC, Sheng W,
Viloria-Petit A, Wang Y, Wu Y, Kerbel RS and Yang BB: Versican/PG-M
G3 domain promotes tumor growth and angiogenesis. FASEB J.
18:754–756. 2004.PubMed/NCBI
|
|
37
|
Zhang Z, Nie F, Chen X, Qin Z, Kang C,
Chen B, Ma J, Pan B and Ma Y: Upregulated periostin promotes
angiogenesis in keloids through activation of the ERK 1/2 and focal
adhesion kinase pathways, as well as the upregulated expression of
VEGF and angiopoietin 1. Mol Med Rep. 11:857–864. 2015.
|
|
38
|
Shao R, Bao S, Bai X, Blanchette C,
Anderson RM, Dang T, Gishizky ML, Marks JR and Wang XF: Acquired
expression of periostin by human breast cancers promotes tumor
angiogenesis through up-regulation of vascular endothelial growth
factor receptor 2 expression. Mol Cell Biol. 24:3992–4003. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zhu M, Fejzo MS, Anderson L, Dering J,
Ginther C, Ramos L, Gasson JC, Karlan BY and Slamon DJ: Periostin
promotes ovarian cancer angiogenesis and metastasis. Gynecol Oncol.
119:337–344. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Kunstman JW, Juhlin CC, Goh G, Brown TC,
Stenman A, Healy JM, Rubinstein JC, Choi M, Kiss N, Nelson-Williams
C, et al: Characterization of the mutational landscape of
anaplastic thyroid cancer via whole-exome sequencing. Hum Mol
Genet. 24:2318–2329. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Kveiborg M, Fröhlich C, Albrechtsen R,
Tischler V, Dietrich N, Holck P, Kronqvist P, Rank F, Mercurio AM
and Wewer UM: A role for ADAM12 in breast tumor progression and
stromal cell apoptosis. Cancer Res. 65:4754–4761. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Cheon DJ, Li AJ, Beach JA, Walts AE, Tran
H, Lester J, Karlan BY and Orsulic S: ADAM12 is a prognostic factor
associated with an aggressive molecular subtype of high-grade
serous ovarian carcinoma. Carcinogenesis. 36:739–747. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Larsen M, Tremblay ML and Yamada KM:
Phosphatases in cell-matrix adhesion and migration. Nat Rev Mol
Cell Biol. 4:700–711. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Murasawa Y, Hayashi T and Wang P-C: The
role of type V collagen fibril as an ECM that induces the motility
of glomerular endothelial cells. Exp Cell Res. 314:3638–3653. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Berchtold S, Grünwald B, Krüger A,
Reithmeier A, Hähl T, Cheng T, Feuchtinger A, Born D, Erkan M,
Kleeff J, et al: Collagen type V promotes the malignant phenotype
of pancreatic ductal adenocarcinoma. Cancer Lett. 356:721–732.
2015. View Article : Google Scholar
|
|
46
|
Fischer H, Stenling R, Rubio C and
Lindblom A: Colorectal carcinogenesis is associated with stromal
expression of COL11A1 and COL5A2. Carcinogenesis. 22:875–878. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Zhou W, Wang Z, Shen N, Pi W, Jiang W,
Huang J, Hu Y, Li X and Sun L: Knockdown of ANLN by lentivirus
inhibits cell growth and migration in human breast cancer. Mol Cell
Biochem. 398:11–19. 2015. View Article : Google Scholar
|
|
48
|
Pandi NS, Manimuthu M, Harunipriya P,
Murugesan M, Asha GV and Rajendran S: In silico analysis of
expression pattern of a Wnt/β-catenin responsive gene ANLN in
gastric cancer. Gene. 545:23–29. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Pardo LA and Stühmer W: The roles of K(+)
channels in cancer. Nat Rev Cancer. 14:39–48. 2014. View Article : Google Scholar
|