Introduction
Colorectal cancer (CRC) is the third most common
type of cancer in men and the second most common type in women,
accounting for ~608,000 deaths annually worldwide (1). The most common cause of death from CRC
is metastasis to distant organs. Although the prognosis of
metastatic colorectal cancer (mCRC) has been improving owing to
chemotherapy and molecular-targeted therapy (2,3), it is
not yet satisfactory. Various immunotherapies for CRC have been
developed and used, such as personalized peptide vaccination
(4) and dendritic cell-based active
immunotherapies (5). Recently,
programmed cell death 1 (PD-1) antibody has also been receiving
increased attention around the world (6). However, useful biomarkers that can
predict good clinical outcomes from immunotherapy have not yet been
identified (7), and there are few
immunological biomarkers, such as the B-cell signature, as
exemplified by the expression of the immunoglobulin G κ chain in
tumor-infiltrating lymphocytes. The development of biomarkers for
immunotherapy is desired for the appropriate selection and
evaluation of a patient population for clinical trials of cancer at
an earlier stage, and for the effective development of cancer
vaccine treatments (7,8).
MicroRNAs (miRNAs) are endogenous single-stranded
RNA molecules consisting of 18–24 nucleotides that regulate the
transcription levels of target genes and are involved in multiple
intracellular processes (9,10). Recently, several studies have
reported a relationship between the immune response and miRNAs. As
such, it is presumed that miRNAs are involved in the immune
response. In addition, the role of miRNAs as crucial regulators of
innate and adaptive immune responses has been coming to light
(11). In the process of tumor
progression enhanced by an antitumor immunity microenvironment,
miRNAs are considered to be one of the key players in tumor cell
escape from immunological surveillance (12,13) in
the induction of antitumor T cells (14) and in the immune-mediated recognition
of tumor cells (15). As such, in
patients in whom the efficacy of vaccine treatment is insufficient,
there may be impairment of the immune response due to upregulated
or downregulated miRNAs.
It has been reported that various miRNAs in plasma
may be useful as non-invasive biomarkers for detecting early CRC or
for predicting prognosis and recurrence (16,17).
Recently, in our institution, a phase I study in which five epitope
peptides [three derived from tumor-associated oncoantigens and two
derived from vascular endothelial growth factor receptors (VEGFRs)]
were applied to advanced-stage colon cancer patients (18). We subsequently performed a phase II
study with the same vaccine regimen in combination with
oxaliplatin-containing chemotherapy and further assessed its safety
and promising potential to induce cytotoxic T lymphocytes (CTLs)
and improve overall survival (OS) (19). In these studies, we found that a
high CTL response after vaccination and a skin reaction at the
injection site were possible biomarkers for the outcome of vaccine
treatment (18). Moreover, a low
neutrophil/lymphocyte ratio and a low plasma interleukin 6 level
(20) were also possible predictive
biomarkers of longer survival in vaccinated patients (19). We also reported the usefulness of
tumor miRNA expression for predicting the efficacy of
immune-chemotherapy (21).
The purpose of the present study was to explore
novel predictive biomarkers that can predict the efficacy of
vaccine treatment; we investigated the plasma miRNAs of mCRC
patients treated with the phase II study protocol in order to
detect liquid biomarkers.
Materials and methods
Summary of the phase II study
To evaluate the clinical benefits of cancer
vaccination treatment, we conducted a phase II trial that was a
non-randomized HLA-A status double-blinded study using five
HLA-A*2402-restricted peptides: RNF43-721 (NSQPVWLCL) (22), TOMM34-299 (KLRQEVKQNL) (23), KOC1 (IMP-3)-508 (KTVNELQNL)
(24), VEGFR1-1084 (SYGVLLWEI)
(25) and VEGFR2-169 (RFVPDGNRI)
(26). The detailed protocol of
this phase II study was previously described (19). Briefly, the therapy consisted of a
cocktail of five therapeutic epitope peptides in addition to
oxaliplatin-containing chemotherapy. Although the peptides used in
this study were HLA-A*2402-restricted peptides, all enrolled
patients, whose HLA-A*2402 status was double-blinded, were
administrated the same regime of peptide cocktail and
oxaliplatin-containing chemotherapy. The cocktail containing 3 mg
of each of the five peptides was mixed with 1.5 ml of incomplete
Freund's adjuvant (IFA) and administered subcutaneously into the
thigh or axilla regions weekly for 13 weeks; thereafter, the
vaccination schedule was reduced to once every 2 weeks.
Patients were eligible for enrollment if they were
≥20 years of age with a histologically confirmed advanced CRC,
chemotherapy-naïve, had adequate functions of critical organs and
had a life expectancy of ≥3 months. Between February 2009 and
November 2012, 96 chemotherapy-naïve CRC patients were enrolled
under the concealment of their HLA-A*2402 status.
Among the 96 patients who were enrolled in this
study, 93 cases were available for miRNA analysis. Written informed
consent for inclusion was obtained from all patients, and the study
protocol was approved by the local ethics committee (H20-102,
UMIN000001791).
Patients and plasma
A total of ninety-three patients
(HLA-A*2402-matched, n=48 and HLA-unmatched, n=45) with mCRC who
were treated in the phase II study had pretreated plasma available
for miRNA analysis. Peripheral blood from each patient was
collected in ethylenediaminetetraacetic acid (EDTA) tubes. The
blood samples were centrifuged at 400 × g for 15 min at 4°C. The
plasma was then aliquoted and stored at −80°C until use.
miRNA microarray
In order to screen for miRNAs involved in the
response to vaccine treatment, microarray analysis of miRNA
expression was performed using 13 plasma samples collected from
mCRC patients prior to vaccine treatment. All of the plasma sample
were from HLA-matched patients: five were from patients who
survived >3 years and eight were from patients who survived
<2 years.
Total RNAs from plasma samples (n=13) were analyzed
by miRNA microarray. Total RNA was extracted from the samples using
3D-Gene RNA extraction reagent from a liquid sample kit (Toray
Industries, Inc., Tokyo, Japan) according to the manufacturers
protocol. A comprehensive miRNA expression analysis was performed
using a 3D-Gene miRNA Labeling kit and a 3D-Gene Human miRNA Oligo
Chip (Toray Industries, Inc.), which was designed to detect 2,555
miRNAs registered in the miRBase database (release 20). Individual
miRNAs were considered to be present if the corresponding
microarray signals were more than the mean ± 2 standard deviation
(SD) of the negative control signals, of which the top and bottom
5% ranked by signal intensity were removed. Once a miRNA was
regarded to be present, the mean signal of the negative control, of
which the top and bottom 5% ranked by signal intensity were
removed, was subtracted from the signal of the miRNA. If the signal
became a negative value (or was undetected) after subtraction of
the background, the value was replaced by the value of the lowest
signal intensity on the microarray minus 0.1 on a log2 scale. In
order to normalize the signals across the different microarrays
tested, quantile normalization was performed (27).
Selection of miRNAs for
validation
Using the fold-change value and the Fisher
criterion, differentially expressed miRNAs between the responders
(OS ≥3 years) and non-responders (OS <2 years) were classified.
OS between 2 years and ≤3 years were excluded to compare a
long-term survivor and a short-term survivor.
Validation using qRT-PCR
From 400-µl samples of plasma, total RNA was
purified using a miRNeasy Serum/Plasma kit (Qiagen, Tokyo, Japan)
according to the manufacturers protocol. miR-191 was used as an
endogenous internal control (28,29).
We used TaqMan miRNA probes (Applied Biosystems
Japan Ltd., Tokyo, Japan) to perform the qRT-PCR assay according to
the manufacturers instructions. In each step, from plasma
purification to the qRT-PCR, an equal volume (400 µl) of plasma
sample was processed. The total RNA was reverse-transcribed to
complementary DNA using the TaqMan miRNA Reverse Transcription kit
(Applied Biosystems) and stem-loop RT primers (hsa-miR-135,
hsa-miR-6826, hsa-miR-6835 and hsa-miR-6875, in addition to
hsa-miR-191 for the internal control) (Applied Biosystems). RT-PCR
was performed using the LightCycler® 480 System II
(Roche Diagnostics K.K., Tokyo, Japan). The reactions were
initiated at 95°C for 5 min, followed by 40 cycles of 95°C for 15
sec and 60°C for 1 min. All reactions, including the no template
controls, were run in duplicate. The relative expression levels of
the target miRNAs were normalized to those of miR-191 according to
the ΔΔCt method. For every target miRNA, the relative Ct values
were divided into an OS ≥2-year group (responders) and an OS
<2-year group (non-responders) which were plotted
separately.
Statistical analysis
The obtained values are shown as the mean ± SD. The
values beyond the mean ± 3SD were excluded as outliers for each
miRNA. The expression levels of plasma miRNAs were compared between
the responders and non-responders using Scheffes or Dunnetts
test.
For each miRNA, the cut-off value was set as the
median and a survival curve was obtained by the Kaplan-Meier method
to evaluate the efficacy of vaccine treatment. P-values were
calculated with the log-rank test. The P-value for the relative Ct
value between the responders and non-responders was calculated with
the t-test. For every miRNA, a survival curve for each HLA-A*2402
status was obtained using the Kaplan-Meier method and analyzed
using the log-rank test.
A Coxs proportional hazards model and a logistic
regression model were used to estimate the hazard ratios (HRs) for
the treatment effect in relation to OS and biomarkers or prognostic
clinical information. All statistical analyses were performed with
SPSS Statistics 20.0 (SPSS, Inc., Chicago, IL, USA). A value of
P<0.05 was considered statistically significant.
Results
miRNA microarray
Ten candidate miRNAs as biomarkers were selected by
a comprehensive analysis of the miRNAs, according to the miRNA
expression levels ranked using the Fisher criterion between the
patients who survived >3 years and those who survived <2
years (Table I). Finally, we
selected four miRNAs (miR-135a-3p, miR-6875-5p, miR-6835-5p and
miR-6826-5p) for which the expression difference according to the
absolute value of the log2 ratio was >1.30 between the long-term
survivor and the short-term survivor (Table I).
 | Table I.Selection of the microRNA from the
result of the comprehensive analysis of the microarray. |
Table I.
Selection of the microRNA from the
result of the comprehensive analysis of the microarray.
|
| OS ≥3 years
(n=5) | OS <2 years
(n=8) |
|
|
|---|
|
|
|
|
|
|
|---|
| microRNA name | Mean | SD | Mean | SD | |Log2 ratio| | Fisher ratio |
|---|
|
miR-135a-3p | 148.0 | 131.5 | 49.7 | 39.5 | 1.6 | 1.02 |
|
miR-6875-5p | 451.4 | 431.7 | 184.3 | 213.9 | 1.3 | 0.61 |
| miR-6798-5p | 505.0 | 309.5 | 295.6 | 260.7 | 0.8 | 0.59 |
| miR-1233-5p | 1,477.7 | 1,062.1 | 3,007 | 2,600.6 | 1.0 | 0.57 |
| miR-6124 | 137.9 | 102.4 | 257.7 | 238.6 | 0.9 | 0.57 |
| miR-1275 | 97.2 | 67.0 | 199.4 | 192.8 | 1.0 | 0.54 |
| miR-1229-5p | 126.2 | 85.3 | 244.3 | 234.8 | 1.0 | 0.50 |
| miR-197-5p | 41.1 | 26.2 | 90.6 | 101.9 | 1.1 | 0.45 |
|
miR-6826-5p | 145.3 | 130.3 | 425.9 | 510.7 | 1.6 | 0.44 |
|
miR-6835-5p | 70.9 | 77.4 | 253.5 | 332.6 | 1.8 | 0.43 |
Validation analysis
In the validation phase we defined a responder as OS
≥2 years and a non-responder as OS <2 years. The expression of
miR-6826 was significantly higher in the non-responders than that
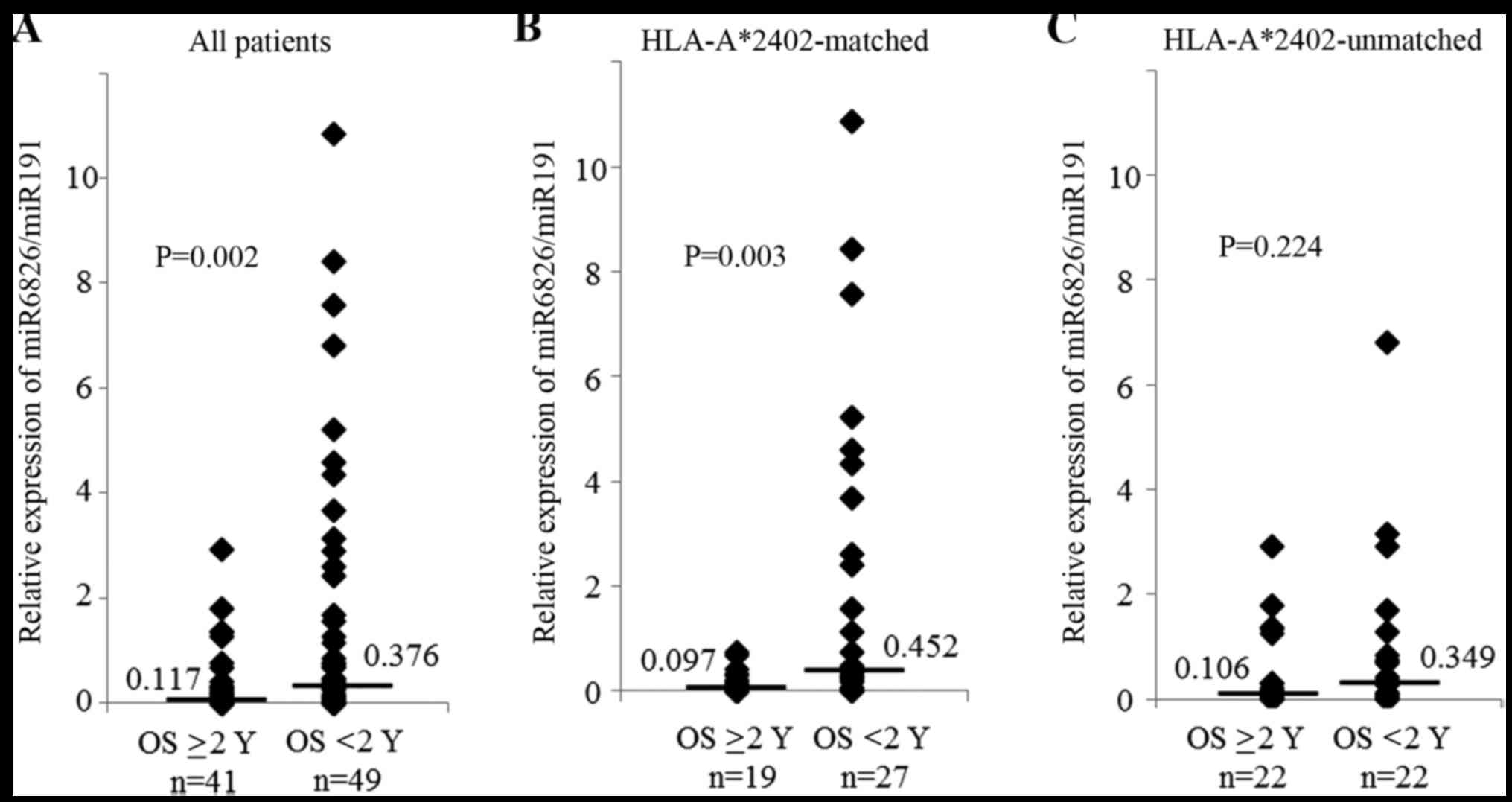
observed in the responders (P=0.002, Fig. 1A). To investigate the efficacy of
vaccine treatment, samples were divided according to two HLA
statuses. The expression of miR-6826 was also significantly higher
in the non-responders than in the responders in the
HLA-A*2402-matched group (P=0.003, Fig.
1B). In contrast, there was no significant difference in the
expression of miR-6826 between the responders and non-responders in
the HLA-A*2402-unmatched group (Fig.
1C). As such, a high expression level of miR-6826 may indicate
that the vaccine treatment will have poor efficacy.
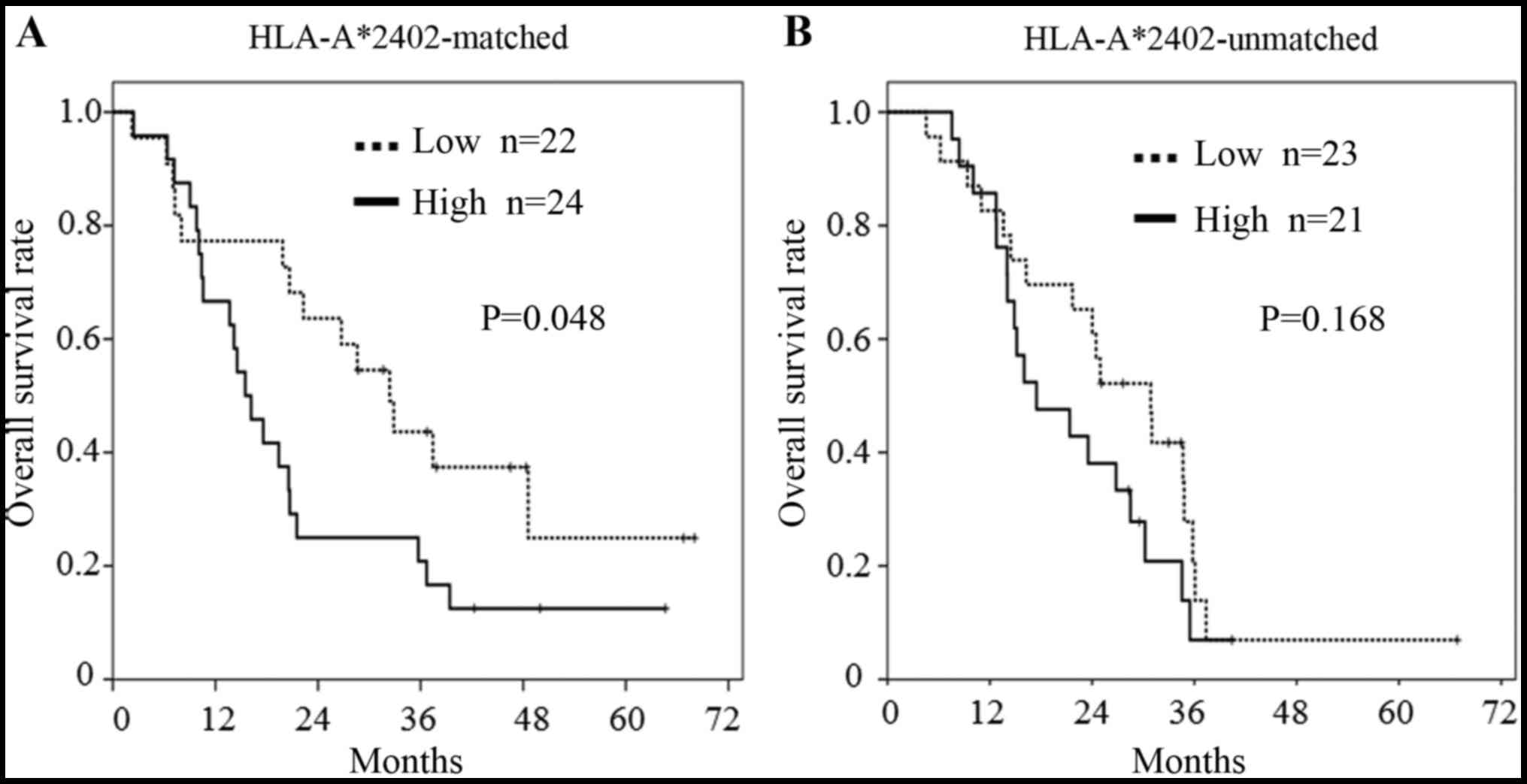
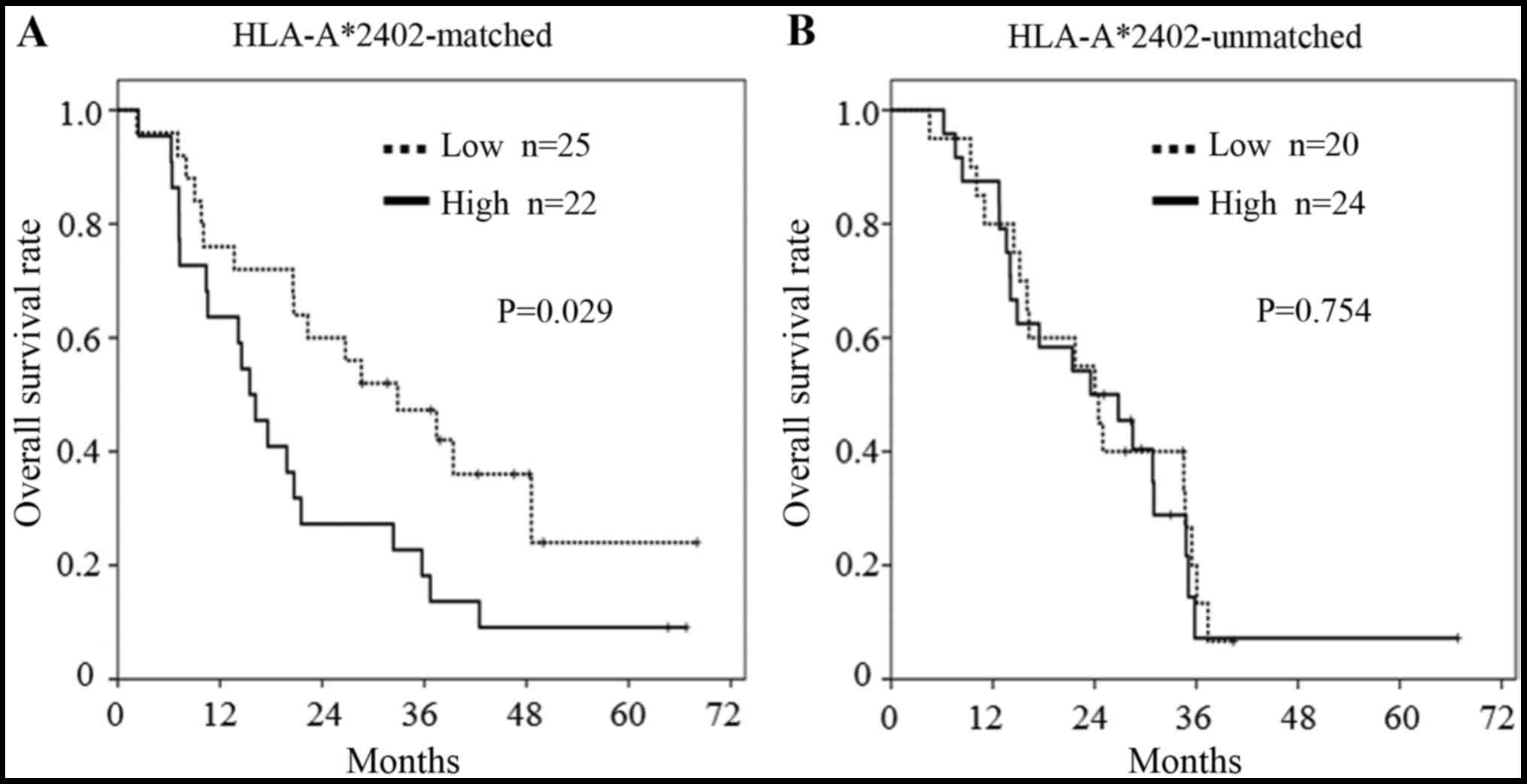
The median of each miRNA Ct value was used as the
cut-off value to discriminate high and low values. In the subgroup
analysis, patients bearing HLA-A*2402, with a lower miR-6826
expression showed a longer OS than patients with a higher miR-6826
expression in the HLA-A*2402-matched group (P=0.048, Fig. 2A). In contrast, in the
HLA-A*2402-unmatched group, there was no difference in OS between
those with a high or low miR-6826 expression (Fig. 2B). This suggested that miR-6826
could be a useful biomarker for predicting the efficacy of vaccine
treatment.
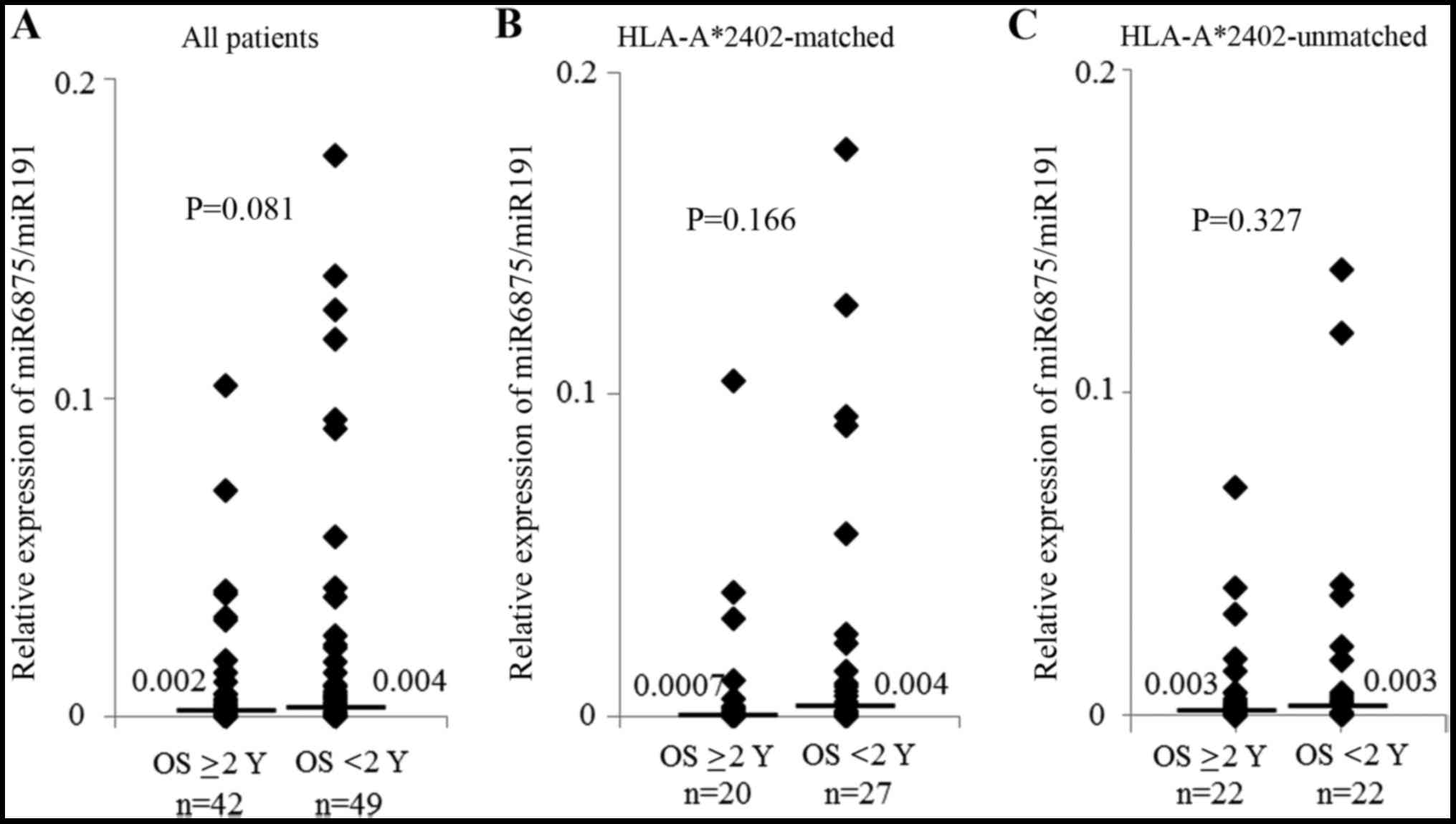
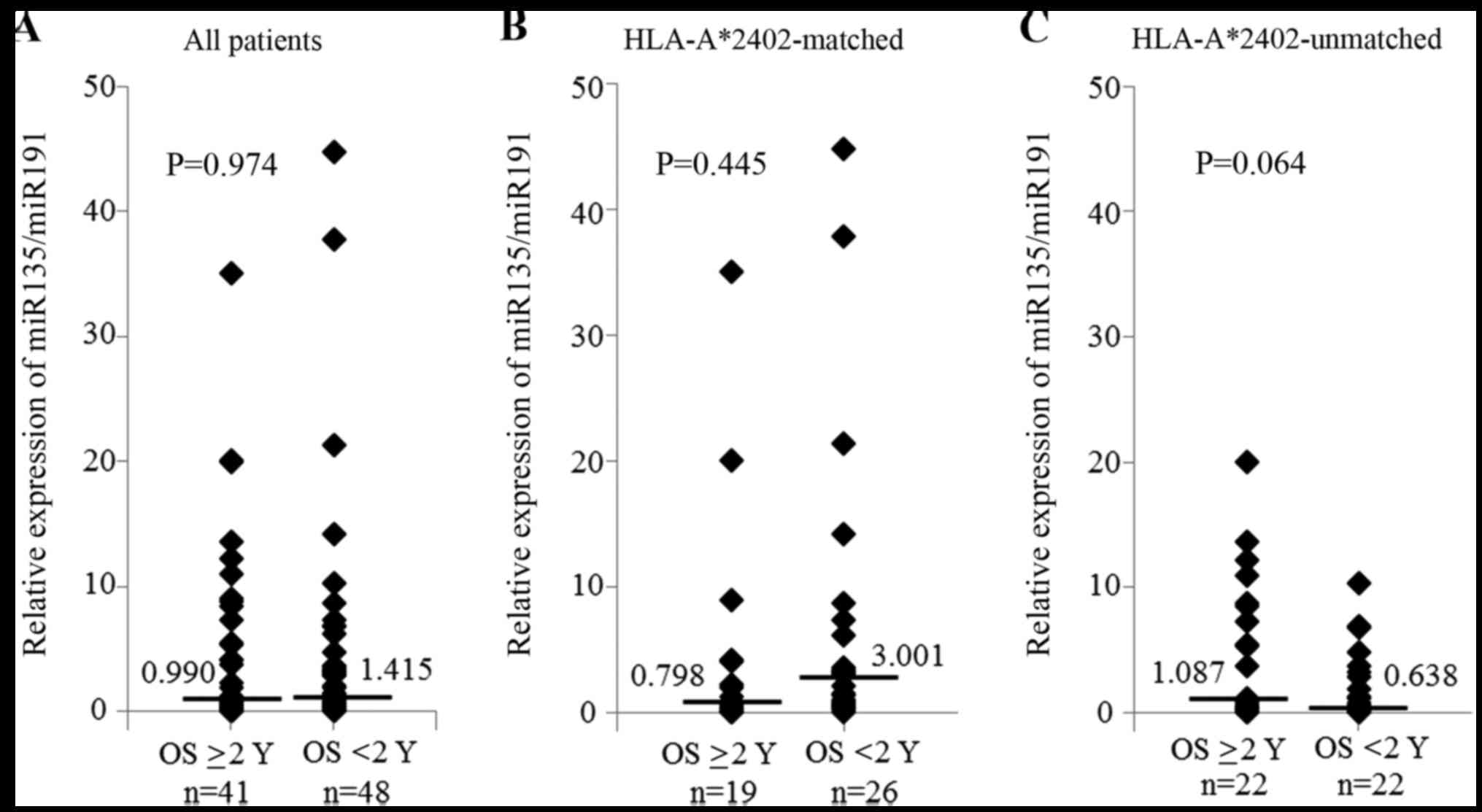
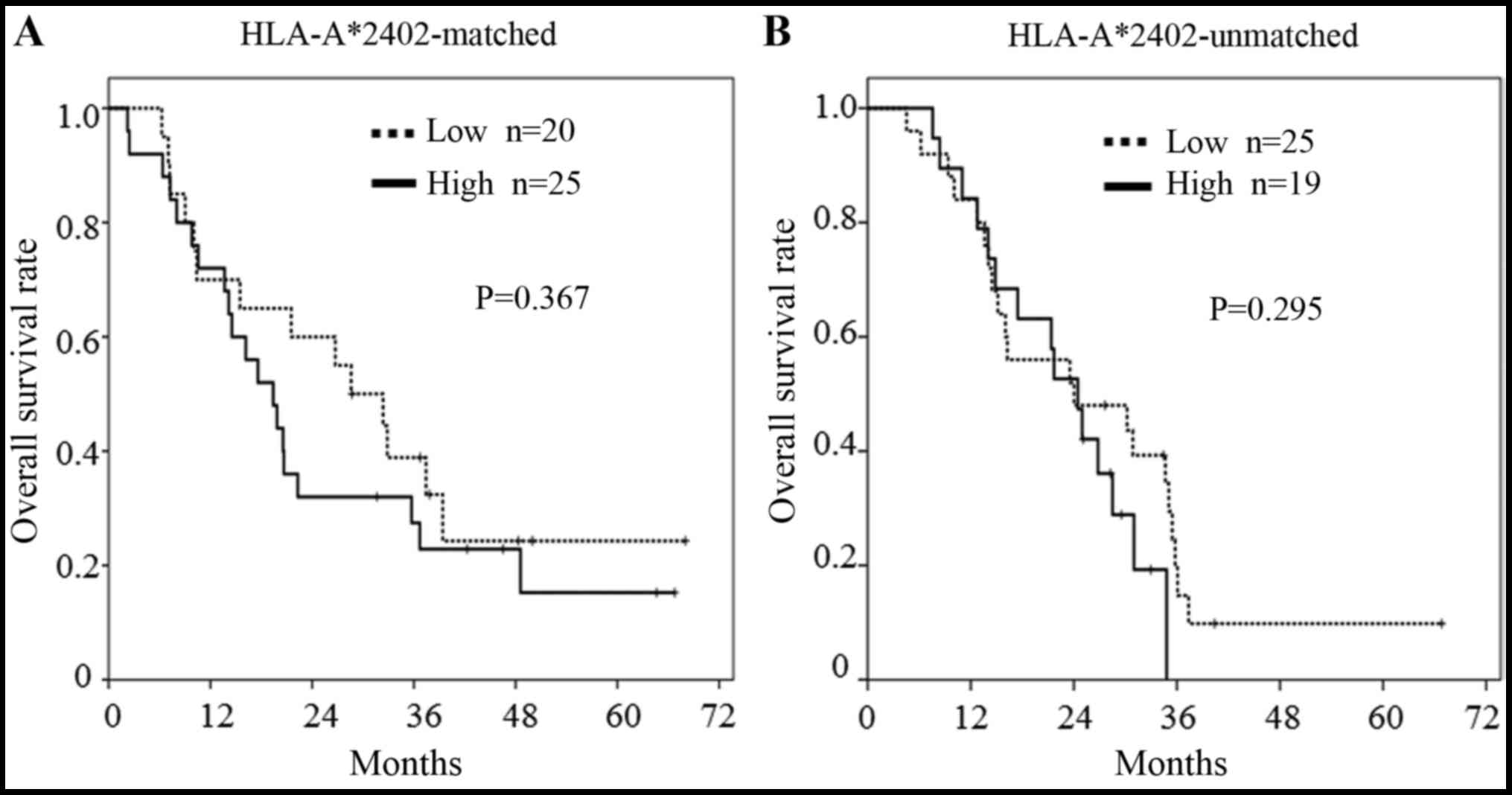
Regarding the expression of miR-6875 and miR-135,
there was no significant difference between the patients who
survived ≥2 years and those who survived <2 years (Figs. 3 and 4). However, in the subgroup analysis of
patients bearing HLA-A*2402, patients with a lower miR-6875
expression had a longer OS than patients with a higher miR-6875
expression (P=0.029, Fig. 5A). In
addition, there was no difference in OS between the patients with a
lower or higher miR-6875 expression in the HLA-A*2402-unmatched
group (Fig. 5B). This suggested
that miR-6875 may also be useful as a biomarker for predicting the
efficacy of vaccine treatment.
There was no significant difference in OS according
to miR-135 expression between the HLA-A*2402-matched group and the
HLA-A*2402-unmatched group (Fig.
6). A high expression of miR-6826 and miR-6875 in plasma may
indicate a poor response to not only vaccine treatment in
combination with chemotherapy, but also to the vaccine treatment by
itself.
The Ct value of miR-6835 could not be measured in
all samples even after >55 cycles, indicating that the quantity
of miR-6835 in plasma was too small (data not shown).
To explore biomarkers for this vaccine therapy, we
analyzed immunological parameters, tumor factors, as well as miRNA
expression levels by a Coxs proportional hazards model and a
logistic regression model. Multivariate analysis of the Cox
regression model indicated that the expression of miR-6826 was the
most significant predictor for OS (P=0.003, HR, 3.670) (Table II). Moreover, the sensitivity of
miR-6826 to predict prolonged OS was 100%, and negative predictive
value was also 100% in the HLA-A*2402 matched patients (Table III).
 | Table II.Univariate and multivariate analyses
of the associations between clinical data and overall survival. |
Table II.
Univariate and multivariate analyses
of the associations between clinical data and overall survival.
|
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|
|---|
|
|
|
| 95% CI |
|
| 95% CI |
|
|---|
|
|
|
|
|
|
|
|
|
|---|
| Factor | Cut-off | HR | Lower | Upper | P-value | HR | Lower | Upper | P-value |
|---|
| CRP | >1 | 1.302 | 0.635 | 2.673 | 0.471 |
|
|
|
|
| NLR | >3 | 1.714 | 0.882 | 3.332 | 0.112 |
|
|
|
|
| CEA | >100 | 1.149 | 0.578 | 2.284 | 0.692 |
|
|
|
|
| CA19-9 | >100 | 1.001 | 0.496 | 2.020 | 0.999 |
|
|
|
|
| No. of involved
organs | Two or more | 1.706 | 0.855 | 3.406 | 0.130 | 2.173 | 1.030 | 4.584 | 0.042 |
| Relative expression
of miR-6826 | >1.00 (mean
value) | 3.510 | 1.551 | 7.942 | 0.003 | 3.670 | 1.569 | 8.581 | 0.003 |
| Relative expression
of miR-6875 | >0.016 (mean
value) | 1.389 | 0.652 | 2.961 | 0.395 |
|
|
|
|
 | Table III.Expression of miR-6826 and overall
survival. |
Table III.
Expression of miR-6826 and overall
survival.
|
| Overall
survival |
|---|
|
|
|
|---|
| Parameters | ≥2 Years | <2 Years |
|---|
| Relative expression
of miR-6826 |
|
|
<1.0 | 19 | 16 |
|
≥1.0 | 0 | 11 |
| Sensitivity | 19/19 | (100%) |
| Specificity | 11/27 | (40.7%) |
| Positive predictive
value | 19/35 | (54.3%) |
| Negative predictive
value | 11/11 | (100%) |
Discussion
Many novel vaccine approaches, such as whole tumor
cell vaccines, peptide vaccines (30), viral vector vaccines and dendritic
cell vaccines, for the treatment of cancer have been developed.
However, useful biomarkers that can predict a good clinical outcome
from immunotherapy have not yet been identified (7) and few immunological or other
biomarkers are available for use in clinical trials of
immunotherapy.
To the best of our knowledge, this is the first
study performed on the measurement of plasma (or serum) miRNAs for
predicting the efficacy of immunotherapy using liquid biopsy.
Firstly, we selected four miRNAs as biomarkers for predicting the
efficacy of the vaccine treatment. Next, we validated the results
of the comprehensive analysis using qPCR of the plasma miRNAs of 93
patients; in the vaccine-treated group, patients with a high
expression of plasma miR-6826 and miR-6875 had a poorer prognosis
than those with a low expression. Hence, we concluded that plasma
miR-6826 and miR-6875 levels are negative predictive biomarkers for
the efficacy of the vaccine treatment. Moreover, multivariate
analysis indicated that the expression of miR-6826 was the most
powerful predictor for OS, among immunological parameters, tumor
factors, and miRNA expression levels. In consideration that the
negative predictive value of miR-6826 was 100%, the high value of
miR-6826 may be an exclusion criteria in upcoming clinical studies
of immunotherapy.
These results also suggested that a high expression
level of miR-6826 or miR-6875 may be related to the suppression of
immune competence, and may be novel molecular targets for
regulating the effects of immunosuppressive factors. miR-6826 was
previously found to be upregulated in the serum of patients with
pancreato-biliary cancer, and was a diagnostic marker for
pancreato-biliary cancer (31). In
addition, miR-6875 was reported to be a tumor marker for detecting
early-stage breast cancer in combination with four other miRNAs
(32), although the roles of
miR-6826 and miR-6875 on the immune system have not yet been
reported and no target mRNA has been reported for miR-6826 or
miR-6875 in the miRBase database (release 18). miRNAs have been
implicated in adaptive immunity by controlling the development and
activation of T and B cells. Dynamic changes in the expression of
miRNAs may be important for the regulation of gene expression
during antigen-induced T cell differentiation.
Regarding such immune-suppressive miRNAs, miR-155 is
implicated in the upregulation of regulatory T cells (Tregs) and
myeloid-derived suppressor cells (33,34),
which have been reported to be potent immunosuppressive cells that
protect cancer cells from the host immune system (11). Overexpression of PD-L1, PD-1, and
upregulation of indoleamine-2,3-dioxygenase (IDO) in the tumor
microenvironment were also found to inhibit CTL function (35). Hence, to overcome these
immune-escape mechanisms, various approaches have been taken in the
last decade (36,37). For successful next-generation
immunotherapy, peptide vaccines should be combined with other
agents to modify immunosuppressive tumor microenvironments.
In conclusion, the expression levels of miR-6826 and
miR-6875 may be applicable as biomarkers for assessing and
identifying patients who can expect poor efficacy from vaccine
treatment. In addition, although further clarification is needed on
the functions of miR-6826 and miR-6875 and on their relationship to
immune-related molecules, these miRNAs are potential targets for
impeding the effects of immunosuppressive factors.
Acknowledgements
This study was performed as a research program of
the Project for the Development of Innovative Research on Cancer
Therapeutics (P-Direct), The Japan Agency for Medical Research and
Development (AMED). This study was edited by a native English
speaker at Forte Science Communications (Tokyo, Japan).
References
|
1
|
Siegel R, Ma J, Zou Z and Jemal A: Cancer
statistics, 2014. CA Cancer J Clin. 64:9–29. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Williet N, Fovet M and Phelip JM:
Management of metastatic colorectal cancer. Rev Prat. 65:793–797.
2015.(In French). PubMed/NCBI
|
|
3
|
Ciombor KK, Wu C and Goldberg RM: Recent
therapeutic advances in the treatment of colorectal cancer. Annu
Rev Med. 66:83–95. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kibe S, Yutani S, Motoyama S, Nomura T,
Tanaka N, Kawahara A, Yamaguchi T, Matsueda S, Komatsu N, Miura M,
et al: Phase II study of personalized peptide vaccination for
previously treated advanced colorectal cancer. Cancer Immunol Res.
2:1154–1162. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hunyadi J, András C, Szabó I, Szántó J,
Szluha K, Sipka S, Kovács P, Kiss A, Szegedi G, Altorjay I, et al:
Autologous dendritic cell based adoptive immunotherapy of patients
with colorectal cancer-A phase I–II study. Pathol Oncol Res.
20:357–365. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Diaz LA Jr and Le DT: PD-1 Blockade in
tumors with mismatch-repair deficiency. N Engl J Med. 373:19792015.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Copier J, Whelan M and Dalgleish A:
Biomarkers for the development of cancer vaccines: current status.
Mol Diagn Ther. 10:337–343. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Whiteside TL: Immune responses to cancer:
Are they potential biomarkers of prognosis? Front Oncol. 3:1072013.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lagos-Quintana M, Rauhut R, Lendeckel W
and Tuschl T: Identification of novel genes coding for small
expressed RNAs. Science. 294:853–858. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lee RC and Ambros V: An extensive class of
small RNAs in Caenorhabditis elegans. Science. 294:862–864. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Facciabene A, Motz GT and Coukos G:
T-regulatory cells: key players in tumor immune escape and
angiogenesis. Cancer Res. 72:2162–2171. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ueda R, Kohanbash G, Sasaki K, Fujita M,
Zhu X, Kastenhuber ER, McDonald HA, Potter DM, Hamilton RL, Lotze
MT, et al: Dicer-regulated microRNAs 222 and 339 promote resistance
of cancer cells to cytotoxic T-lymphocytes by down-regulation of
ICAM-1. Proc Natl Acad Sci USA. 106:10746–10751. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Sonda N, Simonato F, Peranzoni E, Calì B,
Bortoluzzi S, Bisognin A, Wang E, Marincola FM, Naldini L, Gentner
B, et al: miR-142-3p prevents macrophage differentiation during
cancer-induced myelopoiesis. Immunity. 38:1236–1249. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Trifari S, Pipkin ME, Bandukwala HS, Äijö
T, Bassein J, Chen R, Martinez GJ and Rao A: MicroRNA-directed
program of cytotoxic CD8+ T-cell differentiation. Proc
Natl Acad Sci USA. 110:18608–18613. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Min D, Lv XB, Wang X, Zhang B, Meng W, Yu
F and Hu H: Downregulation of miR-302c and miR-520c by
1,25(OH)2D3 treatment enhances the
susceptibility of tumour cells to natural killer cell-mediated
cytotoxicity. Br J Cancer. 109:723–730. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Huang Z, Huang D, Ni S, Peng Z, Sheng W
and Du X: Plasma microRNAs are promising novel biomarkers for early
detection of colorectal cancer. Int J Cancer. 127:118–126. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yuan D, Li K, Zhu K, Yan R and Dang C:
Plasma miR-183 predicts recurrence and prognosis in patients with
colorectal cancer. Cancer Biol Ther. 16:268–275. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hazama S, Nakamura Y, Takenouchi H, Suzuki
N, Tsunedomi R, Inoue Y, Tokuhisa Y, Iizuka N, Yoshino S, Takeda K,
et al: A phase I study of combination vaccine treatment of five
therapeutic epitope-peptides for metastatic colorectal cancer;
safety, immunological response, and clinical outcome. J Transl Med.
12:632014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hazama S, Nakamura Y, Tanaka H, Hirakawa
K, Tahara K, Shimizu R, Ozasa H, Etoh R, Sugiura F, Okuno K, et al:
A phase II study of five peptides combination with
oxaliplatin-based chemotherapy as a first-line therapy for advanced
colorectal cancer (FXV study). J Transl Med. 12:1082014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Hazama S, Takenouchi H, Tsunedomi R, Iida
M, Suzuki N, Iizuka N, Inoue Y, Sakamoto K, Nakao M, Shindo Y, et
al: Predictive biomarkers for the outcome of vaccination of five
therapeutic epitope peptides for colorectal cancer. Anticancer Res.
34:4201–4205. 2014.PubMed/NCBI
|
|
21
|
Shindo Y, Hazama S, Nakamura Y, Inoue Y,
Kanekiyo S, Suzuki N, Takenouchi H, Tsunedomi R, Nakajima M, Ueno
T, et al: miR-196b, miR-378a and miR-486 are predictive biomarkers
for the efficacy of vaccine treatment in colorectal cancer. Oncol
Lett. (In Press).
|
|
22
|
Uchida N, Tsunoda T, Wada S, Furukawa Y,
Nakamura Y and Tahara H: Ring finger protein 43 as a new target for
cancer immunotherapy. Clin Cancer Res. 10:8577–8586. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Shimokawa T, Matsushima S, Tsunoda T,
Tahara H, Nakamura Y and Furukawa Y: Identification of TOMM34,
which shows elevated expression in the majority of human colon
cancers, as a novel drug target. Int J Oncol. 29:381–386.
2006.PubMed/NCBI
|
|
24
|
Suda T, Tsunoda T, Daigo Y, Nakamura Y and
Tahara H: Identification of human leukocyte antigen-A24-restricted
epitope peptides derived from gene products upregulated in lung and
esophageal cancers as novel targets for immunotherapy. Cancer Sci.
98:1803–1808. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ishizaki H, Tsunoda T, Wada S, Yamauchi M,
Shibuya M and Tahara H: Inhibition of tumor growth with
antiangiogenic cancer vaccine using epitope peptides derived from
human vascular endothelial growth factor receptor 1. Clin Cancer
Res. 12:5841–5849. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wada S, Tsunoda T, Baba T, Primus FJ,
Kuwano H, Shibuya M and Tahara H: Rationale for antiangiogenic
cancer therapy with vaccination using epitope peptides derived from
human vascular endothelial growth factor receptor 2. Cancer Res.
65:4939–4946. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Smyth GK: limma: Linear models for
microarray dataBioinformatics and Computational Biology Solutions
Using R and Bioconductor: Statistics for Biology and Health.
Gentleman R, Carey V, Dudoit S, Irizarry R and Huber W: Springer;
New York: pp. 397–420. 2005, View Article : Google Scholar
|
|
28
|
Hu Z, Dong J, Wang LE, Ma H, Liu J, Zhao
Y, Tang J, Chen X, Dai J, Wei Q, et al: Serum microRNA profiling
and breast cancer risk: the use of miR-484/191 as endogenous
controls. Carcinogenesis. 33:828–834. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zheng G, Wang H, Zhang X, Yang Y, Wang L,
Du L, Li W, Li J, Qu A, Liu Y, et al: Identification and validation
of reference genes for qPCR detection of serum microRNAs in
colorectal adenocarcinoma patients. PLoS One. 8:e830252013.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Boon T and van der Bruggen P: Human tumor
antigens recognized by T lymphocytes. J Exp Med. 183:725–729. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kojima M, Sudo H, Kawauchi J, Takizawa S,
Kondou S, Nobumasa H and Ochiai A: MicroRNA markers for the
diagnosis of pancreatic and biliary-tract cancers. PLoS One.
10:e01182202015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Shimomura A, Shiino S, Kawauchi J,
Takizawa S, Sakamoto H, Matsuzaki J, Ono M, Takeshita F, Niida S,
Shimizu C, et al: Novel combination of serum microRNA for detecting
breast cancer in the early stage. Cancer Sci. 107:326–334. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Chen S, Wang L, Fan J, Ye C, Dominguez D,
Zhang Y, Curiel TJ, Fang D, Kuzel TM and Zhang B: Host miR155
promotes tumor growth through a myeloid-derived suppressor
cell-dependent mechanism. Cancer Res. 75:519–531. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zheng Y, Josefowicz SZ, Kas A, Chu TT,
Gavin MA and Rudensky AY: Genome-wide analysis of Foxp3 target
genes in developing and mature regulatory T cells. Nature.
445:936–940. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Gajewski TF, Schreiber H and Fu YX: Innate
and adaptive immune cells in the tumor microenvironment. Nat
Immunol. 14:1014–1022. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
36
|
Okazaki T, Tanaka Y, Nishio R, Mitsuiye T,
Mizoguchi A, Wang J, Ishida M, Hiai H, Matsumori A, Minato N, et
al: Autoantibodies against cardiac troponin I are responsible for
dilated cardiomyopathy in PD-1-deficient mice. Nat Med.
9:1477–1483. 2003. View
Article : Google Scholar : PubMed/NCBI
|
|
37
|
Pardoll DM: The blockade of immune
checkpoints in cancer immunotherapy. Nat Rev Cancer. 12:252–264.
2012. View
Article : Google Scholar : PubMed/NCBI
|