Introduction
Glioma is the most common primary malignant tumor in
the central nervous system, and is associated with poor prognosis
and rapid mortality (1). It
accounts for 30–40% of all intracranial tumors (2). Although the standard treatment for
glioma is based on the combination of surgery, chemotherapy and
radiation therapy, the relapse rate is still very high (3). Based on certain pathological
characteristics, gliomas are classified into 4 grades (WHO grade I,
II, III and IV). Grade I or II tumors are low grade astrocytomas,
grade III tumors are anaplastic astrocytomas, and grade IV tumors
are glioblastoma multiforme (4). To
date, the median survival time of patients with grade IV
glioblastoma multiforme is only 14.7 months (5). A dilemma in this area is further
compounded by the fact that patients with glioblastoma (WHO grade
IV) account for 50% of the new cases of malignant CNS tumors
(5). Recurrence and invasion of the
primary tumor are thought to be key contributors to the incurable
nature of glioma (2). Glioma stem
cells (GSCs) are a subgroup of glioma cells having the ability to
self-renew and to differentiate into mature tumor cells (6,7).
Accumulating evidence suggests that GSCs play an important role in
tumorigenesis, treatment resistance and tumor recurrence (1,7). Thus,
therapy targeting GSCs is of critical importance in the development
of relapse-free treatment for glioma patients.
Recently, long non-coding RNAs (lncRNAs), which
represent non-protein coding transcripts longer than 200
nucleotides, have been implicated in several cellular functions,
including cell proliferation, apoptosis, transformation, metastasis
and differentiation (8,9). Accumulated evidence has proposed that
RNA interference holds great promise for tumor therapy. Among these
lncRNAs, nuclear enriched abundant transcript 1 (NEAT1) has been
advocated as an important mediator of many tumor cells. NEAT1
expression levels have been found to be related with the pathologic
grade in many types of cancer, including esophageal squamous cell
carcinoma (10), colorectal
(11) and prostate cancer, and
gliomas (12,13). Furthermore, NEAT1 knockdown was also
found to induce apoptosis in B-cell lymphoma and LSCC cells
(14,15). In the glioma research field,
knockdown of NEAT1 was reported to reduce glioma cell proliferation
in vitro (16). Since
studies focusing on the NEAT1 function in glioma are limited,
whether NEAT1 is capable of targeting GSCs remains unclear. A
better understanding of the precise function of NEAT1 on GSCs is
important to improve current therapies and to design new treatment
modalities.
In our previous study, we found that NEAT1 knockdown
plays a crucial regulatory role in proliferation, apoptosis and
metastasis of esophageal squamous cell carcinoma (10). We hypothesized that NEAT1 plays a
regulatory role in GSCs. In the present study, we provide
experimental evidence to support this hypothesis and demonstrate a
functional role of NEAT1 in GSCs. We demonstrated that NEAT1
knockdown induced G1 cell cycle arrest, and apoptosis while
reducing proliferation in GSCs. The present study proposes that the
targeting of NEAT1 may represent a novel and important therapeutic
strategy for the treatment of glioma.
Materials and methods
Patients and surgical specimens
Tissue samples from patients with gliomas were used
in the present study. Diagnostic criteria for gliomas were
preoperative magnetic resonance imaging (MRI) and postoperative
histopathological findings. All patients received surgery between
May 2015 and December 2015 at the Department of Neurosurgery, The
First Affiliated Hospital of Henan University of Science and
Technology (Luoyang, China). There were 29 patients with gliomas
(18 men, 11 women; age range 45–69 years, mean age 51.50±16.00
years). For each sample, the tissue was dissected. One half was
sent to the laboratory for primary culture, protein or RNA
extraction, and the other half was fixed and embedded in paraffin
for histology and immunohistochemisty studies. The study protocol
was approved by the local Ethics Committee of the Henan University
of Science and Technology and informed consent was obtained from
all patients in the present study.
Cell lines and culture protocols
The U87 human glioblastoma cell line was purchased
from the American Type Culture Collection (ATCC; Manassas, VA,
USA). Glioma primary cultured cells were prepared in our laboratory
as follows. Resected brain glioma tissues were immediately send to
the laboratory by placing them in 0.9% NaCl. Samples were digested
by incubation in 0.1% type I collagenase (Sigma-Aldrich, St. Louis,
MO, USA) in phosphate-buffered saline (PBS) at 37°C for 2 h, and
then filtered using a 40-µm steel mesh. The cell suspensions were
cultured in the following medium. For regular cell maintenance, the
cells were cultured in Dulbecco's modified Eagle's medium (DMEM)
containing 10% fetal bovine serum (FBS; Gibco, Life Technologies,
Carlsbad, CA, USA) and 100 U/100 µg/ml penicillin-streptomycin in a
humidified incubator at 37°C with 5% CO2.
To induce stem cell proliferation and negatively
select against differentiated cells, the stem cells isolated from
U87 and primary cultured cells were cultured in DMEM-F12
supplemented with 20 ng/µl hEGF, 20 ng/µl bFGF (both from Gibco),
1X B27 and 100 U/100 µg/ml penicillin-streptomycin in a humidified
incubator at 37°C with 5% CO2.
Isolation and identification of glioma
stem cells
The cell suspensions were collected and washed with
PBS and incubated with magnetic microbeads conjugated with the
anti-CD133 antibody (Miltenyi Biotec, Cambridge, MA, USA). The
bead-bound (CD133+) and unbound cells
(CD133−) were separated using the QuadroMACS™ Separation
Unit (Miltenyi Biotec). The purity of the isolated
CD133+ cells was confirmed by flow cytometric analyses.
The isolated CD133+ cells were then cultured in stem
cell media.
Neurospheres
Briefly, 1×103 CD133+ cells
were seeded into 6-well ultra-low attachment culture plates
(Corning Inc., Corning, NY, USA) with DMEM-F12 supplemented with 20
ng/µl hEGF, 20 ng/µl bFGF (both from Gibco), 1X B27 and 100 U/100
µg/ml penicillin-streptomycin. Culture medium was replaced every 4
days. Glioma spheres derived from one mother cell in the subsphere
forming assay were mechanically digested into single cells that
were seeded into standard medium for differentiation (DMEM with 10%
of FBS). Then, the cells were analyzed at different time
points.
Virus transduction
The siNEAT1 and the control lentiviruses
(GFP-lentivirus) were constructed by GeneCopoeia-FulenGen
(Shanghai, China). The sequences of siNEAT1 were as follows:
5′-GUGAGAAGUUGCUUAGAAACUUUCC-3′. CD133+ GSC cells were
transduced as follows. The lentiviruses were diluted in complete
medium incubated with the cells for 12 h at 37°C. Next, the cells
were incubated with freshly prepared Polybrene-DMEM for 24 h. Cells
were selected in medium containing 5 µg/ml puromycin
(Sigma-Aldrich) for 48 h.
Soft agar colony formation assays
The proliferation of the CD133+ GFP
control and CD133+ siNEAT1 cells was measured using the
soft agar colony formation assays. Briefly, ~1×105 cells
were mixed with medium containing 0.4% agar and were spread on top
of a bottom agar layer (0.8% agar in growth medium). Cells were
grown for 2 weeks, and colonies were counted and photographed. The
diameter of the colonies was measured using ImageJ software.
Cell cycle analysis by flow
cytometry
Cells were collected and fixed with cold ethanol at
4°C for 1 h before being stored at −20°C. Fixed cells were washed
and resuspended in 1 ml PBS containing 50 µg/ml RNase A and 50
µg/ml ethidium bromide. After incubating for 20 min at 37°C, the
cells were analyzed for DNA content by flow cytometry (FACSCalibur;
Becton-Dickinson Immunocytometry Systems, San Jose, CA, USA). For
each sample, 20,000 events were acquired, and cell cycle
distribution was determined using cell cycle analysis software
(ModFit LT for Mac version 3.0).
Apoptosis assay by flow cytometry
The cells were washed twice with cold 10 mM PBS and
resuspended in 1X binding buffer (BD Biosciences, San Jose, CA,
USA) at a density of 1×106 cells/ml. Cells were stained
with Annexin V/APC and propidium iodide (PI), using the Annexin V
apoptosis detection kit (BD Biosciences).
Quantitative real-time PCR
Total RNA containing miRNA was extracted from cells
using a Qiagen miRNeasy Mini kit (Qiagen, Mississauga, ON, USA)
following the manufacturer's instructions. cDNA was reverse
transcribed using All-in-One miRNA RT-qPCR kit (GeneCopoeia,
Rockville, MD, USA). Real-time PCR was performed with gene-specific
primers using a Bio-Rad CFX96 Real-Time PCR System (Bio-Rad,
Hercules, CA, USA). Primer sequences used in the present study were
the following: NEAT1 forward, 5′-CTTCCTCCCTTTAACTTATCCATTCAC-3′ and
reverse, 5′-CTCTTCCTCCACCATTACCAACAATAC-3′; miR-107 forward,
5′-ATGATGAGCAGCATTGTACAGG-3′ and reverse, 5′-GCAGGGTCCGAGGTATTC-3′.
GAPDH was used as an NEAT1 internal control and U6 small nuclear
RNA was used as an miR-107 internal control. Samples were run in
triplicate.
Western blot analysis
Western blotting was performed as previously
described (17,18). Protein extracts were electrophoresed
on a 12% SDS-PAGE and transferred to nitrocellulose membrane. The
following antibodies were used: rabbit anti-CDK6 (1:1,000; Abcam,
Cambridge, MA, USA) and mouse anti-actin (1:3,000; Cell Signaling
Technology, Danvers, MA, USA). Signals were visualized using
enhanced chemiluminescence (ECL) reagents (GE Healthcare,
Barrington, IL, USA) and captured with a FluorChem imager from
Bio-Rad.
Statistical analysis
Statistical analysis was conducted using the SPSS
16.0 software. The results are presented as mean ± SD and analyzed
with the Student's t-test. P<0.05 was denoted as statistically
significant.
Results
Isolation and identification of
GSCs
In the present study, we first isolated GSCs from
human glioma tissues. As CD133 is the most accredited marker for
GSCs (19–22), CD133 was employed to identify GSCs
in the present study. The resected brain glioma tissues were
primary cultured by collagenase I digestion method. We first
analyzed the ratio of CD133+ cells in the primary
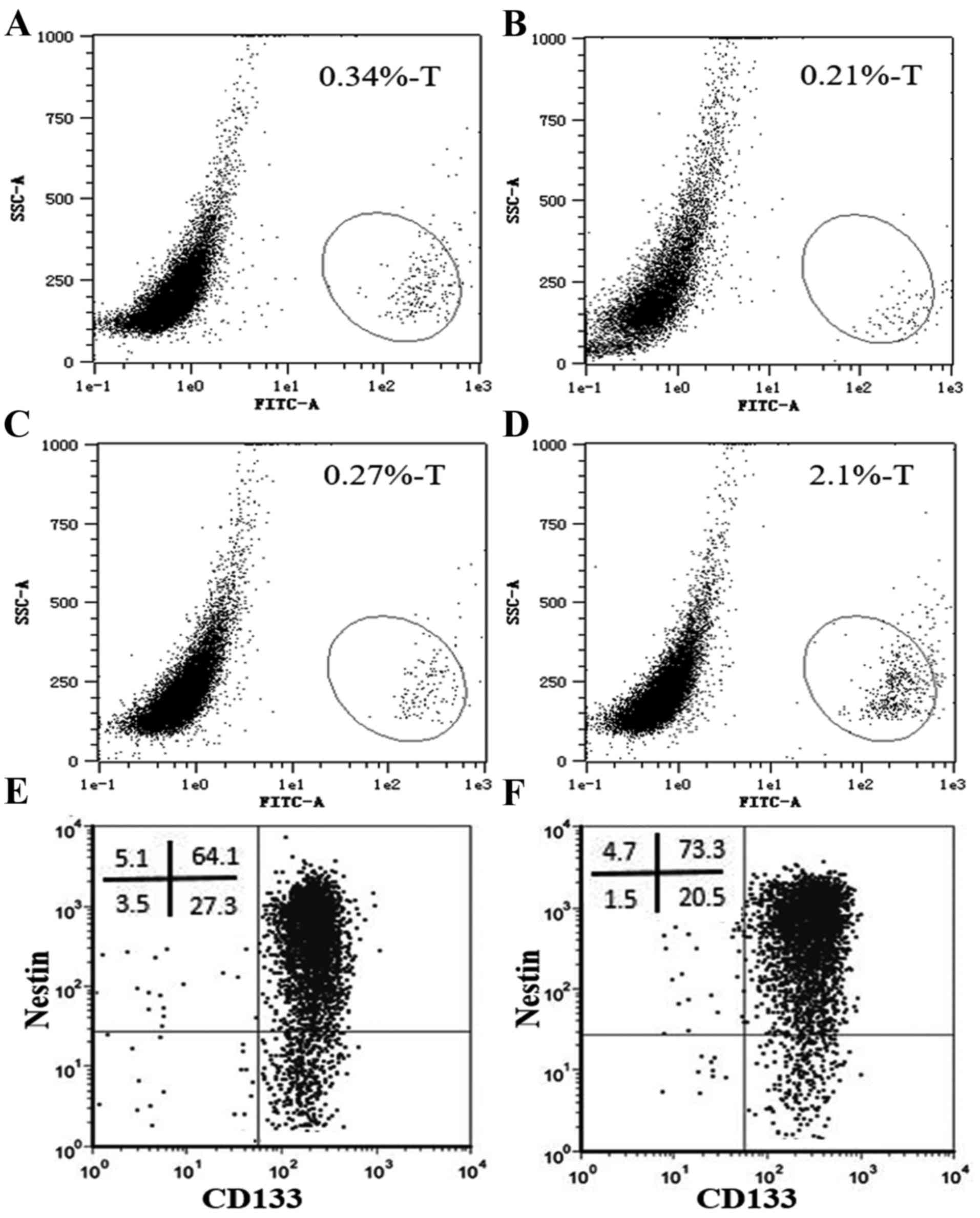
cultured human glioma (Fig. 1A-C)
and U87 cells (Fig. 1D). As shown
in Fig. 1A-C, the ratio of
CD133+ cells in the primary cultured human WHO grade IV
glioma cells was very low and varied greatly in the 3 different
patient tissues. In order to increase cell homogeneity, the U87
human glioma cells and the primary cultured human glioma cells were
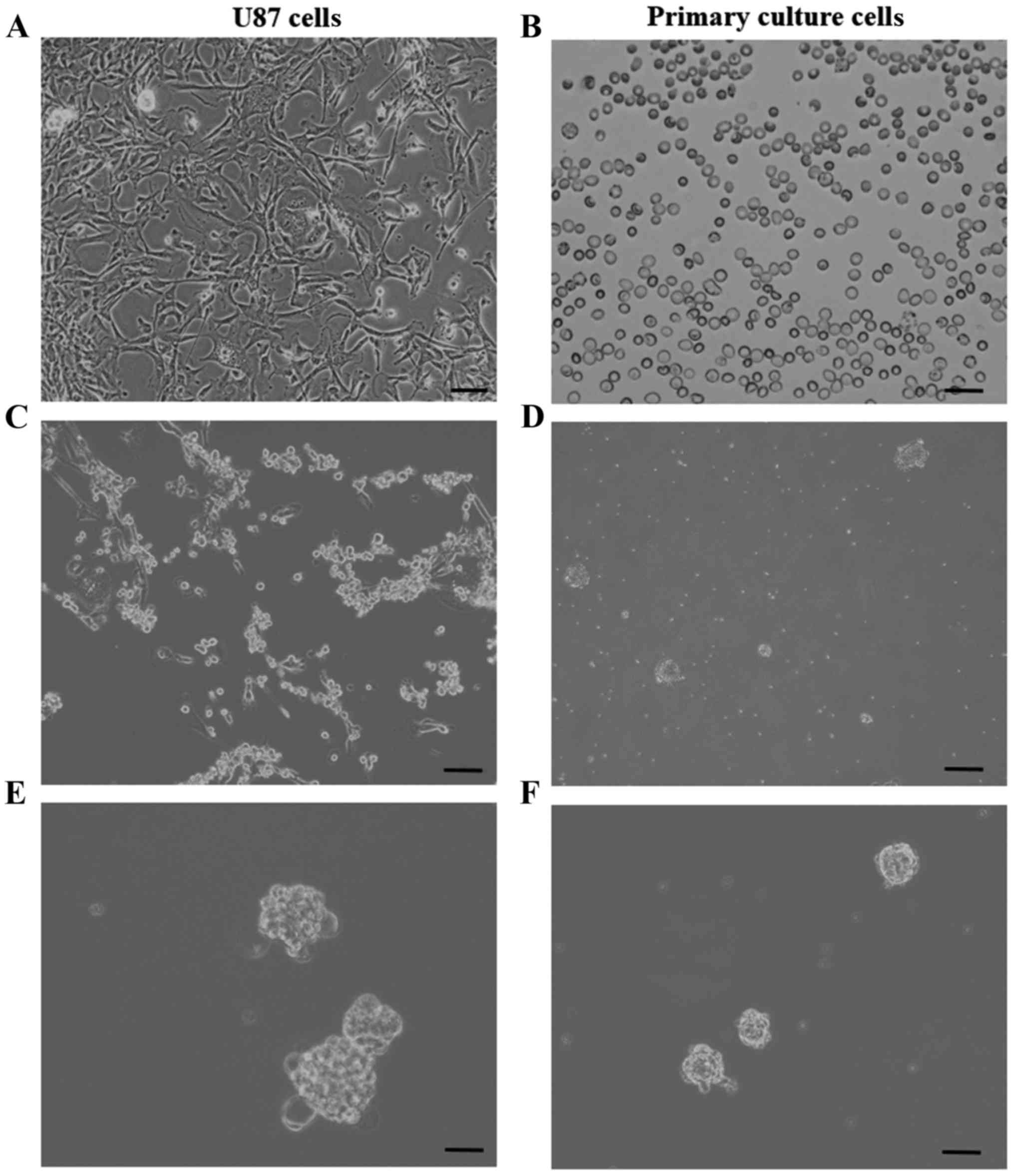
used to isolate GSCs (Fig. 2A and
B). We primary cultured human glioma and U87 cells in
serum-free media containing epidermal growth factor and basic
fibroblast growth factor to preferentially induce stem cell
proliferation and negatively select against differentiated cells
(Fig. 2C and D). As shown in our
previous studies (17,23), the immunomagnetic bead sorting
technique was used to isolate cancer stem cells from human glioma
primary cultured cells and U87 glioma cell line in the present
study.
After isolation, the purity of the isolated
CD133+ cells was confirmed (Fig. 1E and F) and the glioma stem cell
properties were identified. Most of the isolated cells also
displayed nestin expression (Fig. 1E
and F). The neurospheres emerged after one week of culture
(Fig. 2E and F). Furthermore, the
neurospheres had high frequencies of secondary neurosphere
formation. Moreover, when spheroid cells were cultured under
differentiation conditions for 3 days, the neurosphere cells were
adherent to the culture plate and grew in monolayer. On the basis
of this evidence and morphological aspect, most of the
CD133+ isolated cells displayed glioma stem cell
properties: self-renewal and self-differentiation abilities.
CD133+ human glioma primary
culture stem cells display higher NEAT1 expression levels
NEAT1 has been reported to be correlated with higher
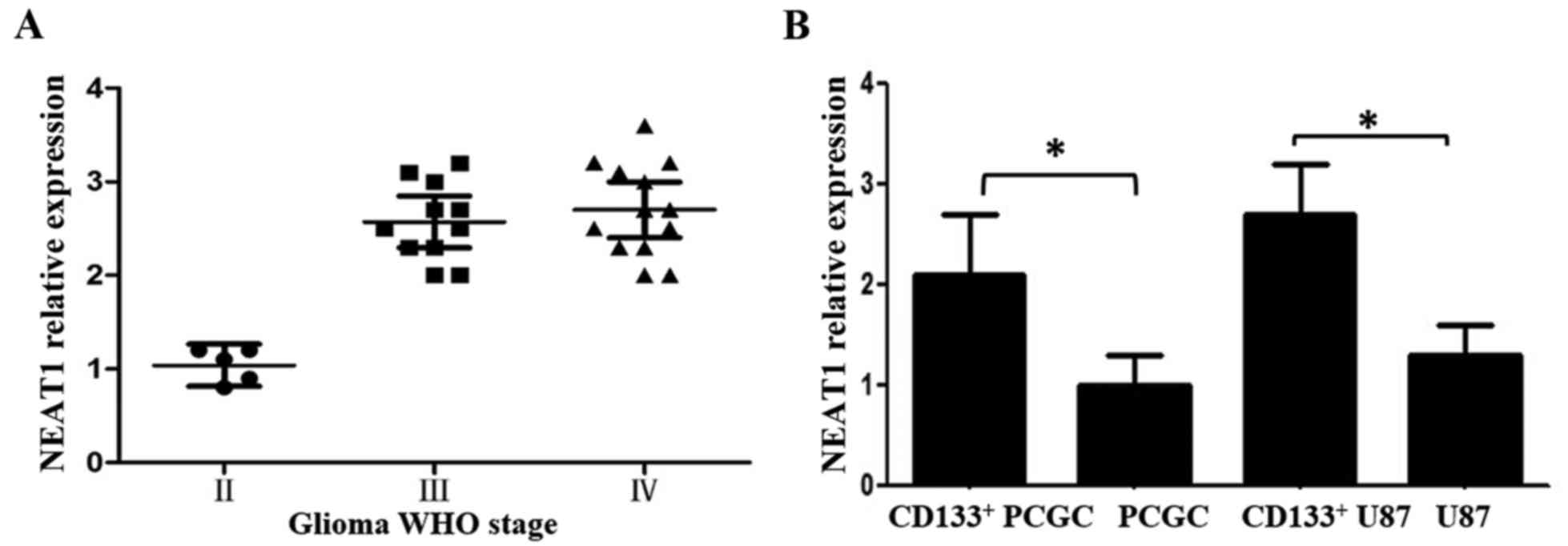
WHO grade human glioma tissues (12). Therefore, in the present study, we
examined the NEAT1 expression levels in 29 human glioma tissues (5
WHO II, 11 WHO III, 13 WHO IV grade) using PCR. The WHO III and IV
glioma tissues displayed higher NEAT1 levels than the WHO II grade
tissues (Fig. 3A). However,
contrary to what we expected, there was no significant difference
between the WHO III and IV glioma tissues, which was inconsistent
with previous studies (12,16,24).
Next, we determined the NEAT1 expression levels in
CD133+ human GSCs and human glioma primary culture
cells. We found that the CD133+ human GSCs displayed
significantly higher NEAT1 levels than the human glioma primary
culture cells. Being similar to the human tissues, the
CD133+ U87 cells displayed higher NEAT1 levels than the
regular U87 cells (Fig. 3B).
NEAT1 knockdown reduces proliferation
while induces cell cycle arrest and apoptosis of CD133+
U87 cells
In order to define the role of NEAT1 in the
CD133+ GSCs, we transfected the CD133+ human
GSCs with siNEAT1 and scramble siRNA. However, we found that the
CD133+ human GSCs from different tissues displayed
variable transfection efficiency. Due to this reason, the
CD133+ U87 cells were transfected and the efficiency was
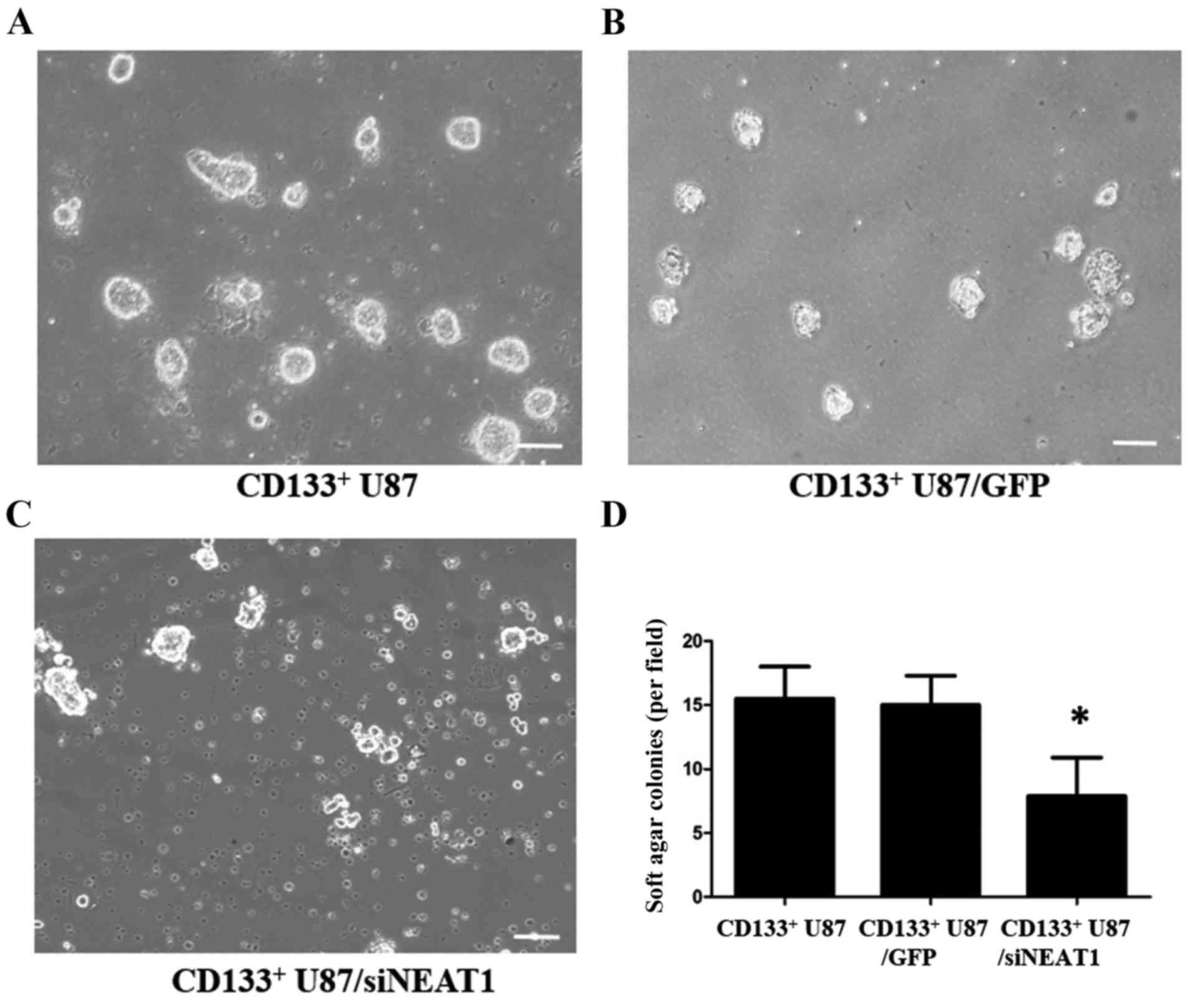
confirmed. Then, we conducted the soft agar colony formation
assays. We found that the proliferation of NEAT1-knockdown
CD133+ U87 cells was significantly lower than the
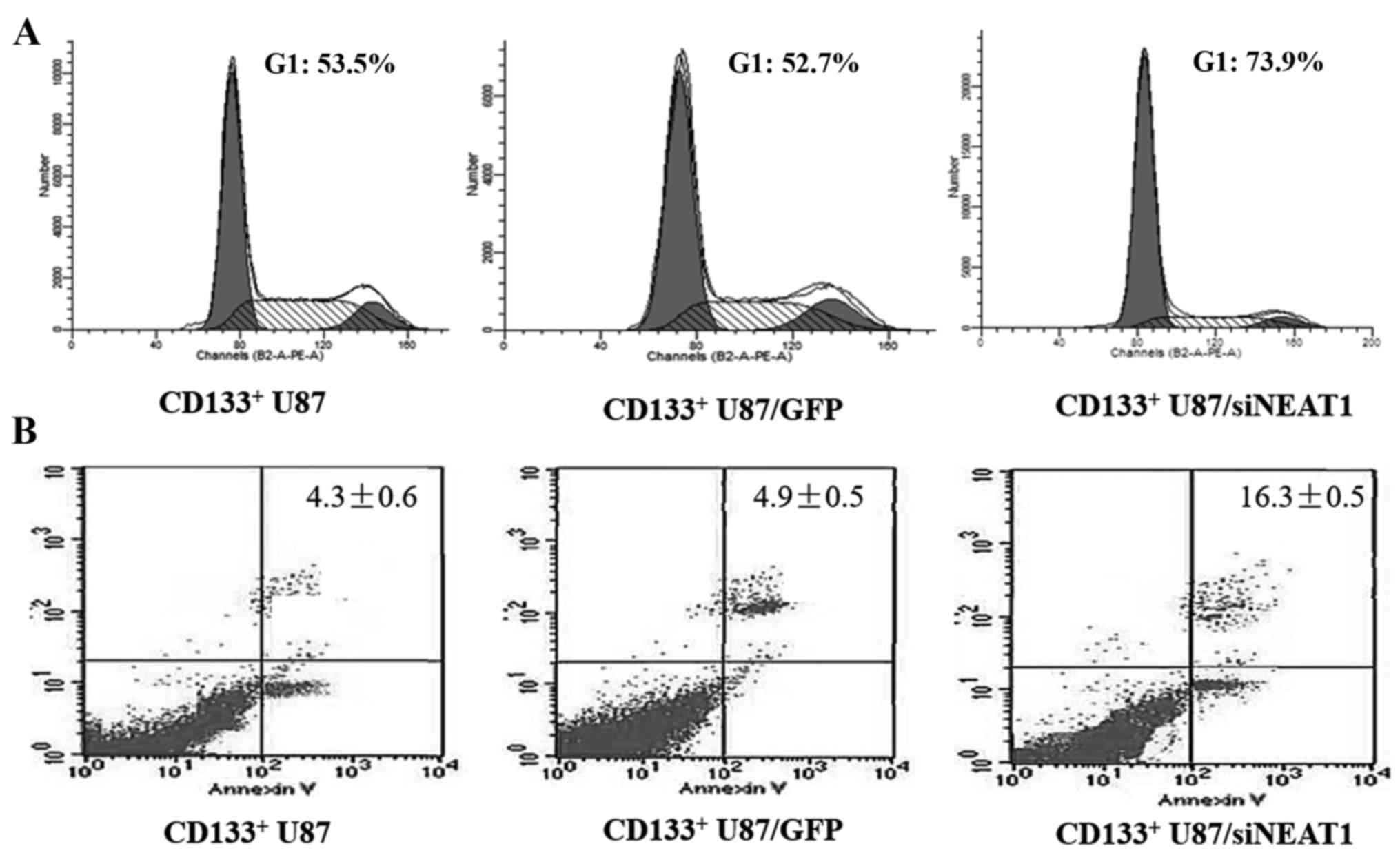
relative control group (Fig. 4). To
further investigate the influence of NEAT1 on the GSCs, flow
cytometry was also used to examine the cell cycle distribution and
cell apoptosis. We found an increased apoptosis rate and an
increased G1 phase accumulation in the NEAT1-knockdown
CD133+ U87 cells compared with normal CD133+
U87 and control CD133+ U87/GFP cells (Fig. 5A and B).
NEAT1 regulates CDK6 expression by
modulating miR-107
Recent study have shown that NEAT1 may regulate
miR-107, which in turn directly modulates CDK6 expression by
binding to the miR-107 seed complementary site located in 3′UTR of
CDK6 (15). CDK6 is known to be an
important regulator of cell cycle progression, modulating cell
cycle G1 phase progression and G1/S transition (25). In order to further explore the
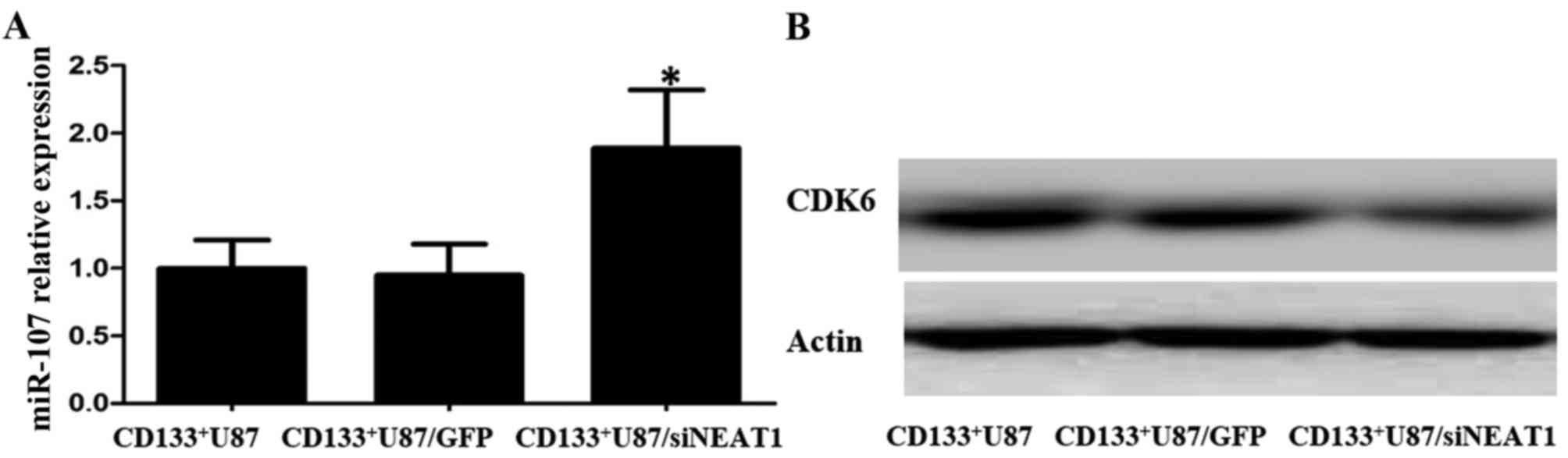
mechanistic base of NEAT1 in GSCs, we verified the miR-107 RNA and
CDK6 protein levels in NEAT1-knockdown CD133+ U87 cells.
We found increased miR-107 RNA levels and decreased CDK6 protein
levels in the NEAT1-knockdown CD133+ U87 cells compared
with these levels in the normal CD133+ U87 and control
CD133+ U87/GFP cells (Fig.
6A and B). These results suggest that the NEAT1/miR-107/CDK6
axis may also exist in U87 GSCs.
Discussion
Glioma is a highly fatal disease and a major
contributor to death in patients with central nervous system tumors
(2,3,5,26).
Glioma is asymptomatic in the early stage and is generally detected
with extensive invasion into adjacent normal brain tissue (26). Accumulating evidence shows that
glioma stem cells (GSCs) are responsible for tumor initiation,
relapse and therapeutic resistance (27–30).
Thus, novel therapeutic approaches targeting GSCs are urgently
needed. Recently, several studies have demonstrated the regulation
of specific lncRNAs in cancer biology including proliferation,
apoptosis, tumor metabolism and invasiveness (9,31–34).
Among these lncRNAs, NEAT1 has earned the reputation as a
transcriptional regulator for numerous genes. Neat1 plays an
important role in maintaining the integrity of subnuclear
paraspeckles (35). NEAT1 regulates
HIF-2α in breast cancer cells under hypoxia (36). NEAT1 regulates expression of
prostate cancer genes by altering the epigenetic landscape at
target gene promoters to favor transcription (13). NEAT1 induces tumor cell migration
and invasion in ESCC (10).
Moreover, the miR-449/NEAT1 axis exists in lung cancer cells
regulating the cell growth and apoptosis of lung cancer cell lines
(37). In gliomas, NEAT1 has been
reported to be correlated to larger tumor size, higher WHO grade
and recurrence (12,16,24).
NEAT1 upregulates c-MET promoting cell growth in gliomas (24). However, its possible role in GSCs
has never been explored. Previous studies in our laboratory have
shown that NEAT1 knockdown can also induce the apoptosis of ESCC
cells (10). These findings
prompted us to evaluate the possible role of NEAT1 in GSCs using
the same siRNAs of NEAT1/scramble siRNA system.
In the present study, we first evaluated the NEAT1
levels in 29 human glioma tissues (5 WHO II, 11 WHO III, 13 WHO IV
grade). We found higher NEAT1 levels in WHO grade III and IV glioma
tissues, which is consistent with previous studies (12,16).
However, there was no difference in NEAT1 levels between WHO grade
III and IV glioma tissues. This observation may be due to the
deviation during the process of obtaining samples from specimens,
since the NEAT1 levels in glioma may be not homogeneous. Moreover,
the number of samples was too small to show the real state. More
samples are needed to determine this issue. In addition, many
researchers obtained RNA from wax-embedded tissue specimen. RNA may
be contaminated and damaged during the different steps of the
processing of tissue sections. Freshly resected glioma tissue is
recommended for further research.
To examine the NEAT1 role in GSCs, we first tried to
isolate GSCs from human glioma samples. There are many markers used
to identify and isolate stem cells in glioma, including CD133,
Nestin, CD15, Sox2 and Msi-1 (6,7). At
present, CD133 remains the most generally accepted marker for GSC
isolation (19–22,38,39).
Although no single marker has been shown to be sufficient to confer
stem cell-like properties, most CD133+ cells in GBM
exhibit stem cell properties (20,22,38).
In a previous study in our laboratory, we isolated cancer stem
cells from prostate cancer cells and GSCs using microbead selection
(17,23). In the present study, we used
microbeads to isolate CD133 and nestin GSCs. However, due to our
limited isolation technologies, we only successfully isolated
CD133+ cells from human glioma primary culture cells. As
nestin is another well accepted marker for GSCs (20), we also examined nestin expression in
the CD133+ cells isolated from our human glioma primary
culture cells. We found that most of the CD133+ cells
were also nestin-positive cells. In addition, the CD133+
cells from human glioma primary culture cells still exhibited stem
cell properties including glioma sphere formation, glioma sphere
proliferation/generation and cell differentiation. After isolation,
we found that CD133+ GSCs had higher NEAT1 levels than
the human glioma primary culture cells. This observation suggests
the NEAT1 may play a role in maintaining glioma stem cell
properties.
Based on these observation, we further investigated
the NEAT1 influence on the GSCs. We first tried to use GSCs from
primary culture of glioma tissue. However, we found that after
NEAT1 knockdown was carried out in GSCs from the primary culture
cells, most of the GSCs were unable to generate glioma sphere and
many cells grew into differentiated cells. These indicates that
NEAT1 is needed for maintained stemness. Another explanation for
these phenomena may be our immature GSC transfection technology.
However, as aforementioned, we also found that the transfection
efficiency in GSCs from different patients varied greatly. Due to
the great heterogeneity, we isolated GSCs from U87 glioma cells to
obtain homogeneity. After isolation and confirmation, we
transfected U87 GSCs with siNEAT1 or scramble siRNA in serum-free
medium. We found that NEAT1 knockdown reduced cell proliferation.
NEAT1 knockdown also reduced the speed of glioma sphere formation,
and some of the NEAT1-knockdown cells grew into differentiated
cells. This observation is in accordance with that of the
CD133+ human glioma primary culture cells, indicating
the possible role of NEAT1 in maintaining stemness in GSCs.
A previous study showed that NEAT1 can modulate CDK6
through miR-107, which in turn regulates the cell cycle in human
laryngeal squamous cell cancer (15). We found decreased CDK6 protein
levels and increased miR-107 RNA levels in the NEAT1-knockdown
cells. These results suggest the existence of the
NEAT1/miR-107/CDK6 axis in GSCs. Since CDK6 is a regulator of the
cell cycle, we further examined the cell cycle in NEAT1-knockdown
cells. We found G1 phase arrest in most of the NEAT1-knockdown
GSCs. This may explain the reason why NEAT1 knockdown reduced the
proliferation of GSCs and promoted GSC to grow into differentiated
cells.
To date, most patients with glioma receive combined
therapy with little efficacy (2).
Since GSCs are responsible for tumor maintenance, recurrence and
resistance to conventional treatments, therapy targeting NEAT1 may
hold great promise for glioma treatment (5,26). In
the present study, we found that GSCs had higher NEAT1 levels than
the relative differentiated cells. We demonstrated for the first
time that NEAT1 may play a role in maintaining stemness in GSCs.
Consistent with previous study, NEAT1-knockdown cells displayed
increased miR-107 RNA levels and decreased CDK6 protein levels.
These verified the NEAT1/miR-107/CDK6 axis in GSCs, indicating that
NEAT1 may modulate CDK6 by miR-107, which in turn regulates the
cell cycle in GSCs. Previous studies showed that NEAT1 altered the
epigenetic landscape of target gene promoters (11,15,36,37).
However, in addition to the miR-107/CDK6 axis, there must be many
other target genes regulated by NEAT1, which need further
study.
There are many inherent limitations in the present
study. We only isolated GSCs via CD133 selection. Previous data
advocate several cell markers for GSCs. Although in accordance with
previous studies (19,20,22),
CD133+ human glioma primary culture and
CD133+ U87 cancer stem cells exhibit GSC properties,
further cell marker selection may improve the purity and stemness
of the isolated GSCs. NEAT1 has been reported to alter many gene
promoters, which regulate cancer biology (36). However, in addition to the
miR-107/CDK6 axis, many other target genes are needed to be
explored by further research. In addition, the present study is
lacking direct evidence in vivo, which is ongoing in our
laboratory. Nevertheless, the present study demonstrated for the
first time that GSCs have higher NEAT1 levels than the relative
differentiated cells and NEAT1 may play a role in maintaining
stemness in GSCs. The NEAT1/miR-107/CDK6 axis may be involved in
mediating the cell cycle of GSCs. The signaling pathway involved in
the regulation of cancer biology by NEAT1 in GSCs warrants further
exploration.
Acknowledgements
The present study was supported by joint funds of
The National Natural Science foundation of China (grant no.
U1404822).
References
|
1
|
Khan IS and Ehtesham M: Emerging
strategies for the treatment of tumor stem cells in central nervous
system malignancies. Adv Exp Med Biol. 853:167–187. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chien LN, Ostrom QT, Gittleman H, Lin JW,
Sloan AE, Barnett GH, Elder JB, McPherson C, Warnick R, Chiang YH,
et al: International differences in treatment and clinical outcomes
for high grade glioma. PLoS One. 10:e01296022015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Mannas JP, Lightner DD, Defrates SR,
Pittman T and Villano JL: Long-term treatment with temozolomide in
malignant glioma. J Clin Neurosci. 21:121–123. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Perry A and Wesseling P: Histologic
classification of gliomas. Handb Clin Neurol. 134:71–95. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Reulen HJ, Poepperl G, Goetz C, Gildehaus
FJ, Schmidt M, Tatsch K, Pietsch T, Kraus T and Rachinger W:
Long-term outcome of patients with WHO Grade III and IV gliomas
treated by fractionated intracavitary radioimmunotherapy. J
Neurosurg. 123:760–770. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Modrek AS, Bayin NS and Placantonakis DG:
Brain stem cells as the cell of origin in glioma. World J Stem
Cells. 6:43–52. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Dietrich J, Diamond EL and Kesari S:
Glioma stem cell signaling: Therapeutic opportunities and
challenges. Expert Rev Anticancer Ther. 10:709–722. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Qi P and Du X: The long non-coding RNAs, a
new cancer diagnostic and therapeutic gold mine. Mod Pathol.
26:155–165. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Li X, Wu Z, Fu X and Han W: Long noncoding
RNAs: Insights from biological features and functions to diseases.
Med Res Rev. 33:517–553. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chen X, Kong J, Ma Z, Gao S and Feng X: Up
regulation of the long non-coding RNA NEAT1 promotes esophageal
squamous cell carcinoma cell progression and correlates with poor
prognosis. Am J Cancer Res. 5:2808–2815. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Li Y, Li Y, Chen W, He F, Tan Z, Zheng J,
Wang W, Zhao Q and Li J: NEAT expression is associated with tumor
recurrence and unfavorable prognosis in colorectal cancer.
Oncotarget. 6:27641–27650. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
He C, Jiang B, Ma J and Li Q: Aberrant
NEAT1 expression is associated with clinical outcome in high grade
glioma patients. APMIS. 124:169–174. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Chakravarty D, Sboner A, Nair SS,
Giannopoulou E, Li R, Hennig S, Mosquera JM, Pauwels J, Park K,
Kossai M, et al: The oestrogen receptor alpha-regulated lncRNA
NEAT1 is a critical modulator of prostate cancer. Nat Commun.
5:53832014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Blume CJ, Hotz-Wagenblatt A, Hüllein J,
Sellner L, Jethwa A, Stolz T, Slabicki M, Lee K, Sharathchandra A,
Benner A, et al: p53-dependent non-coding RNA networks in chronic
lymphocytic leukemia. Leukemia. 29:2015–2023. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wang P, Wu T, Zhou H, Jin Q, He G, Yu H,
Xuan L, Wang X, Tian L, Sun Y, et al: Long noncoding RNA NEAT1
promotes laryngeal squamous cell cancer through regulating
miR-107/CDK6 pathway. J Exp Clin Cancer Res. 35:222016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Liu Q, Sun S, Yu W, Jiang J, Zhuo F, Qiu
G, Xu S and Jiang X: Altered expression of long non-coding RNAs
during genotoxic stress-induced cell death in human glioma cells. J
Neurooncol. 122:283–292. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang X, Ma Z, Xiao Z, Liu H, Dou Z and Shi
H Fengxand: Chk1 knockdown confers radiosensitization in prostate
cancer stem cells. Oncol Rep. 28:2247–2254. 2012.PubMed/NCBI
|
|
18
|
Chen W, Xiao Z, Zhao Y, Huang L and Du G:
HIF-1α inhibition sensitizes pituitary adenoma cells to
temozolomide by regulating MGMT expression. Oncol Rep.
30:2495–2501. 2013.PubMed/NCBI
|
|
19
|
Lathia JD, Hitomi M, Gallagher J, Gadani
SP, Adkins J, Vasanji A, Liu L, Eyler CE, Heddleston JM, Wu Q, et
al: Distribution of CD133 reveals glioma stem cells self-renew
through symmetric and asymmetric cell divisions. Cell Death Dis.
2:e2002011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Dahlrot RH, Hansen S, Jensen SS, Schrøder
HD, Hjelmborg J and Kristensen BW: Clinical value of CD133 and
nestin in patients with glioma: A population-based study. Int J
Clin Exp Pathol. 7:3739–3751. 2014.PubMed/NCBI
|
|
21
|
Clément V, Dutoit V, Marino D, Dietrich PY
and Radovanovic I: Limits of CD133 as a marker of glioma
self-renewing cells. Int J Cancer. 125:244–248. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Griguer CE, Oliva CR, Gobin E, Marcorelles
P, Benos DJ, Lancaster JR Jr and Gillespie GY: CD133 is a marker of
bioenergetic stress in human glioma. PLoS One. 3:e36552008.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wu J, Lai G, Wan F, Xiao Z, Zeng L, Wang
X, Ye F and Lei T: Knockdown of checkpoint kinase 1 is associated
with the increased radiosensitivity of glioblastoma stem-like
cells. Tohoku J Exp Med. 226:267–274. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhen L, Yun-Hui L, Hong-Yu D, Jun M and
Yi-Long Y: Long noncoding RNA NEAT1 promotes glioma pathogenesis by
regulating miR-449b-5p/c-Met axis. Tumour Biol. 37:673–683. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Rader J, Russell MR, Hart LS, Nakazawa MS,
Belcastro LT, Martinez D, Li Y, Carpenter EL, Attiyeh EF, Diskin
SJ, et al: Dual CDK4/CDK6 inhibition induces cell-cycle arrest and
senescence in neuroblastoma. Clin Cancer Res. 19:6173–6182. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Brzozowska A, Toruń A and Mazurkiewicz M:
The impact of surgery on the efficacy of adjuvant therapy in
glioblastoma multiforme. Adv Clin Exp Med. 24:279–287. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Jung J, Gilbert MR and Park DM: Isolation
and propagation of glioma stem cells from acutely resected tumors.
Methods Mol Biol. 1516:361–369. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Codrici E, Enciu AM, Popescu ID, Mihai S
and Tanase C: Glioma stem cells and their microenvironments:
Providers of challenging therapeutic targets. Stem Cells Int.
2016:57284382016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Liebelt BD, Shingu T, Zhou X, Ren J, Shin
SA and Hu J: Glioma stem cells: Signaling, microenvironment, and
therapy. Stem Cells Int. 2016:78498902016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Kim SS, Pirollo KF and Chang EH: Isolation
and culturing of glioma cancer stem cells. Curr Protoc Cell Biol.
67:23.10.1–23.10.10. 2015.doi: 10.1002/0471143030.cb2310s67.
View Article : Google Scholar
|
|
31
|
Han L, Zhang K, Shi Z, Zhang J, Zhu J, Zhu
S, Zhang A, Jia Z, Wang G, Yu S, et al: LncRNA profile of
glioblastoma reveals the potential role of lncRNAs in contributing
to glioblastoma pathogenesis. Int J Oncol. 40:2004–2012.
2012.PubMed/NCBI
|
|
32
|
Sørensen KP, Thomassen M, Tan Q, Bak M,
Cold S, Burton M, Larsen MJ and Kruse TA: Long non-coding RNA
expression profiles predict metastasis in lymph node-negative
breast cancer independently of traditional prognostic markers.
Breast Cancer Res. 17:552015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhang X, Sun S, Pu JK, Tsang AC, Lee D,
Man VO, Lui WM, Wong ST and Leung GK: Long non-coding RNA
expression profiles predict clinical phenotypes in glioma.
Neurobiol Dis. 48:1–8. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Li R, Qian J, Wang YY, Zhang JX and You
YP: Long noncoding RNA profiles reveal three molecular subtypes in
glioma. CNS Neurosci Ther. 20:339–343. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Clemson CM, Hutchinson JN, Sara SA,
Ensminger AW, Fox AH, Chess A and Lawrence JB: An architectural
role for a nuclear noncoding RNA: NEAT1 RNA is essential for the
structure of paraspeckles. Mol Cell. 33:717–726. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Choudhry H, Albukhari A, Morotti M, Haider
S, Moralli D, Smythies J, Schödel J, Green CM, Camps C, Buffa F, et
al: Tumor hypoxia induces nuclear paraspeckle formation through
HIF-2α dependent transcriptional activation of NEAT1 leading to
cancer cell survival. Oncogene. 34:45462015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
You J, Zhang Y, Liu B, Li Y, Fang N, Zu L,
Li X and Zhou Q: MicroRNA-449a inhibits cell growth in lung cancer
and regulates long noncoding RNA nuclear enriched abundant
transcript 1. Indian J Cancer. 51:(Suppl 3). e77–e81. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Campos B and Herold-Mende CC: Insight into
the complex regulation of CD133 in glioma. Int J Cancer.
128:501–510. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zhang M, Song T, Yang L, Chen R, Wu L,
Yang Z and Fang J: Nestin and CD133: Valuable stem cell-specific
markers for determining clinical outcome of glioma patients. J Exp
Clin Cancer Res. 27:852008. View Article : Google Scholar : PubMed/NCBI
|