Introduction
Acute lymphoblastic leukemia (ALL) is one of the
most common childhood (0–15 years of age) hematologic malignancies
(1,2). In recent years, with more and more
in-depth studies of ALL, the survival rate of ALL patients has been
significantly improved (3).
Complete remission (CR) has been attained in more than 95% of cases
and the 5-year event-free survival (EFS) has reached 63–83% in
pediatric ALL patients (4), while,
the CR and 3–5-year EFS of adult ALL patients have reached 75–89%
and 40%, respectively (5).
Nevertheless, numerous patients still suffer from the adverse
events caused by conventional treatment and die from relapse
(6). Therefore, a better
understanding of the mechanism underlying ALL and development of
new strategies for improving efficiency of ALL therapy are
required. Emerging evidence indicates that insulin-like growth
factor binding protein 3 (IGFBP3) is inversely associated with
leukemia (7). Low IGFBP3 is related
to the high-risk of events such as recurrence and decreased
remission at the time of diagnosis (6), suggesting that the downregulation of
expression of IGFBP3 plays an important role in the development of
ALL.
MicroRNAs (miRNAs) are a family of endogenous,
conserved, small non-coding RNAs (20–25 nucleotides in length). The
complementary messenger RNAs (mRNAs) can be directly targeted on
the 3′-untranslated regions (3′-UTRs) and suppressed by miRNAs in
eukaryotes (8,9). Altered expression of miRNAs
participates in a variety of biological processes such as
carcinogenesis, immunity, infection, endocrine homeostasis,
differentiation and apoptosis (10,11).
By targeting complementary genes to control the expression of
tumor-suppressor or oncogenic proteins, miRNAs are considered to
play a significant role in the biology of cancers and to regulate
cell proliferation, migration, invasion and apoptosis in cancers
(12), thereby suggesting that a
promising alternative novel approach for cancer treatment may be
provided by miRNAs. It was reported that miR-196b is one of the
most upregulated miRNAs in T-cell ALL (T-ALL) (13,14).
In addition, regardless of treatment protocol, miR-1290 is capable
to serve as a new biomarker in childhood B-cell ALL (B-ALL)
patients for outcome (3). However,
the detailed regulatory mechanism of miR-196b or miR-1290 in ALL is
still not well understood.
Numerous chemotherapeutic and chemopreventive
compounds have been developed from natural sources and offer
potential new alternatives to treat cancers (15). Resveratrol
(3,5,40-trihydroxy-trans-stilbene), a natural polyphenol, is
widely used in Traditional Chinese medicines (TCMs; such as
Polygonum cuspidatum and Rheum officinale Baill.) and
is found in peanuts, blueberries, cranberries, red wine and grape
skin (16,17). Accumulating research suggests that
resveratrol has a number of important pharmacological properties
such as antiproliferative, antioxidant, cardio-protective and
anti-inflammatory activities (18–20).
Resveratrol also displays anticancer activities by disturbing the
three stages of carcinogenesis: initiation, promotion and
progression (21). Previous studies
have demonstrated that resveratrol inhibited the cell growth and
induced apoptosis in several ALL cell lines, suggesting the
anti-ALL effect of this agent (22–25).
Nevertheless, the molecular mechanism of resveratrol-mediated
anti-ALL activity has not been fully elucidated.
The present study aimed to ascertain whether
miR-196b and miR-1290 serve as novel targets involved in the
antitumor effect of resveratrol in ALL and to explore the probable
common regulatory mechanism focusing on IGFBP3.
Materials and methods
Clinical samples
Peripheral blood and bone marrow samples were
collected from 15 pairs of ALL patients and healthy volunteers at
the Department of Hematology, Guangzhou First People's Hospital,
Guangzhou Medical University, Guangzhou, Guangdong, China. Density
gradient separation was used to isolate the human peripheral blood
mononuclear cells (PBMCs) from whole blood by Ficoll-Paque Plus (GE
Healthcare Bio-Sciences AB, Uppsala, Sweden) and the samples were
then cryopreserved in liquid nitrogen with 90% fetal bovine serum
(FBS) (Gibco, Carlsbad, CA, USA) and 10% dimethyl sulfoxide (DMSO)
until analyzed.
Ethics statements
Permission to use the human bone marrow and
peripheral blood samples for the present study was approved by the
Ethics Committee of Guangzhou First People's Hospital (Guangdong,
China).
Cell lines and cell culture
American Type Culture Collection (ATCC) (Manassas,
VA, USA) provided the human embryonic kidney 293T, T-ALL TALL-104
and B-ALL SUP-B15 cells. 293T cells were cultured in Dulbecco's
modified Eagle's medium (DMEM) with 10% FBS and 2 mM L-glutamine
(Invitrogen Life Technologies, Carlsbad, CA, USA). TALL-104 cells
were cultured in complete medium [ATCC-formulated Iscove's modified
Dulbecco's medium (IMDM) with 20% FBS, supplemented with 2.5 mg/ml
human albumin, 0.5 mg/ml D-mannitol and 100 U recombinant human
IL-2 (all from Sigma-Aldrich, St. Louis, MO, USA)]. SUP-B15 cells
were cultured in IMDM, supplemented with 10% FBS, 2 mM L-glutamine,
0.05 µM 2-β-mercaptoethanol (Sigma-Aldrich), 100 µg/ml streptomycin
and 100 U/ml penicillin (Gibco). The cells were maintained at 37°C
in a humidified atmosphere containing 5% CO2.
Reagents
Resveratrol was obtained from Sigma-Aldrich. IGFBP3
siRNA and negative control siRNA were purchased from GenePharma
(Shanghai, China). The antibody against IGFBP3, caspase-3 and GAPDH
were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA).
The Cell Counting Kit-8 (CCK-8) was purchased from Dojindo
Molecular Technologies (Kumamoto, Japan).
Cell proliferation assay
The cells were seeded into 96-well cell culture
plates, and were incubated at 37°C for 0, 24, 48 or 72 h in 5%
CO2. Cell proliferation was assessed via the CCK-8
assay. The numerical values obtained on an enzyme-labeled
instrument (Thermo Fisher Scientific, Germany) with 450 nm
wavelength were used to compare the cell viability.
Flow cytometry
Cells were collected, washed in ice-cold
phosphate-buffered saline (PBS) and fixed in ice-cold 70% ethanol
(4°C, overnight). After centrifugation (1,000 rpm, 5 min), the
cells were diluted with PBS and re-centrifuged. For the cell cycle
assay, the cells were stained using a cell cycle kit (LiankeBio,
Zhejiang, China) and incubated in the dark at 37°C for 30 min. For
analysis of apoptosis, the cells were stained using the Annexin
V-FITC apoptosis detection kit (LiankeBio) and incubated in the
dark at room temperature for 15 min. Stained cells were detected
via flow cytometry with a BD FACSCalibur (BD Biosciences,
Heidelberg, Germany).
Cell migration assay
The migration of cells was performed in a Boyden
Transwell chamber (Millipore, Bedford, MA, USA) containing a
polycarbonate filter with a pore size of 8-µm. A cell suspension
(0.2 ml) (1×105 cells/ml) was added to the upper
compartment of each chamber lined with an uncoated membrane. The
bottom chamber was filled with 0.6 ml IMDM containing 10% FBS as a
chemoattractant. After incubation for 48 h at 37°C with 5%
CO2, the non-filtered cells were gently removed with a
cotton swab and fixed with 4% paraformaldehyde. Filtered cells on
the lower surface of the chamber were stained with 0.1% crystal
violet (Sigma-Aldrich) and quantified manually in five random
fields under a microscope (Olympus, Tokyo, Japan).
Quantitative real-time RT-PCR
(qRT-PCR)
Total RNA was extracted from the cells with TRIzol
reagent (Invitrogen, Carlsbad, CA, USA) according to the
manufacturer's instructions. The mRNA expression of IGFBP3,
miR-196b or miR-1290 was detected by qRT-PCR using the standard
SYBR-Green RT-PCR kit (Takara, Tokyo, Japan) following the
manufacturer's manual. Real-time RT-PCR was performed using a
sequence detector (Sigma-Aldrich). Specific primers were obtained
from Genepharma: IGFBP3 forward, 5′-ATAATCATCATCAAGAAAGGGCA-3′ and
reverse, 5′-AGTTCTGGGTATCTGTGCTCTGA-3′; miR-196b forward,
5′-ACACTCCAGCTGGGTAGGTAGTTTCATG-3′ and reverse,
5′-CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGCCCAACAA-3′; miR-1290
forward, 5′-ACACTCCAGCTGGGTGGATTTTTGGATC-3′ and reverse,
5′-CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGTCCCTG-3′. The relative
expression levels were calculated using the 2−ΔΔCt
method.
Western blotting
Protein was extracted from the peripheral blood or
cells using RIPA lysis buffer with a proteinase inhibitor. The
protein concentration in the lysates was quantitated with the BCA
Protein Assay kit (Bio-Rad, Hercules, CA, USA). Proteins were
resolved on 10% SDS-PAGE gels under reducing conditions, followed
by electrophoretic transfer onto polyvinylidene difluoride
membranes (Millipore). Immunoblots were incubated with primary
antibodies against IGFBP3 (1:2,000) or caspase-3 (1:1,000) (both
from Abcam, Cambridge, USA) at 4°C overnight. Immunoreactive bands
were detected using horseradish peroxidase (HRP)-conjugated
secondary antibodies (1:20,000; Boster, Wuhan, China) with the
Western Lightning Chemiluminescence Plus reagent (Perkin-Elmer Life
Sciences, Boston, MA, USA). GAPDH was selected as the reference
protein.
Dual-luciferase reporter assay
Cells were co-transfected with psiCHECK2-IGFBP3
3′-UTR or psiCHECK2-IGFBP3 3′-UTR mutant and miR-196b/miR-1290
mimics. Cells were lysed and the firefly luciferase activity was
detected. Renilla luciferase activity was used for
normalization. The lysate was detected using Dual-Luciferase
Reporter Assay System (Promega, Madison, WI, USA) with a
luminometer (Turner Designs, Sunnyvale, CA, USA).
Immunohistochemistry
Specimens were embedded in paraffin and a rotary
microtome was used (HM355; Microm, Walldorf, Germany) to prepare
serial sections with 3-µm thickness. Some sections were stained
with hematoxylin and eosin (H&E) according to the
manufacturer's protocol (Sigma-Aldrich). Before immunostaining,
antigen retrieval was carried out via the treatment of 0.1% pepsin
with 10 mM HCl at 37°C for 10 min. The slides were incubated with
the monoclonal mouse anti-human IGFBP3 (1:500; Sigma-Aldrich), and
then anti-mouse IgG conjugated to HRP (Santa Cruz Biotechnology)
for immunohistochemistry. The slides were exoposed to
diaminobenzidine for 5 min and counterstained with hematoxylin
(both from Sigma-Aldrich). A microscope (Olympus) was used to
obtain the images.
Short interfering (si)RNA
transfection
Synthetic IGFBP3 siRNA (20 ng) (Ambion, Austin, TX,
USA) and the respective negative control were delivered into
TALL-104 or SUP-B15 cells using Lipofectamine™ RNAiMAX (Life
Technologies Corp., Carlsbad, CA, USA). Briefly, the cells were
seeded into 6-well plates at 30% confluency. On the following day,
IGFBP3 siRNA and the negative control were diluted in serum-free
medium, and incubated with Lipofectamine™ RNAiMAX transfection
reagent for 20 min at room temperature. The plates were gently
swirled when adding the transfection complexes to the cell
cultures. Fresh media were used to replace the culture media after
6 h and then the cells were incubated for 48 h.
Statistical analysis
All data are expressed as the mean ± SD. Student's
t-test was used to evaluate the differences between two groups. For
multiple comparisons, statistically significant differences were
assessed via one-way ANOVA. P-value <0.05 was considered to
indicate a statistically significant.
Results
IGFBP3 expression is decreased in ALL
patients
To explore the role of IGFBP3 in ALL, we initially
examined the protein expression of IGFBP3 in 15 pairs of bone
marrow from ALL patients and the healthy volunteers by
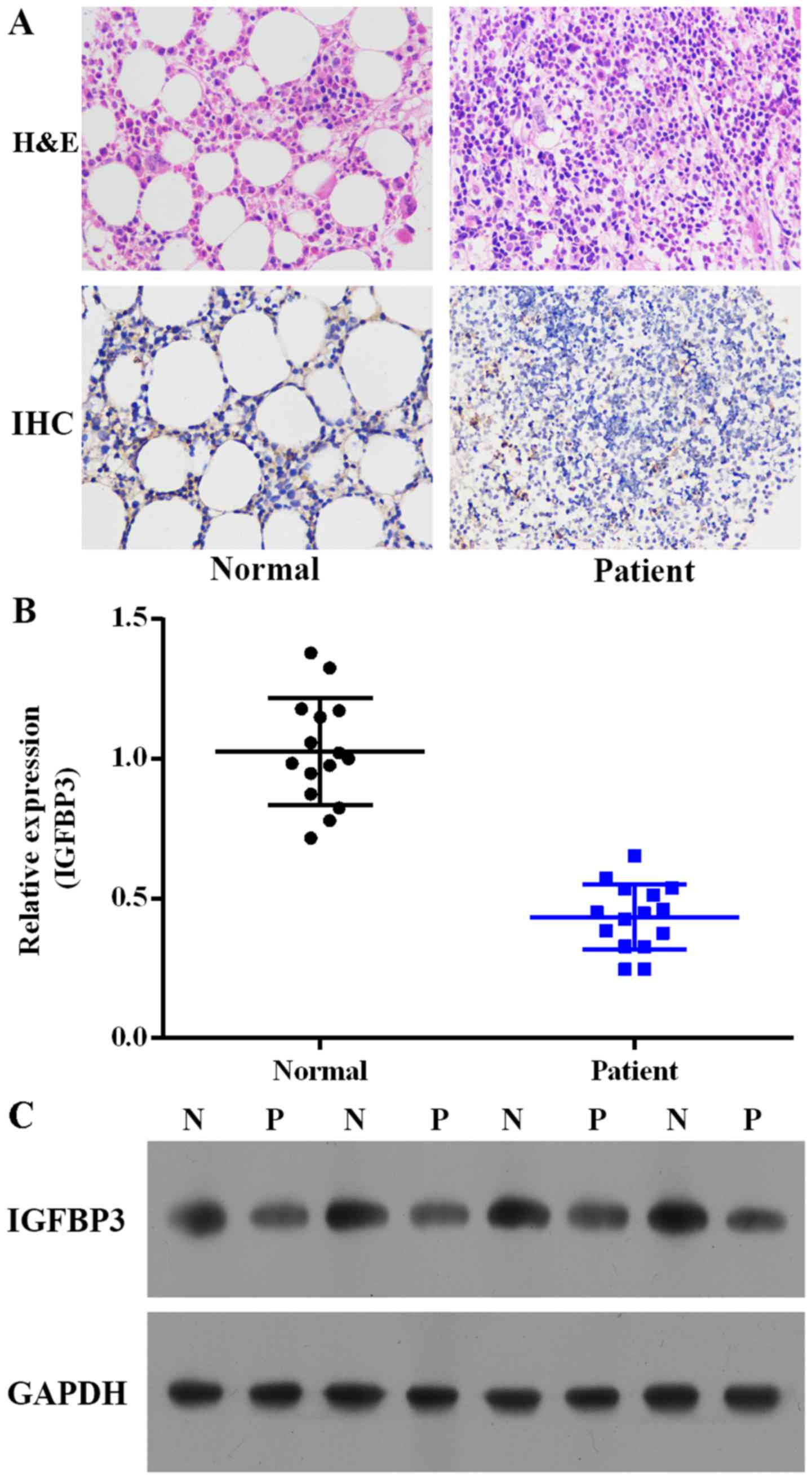
immunohistochemistry. As depicted in Fig. 1A, the IGFBP3 expression in ALL
patients was notably decreased compared with the level in the
healthy volunteers. We further investigated the expression levels
of IGFBP3 mRNA and protein in peripheral blood from the ALL
patients and the healthy volunteers. As depicted in Fig. 1B and C, the mRNA and protein
expression levels of IGFBP3 were decreased in the ALL patients
compared with these levels in the healthy volunteers.
Resveratrol exerts an antitumor effect
by the regulation of miR-196b/miR-1290 in ALL cells
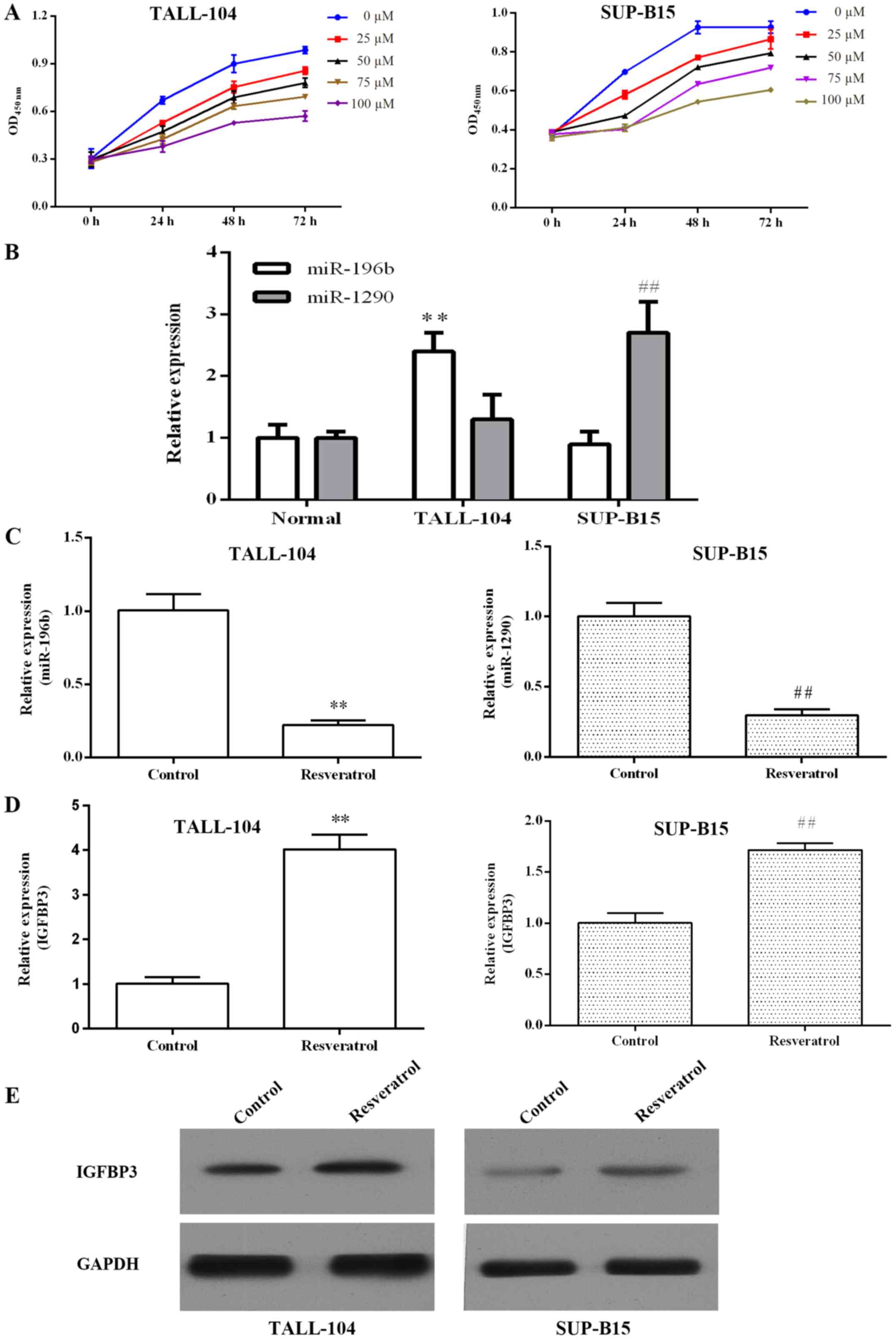
Resveratrol dose- and time-dependently inhibited the
proliferation of TALL-104 and SUP-B15 cells (Fig. 2A). Previous miRNA microarray
profiling indicated that miR-196b was upregulated in T-ALL and
miR-1290 was upregulated in B-ALL. As shown in Fig. 2B, we confirmed that the miR-196b
expression level was significantly increased in TALL-104 cells
compared with the level in the PBMCs (P<0.01), and miR-1290 was
overexpressed in the SUP-B15 cells (P<0.01). qRT-PCR was
performed to investigate whether resveratrol regulates
miR-196b/miR-1290 in ALL cells. As shown in Fig. 2C, resveratrol markedly inhibited
miR-196b/miR-1290 expression in TALL-104/SUP-B15 cells,
respectively. Furthermore, we found that resveratrol elevated
IGFBP3 mRNA and protein expression in both TALL-104 and SUP-B15
cells (Fig. 2D and E).
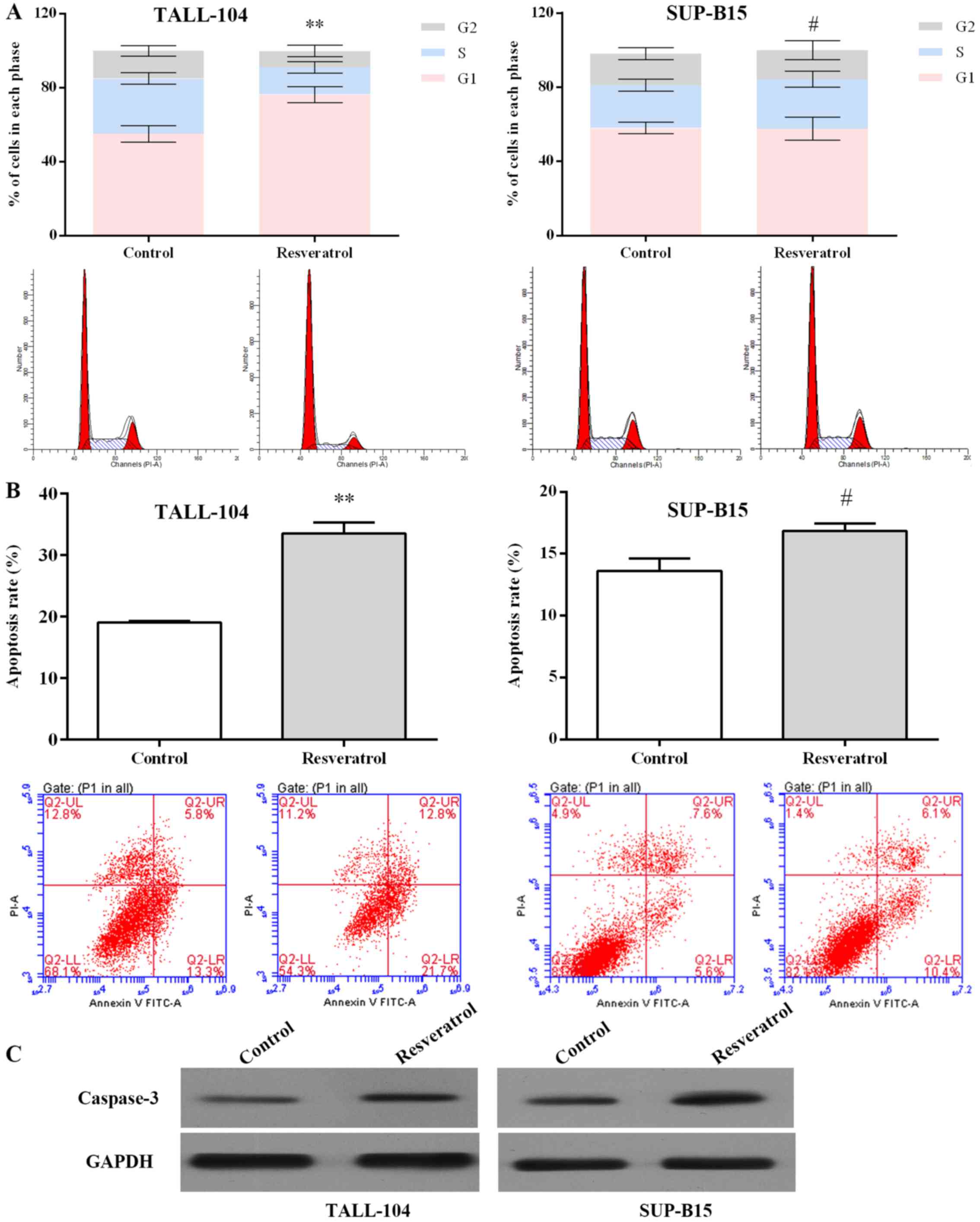
As an miR-196b/miR-1290 inhibitor, resveratrol was
further examined in regards to its antitumor effect. As displayed
in Fig. 3A, resveratrol arrested
the cell cycle at the G1 phase in TALL-104 cells (P<0.01), and
arrested the cell cycle at S phase in SUP-B15 cells (P<0.05).
Resveratrol increased the apoptotic rate in the TALL-104 and
SUP-B15 cells notably when compared with the rate in the control
group (Fig. 3B). Activation of
caspase-3 is significant in apoptosis (26). As shown in Fig. 3C, resveratrol markedly upregulated
the caspase-3 expression in both TALL-104 and SUP-B15 cells.
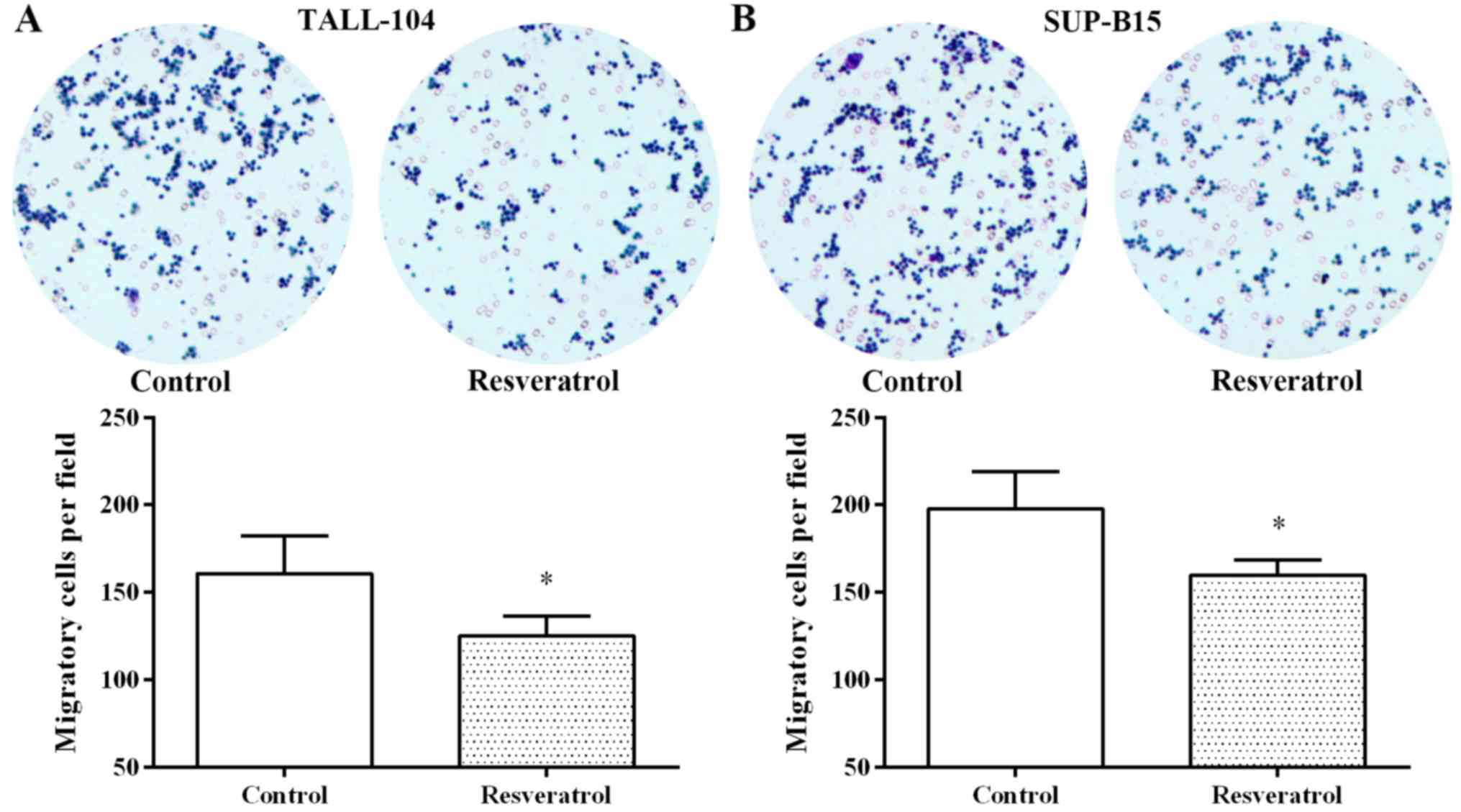
Furthermore, resveratrol also notably inhibited cell migration in
the TALL-104 and SUP-B15 cells (Fig.
4). These findings suggest that resveratrol exerted an anti-ALL
effect by regulating miR-196b/miR-1290.
Both miR-196b and miR-1290 target
IGFBP3 in ALL cells
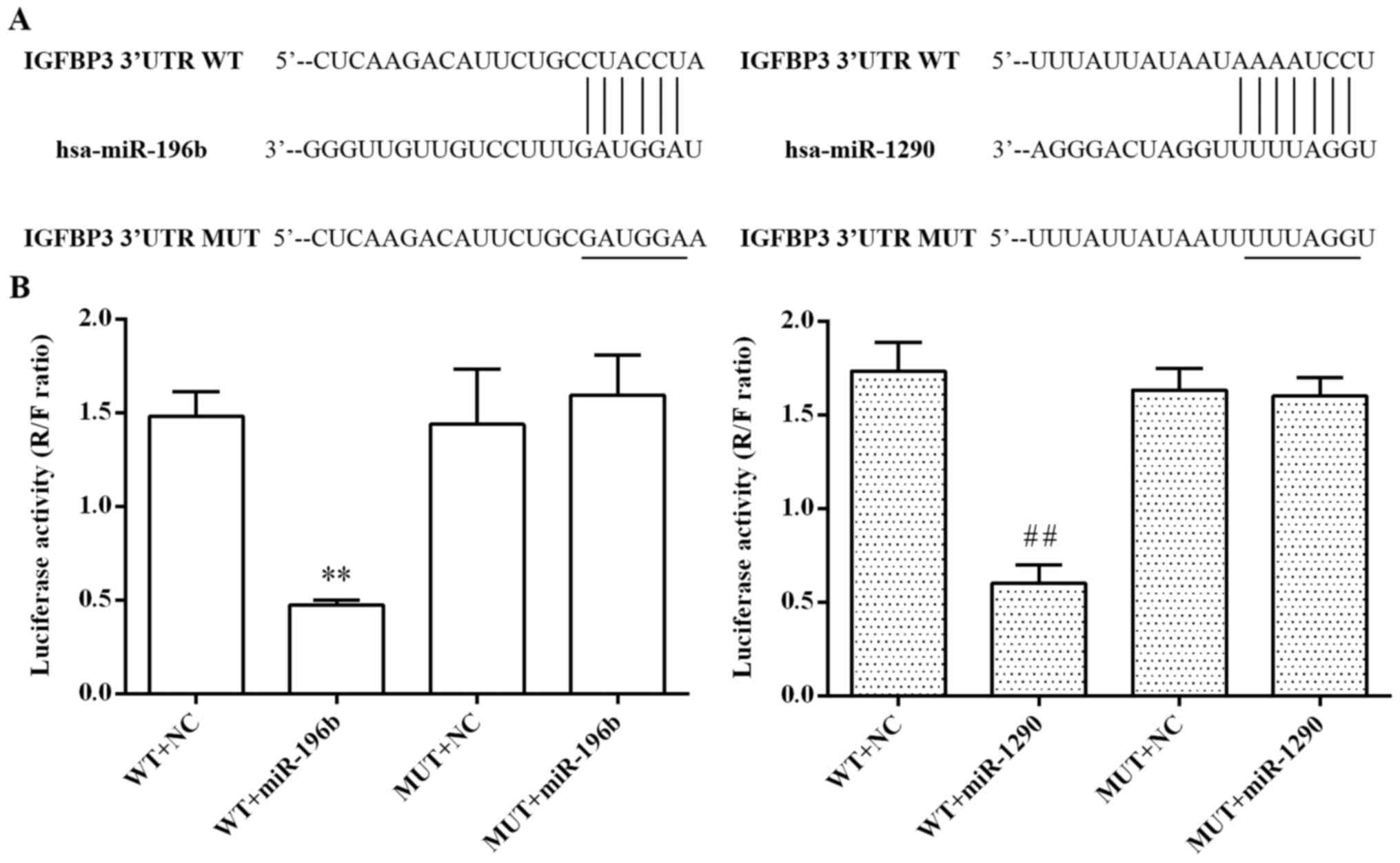
As shown in Fig. 5A,
the predicted binding sites of miR-196b and miR-1290 within the
3′-UTR of the IGFBP3 gene are indicated. To confirm their
relationship, we further performed dual-luciferase reporter assay.
As shown in Fig. 5B, the relative
luciferase activity was markedly decreased after co-transfection
with the wild-type 3′-UTR of IGFBP3 and miR-196b or miR-1290 in
293T cells (P<0.01, respectively), while the mutant 3′-UTR of
IGFBP3 showed slight inhibitory function on the luciferase
activity, suggesting that both miR-196b and miR-1290 suppressed the
transcription activity of the IGFBP3 gene by directly targeting the
binding site in the 3′-UTR of IGFBP3 mRNA.
siRNA against IGFBP3 attenuates the
antitumor effect of resveratrol on ALL cells
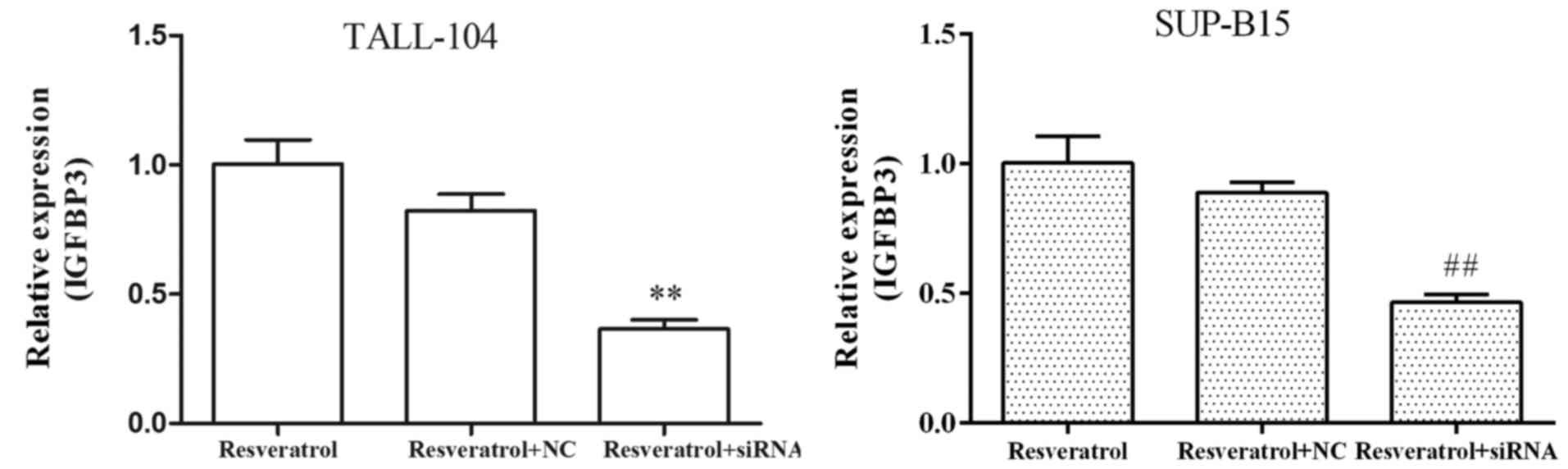
Resveratrol markedly upregulated the expression
levels of IGFBP3 in both TALL-104 and SUP-B15 cells (Fig. 2D and E), indicating that resveratrol
exhibited a common response in the different types of ALL cell
lines. To determine the role of miR-196b/miR-1290 in the antitumor
efficacy of resveratrol against ALL, we first transfected both
TALL-104 and SUP-B15 cells with IGFBP3 siRNA, and then examined
whether IGFBP3 siRNA affects the antitumor actions of resveratrol
in ALL cells. As shown in Fig. 6,
targeting IGFBP3 by siRNA resulted in marked attenuation of the
absolute induction of mRNA expression levels of IGFBP3 observed
following treatment of resveratrol. Furthermore, IGFBP3 siRNA
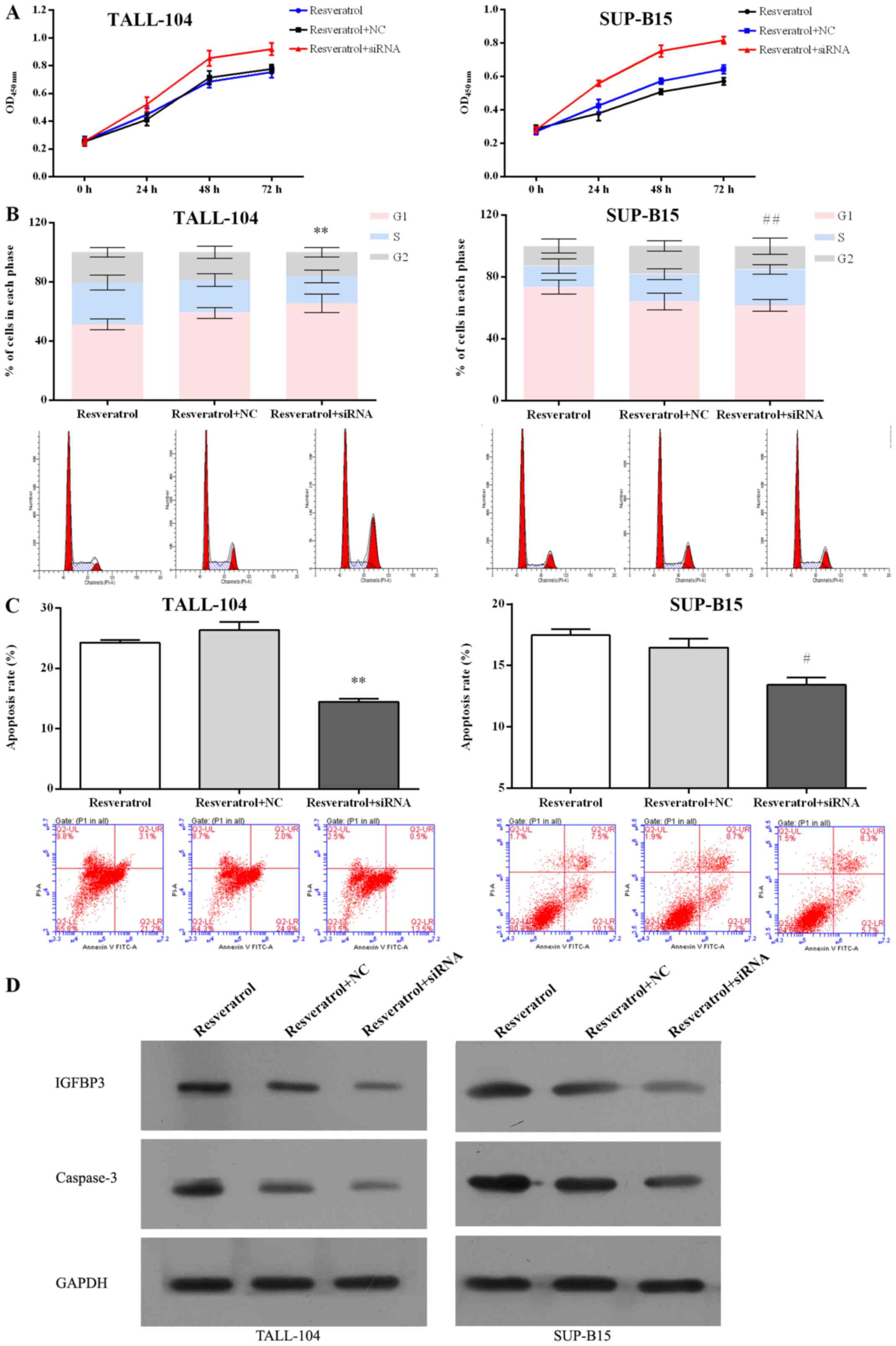
blocked the inhibitory effect pf proliferation mediated by
resveratrol (Fig. 7A) in both
TALL-104 and SUP-B15 cells. IGFBP3 siRNA also attenuated the
ability of resveratrol to induce cell cycle arrest (Fig. 7B) and cell apoptosis (Fig. 7C and D) in both TALL-104 and SUP-B15
cells. These data indicate that downregulation of IGFBP3 attenuated
the anti-ALL effect of resveratrol, suggesting that
miR-196b/miR-1290 play a pivotal role in the antitumor effect of
resveratrol in ALL cells.
Discussion
Accumulating evidence suggests that miRNAs may
function as oncogenes or tumor suppressors in human cancer
development (27,28). In acute lymphoblastic leukemia
(ALL), different miRNAs have been reported to play critical roles
in T-ALL and B-ALL (29). For
example, miRNA-193b-3p was reported to be a potential
tumor-suppressor in T-ALL (30) and
miRNA-17–92 was found to play a critical role in B-ALL (31). In the present study, we identified
that miR-196b and miR-1290 were overexpressed in T-ALL and B-ALL
cells, respectively. However, the function of the two cellular
miRNAs in ALL and their potential contribution to ALL therapy are
still not well clarified.
It is well known that miRNAs function by regulating
the expression of complementary genes. We hypothesized whether
there is a key target co-regulated by the different miRNAs in T-ALL
and B-ALL. We found various studies concerning IGFBP3, which is
downregulated and acts as a key target in ALL (7,32). In
the present study, we validated that the expression of IGFBP3 was
decreased in both bone marrow and peripheral blood of the 15 ALL
patients, which was in accordance with previous studies. Then, we
further explored whether IGFBP3 can be co-regulated by different
miRNAs and its role in T-ALL and B-ALL. The results indicate that
both miR-196b and miR-1290 directly bind to the 3′-UTR of IGFBP3,
suggesting the negative regulation of IGFBP3 expression in T-ALL
and B-ALL cells by miR-196b and miR-1290, respectively.
Resveratrol has been reported to possess antitumor
effects via regulation of specific miRNAs and alteration of the
crucial gene expression they target in colorectal (33), pancreatic (27) and bladder cancer (34), and glioma (15). However, the regulation of miRNAs by
resveratrol in ALL warrants further investigation. We initially
used two ALL cell lines: T-ALL TALL-104 and B-ALL SUP-B15 to
examine the potential antiproliferation effect of resveratrol.
Resveratrol exhibited similar antiproliferative activities in
TALL-104 and SUP-B15 cells. Moreover, resveratrol markedly
decreased the overexpression of miR-196b and miR-1290. Numerous
published studies in recent years have demonstrated the fundamental
roles of miRNAs in carcinogenesis, cell proliferation, migration,
invasion and apoptosis (35). As an
miR-196b/miR-1290 inhibitor, resveratrol was further found to
induce cell cycle arrest, apoptosis, and inhibit migration in ALL
cells. The data suggest that miR-196b and miR-1290 may participate
in the anti-ALL effect of resveratrol, which needs more
confirmation.
By applying IGFBP3 siRNA, we found that knockdown of
IGFBP3 reversed the antiproliferation, cell cycle arrest, apoptosis
induction abilities of resveratrol in both T-ALL and B-ALL cells.
According to the pieces of evidence, we conclude that resveratrol
exhibits anticancer activity in T-ALL and B-ALL by targeting
miR-196b and miR-1290, respectively. However, it should be noted
that each miRNA targets a diversity of genes. Li et al
reported that miR-196b directly targets both FAS tumor-suppressor
and HOXA9/MEIS1 oncogenes in MLL-rearranged leukemia (36). Endo et al revealed that
miR-1290 decreased the expression of forkhead box A1 and
N-acetyltransferase-1 in ER-positive breast cancer (37). The complex functions of miR-196b
(36,38) and miR-1290 (37,39) in
tumors indicate that resveratrol may exert antitumor activity
against other cancers as an miR-196b/miR-1290 inhibitor and the
modulating mechanism of miR-196b/miR-1290 in ALL warrants further
exploration.
In summary, miR-196b and miR-1290 were identified as
new targets of resveratrol. miR-196b and miR-1290 mediated the
inhibition of T-ALL and B-ALL cell growth, survival and migration
achieved by resveratrol. The findings also support that both
miR-196b and miR-1290 target the IGFBP3 3′-UTR and may be potential
therapeutic targets for ALL.
Acknowledgements
This study was supported by grants from the National
Natural Science Foundation of China (no. 81600147) and the Health
Collaborative Innovation Major Projects of Guangzhou City
(201508020254).
References
|
1
|
Inaba H, Greaves M and Mullighan CG: Acute
lymphoblastic leukaemia. Lancet. 381:1943–1955. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Luan C, Yang Z and Chen B: The functional
role of microRNA in acute lymphoblastic leukemia: Relevance for
diagnosis, differential diagnosis, prognosis, and therapy. Onco
Targets Ther. 8:2903–2914. 2015.PubMed/NCBI
|
|
3
|
Avigad S, Verly IR, Lebel A, Kordi O,
Shichrur K, Ohali A, Hameiri-Grossman M, Kaspers GJ, Cloos J,
Fronkova E, et al: miR expression profiling at diagnosis predicts
relapse in pediatric precursor B-cell acute lymphoblastic leukemia.
Genes Chromosomes Cancer. 55:328–339. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Pui CH and Evans WE: Treatment of acute
lymphoblastic leukemia. N Engl J Med. 354:166–178. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Adil SN and Usman M: Adult acute
lymphoblastic leukemia. J Pak Med Assoc. 54:4442004.PubMed/NCBI
|
|
6
|
Vorwerk P, Mohnike K, Wex H, Röhl FW,
Zimmermann M, Blum WF and Mittler U: Insulin-like growth factor
binding protein-2 at diagnosis of childhood acute lymphoblastic
leukemia and the prediction of relapse risk. J Clin Endocrinol
Metab. 90:3022–3027. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Petridou E, Dessypris N, Spanos E,
Mantzoros C, Skalkidou A, Kalmanti M, Koliouskas D, Kosmidis H,
Panagiotou JP, Piperopoulou F, et al: Insulin-like growth factor-I
and binding protein-3 in relation to childhood leukaemia. Int J
Cancer. 80:494–496. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Inui M, Martello G and Piccolo S: MicroRNA
control of signal transduction. Nat Rev Mol Cell Biol. 11:252–263.
2010. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Leal JA and Lleonart ME: MicroRNAs and
cancer stem cells: Therapeutic approaches and future perspectives.
Cancer Lett. 338:174–183. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yi B, Piazza GA, Su X and Xi Y: MicroRNA
and cancer chemoprevention. Cancer Prev Res. 6:401–409. 2013.
View Article : Google Scholar
|
|
13
|
Ghisi M, Corradin A, Basso K, Frasson C,
Serafin V, Mukherjee S, Mussolin L, Ruggero K, Bonanno L, Guffanti
A, et al: Modulation of microRNA expression in human T-cell
development: Targeting of NOTCH3 by miR-150. Blood. 117:7053–7062.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Cammarata G, Augugliaro L, Salemi D,
Agueli C, La Rosa M, Dagnino L, Civiletto G, Messana F, Marfia A,
Bica MG, et al: Differential expression of specific microRNA and
their targets in acute myeloid leukemia. Am J Hematol. 85:331–339.
2010.PubMed/NCBI
|
|
15
|
Wang G, Dai F, Yu K, Jia Z, Zhang A, Huang
Q, Kang C, Jiang H and Pu P: Resveratrol inhibits glioma cell
growth via targeting oncogenic microRNAs and multiple signaling
pathways. Int J Oncol. 46:1739–1747. 2015.PubMed/NCBI
|
|
16
|
Su YC, Li SC, Wu YC, Wang LM, Chao KS and
Liao HF: Resveratrol downregulates interleukin-6-stimulated sonic
hedgehog signaling in human acute myeloid leukemia. Evid Based
Complement Alternat Med. 2013:5474302013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Chen BY, Kuo CH, Liu YC, Ye LY, Chen JH
and Shieh CJ: Ultrasonic-assisted extraction of the botanical
dietary supplement resveratrol and other constituents of Polygonum
cuspidatum. J Nat Prod. 75:1810–1813. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wang ZH, Zhang JL, Duan YL, Zhang QS, Li
GF and Zheng DL: MicroRNA-214 participates in the neuroprotective
effect of resveratrol via inhibiting α-synuclein expression in
MPTP-induced Parkinson's disease mouse. Biomed Pharmacother.
74:252–256. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Cullberg KB, Foldager CB, Lind M,
Richelsen B and Pedersen SB: Inhibitory effects of resveratrol on
hypoxia-induced inflammation in 3T3-L1 adipocytes and macrophages.
J Funct Foods. 7:171–179. 2014. View Article : Google Scholar
|
|
20
|
Frémont L: Biological effects of
resveratrol. Life Sci. 66:663–673. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Jang M, Cai L, Udeani GO, Slowing KV,
Thomas CF, Beecher CW, Fong HH, Farnsworth NR, Kinghorn AD, Mehta
RG, et al: Cancer chemopreventive activity of resveratrol, a
natural product derived from grapes. Science. 275:218–220. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ferry-Dumazet H, Garnier O, Mamani-Matsuda
M, Vercauteren J, Belloc F, Billiard C, Dupouy M, Thiolat D, Kolb
JP, Marit G, et al: Resveratrol inhibits the growth and induces the
apoptosis of both normal and leukemic hematopoietic cells.
Carcinogenesis. 23:1327–1333. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Azimi A, Hagh MF, Talebi M, Yousefi B,
Feizi AA Hossein Pour, Baradaran B, Movassaghpour AA, Shamsasenjan
K, Khanzedeh T, Ghaderi AH, et al: Time- and
concentration-dependent effects of resveratrol on miR 15a and
miR16-1 expression and apoptosis in the CCRF-CEM acute
lymphoblastic leukemia cell line. Asian Pac J Cancer Prev.
16:6463–6468. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ge J, Liu Y, Li Q, Guo X, Gu L, Ma ZG and
Zhu YP: Resveratrol induces apoptosis and autophagy in T-cell acute
lymphoblastic leukemia cells by inhibiting Akt/mTOR and activating
p38-MAPK. Biomed Environ Sci. 26:902–911. 2013.PubMed/NCBI
|
|
25
|
Ghorbani A, Zand H, Jeddi-Tehrani M,
Koohdani F, Shidfar F and Keshavarz SA: PTEN over-expression by
resveratrol in acute lymphoblastic leukemia cells along with
suppression of AKT/PKB and ERK1/2 in genotoxic stress. J Nat Med.
69:507–512. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Riedl SJ and Shi Y: Molecular mechanisms
of caspase regulation during apoptosis. Nat Rev Mol Cell Biol.
5:897–907. 2004. View
Article : Google Scholar : PubMed/NCBI
|
|
27
|
Liu P, Liang H, Xia Q, Li P, Kong H, Lei
P, Wang S and Tu Z: Resveratrol induces apoptosis of pancreatic
cancers cells by inhibiting miR-21 regulation of BCL-2 expression.
Clin Transl Oncol. 15:741–746. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Adams BD, Kasinski AL and Slack FJ:
Aberrant regulation and function of microRNAs in cancer. Curr Biol.
24:R762–R776. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Schotte D, Chau JC, Sylvester G, Liu G,
Chen C, van der Velden VH, Broekhuis MJ, Peters TC, Pieters R and
den Boer ML: Identification of new microRNA genes and aberrant
microRNA profiles in childhood acute lymphoblastic leukemia.
Leukemia. 23:313–322. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Mets E, Van der Meulen J, Van Peer G,
Boice M, Mestdagh P, Van de Walle I, Lammens T, Goossens S, De
Moerloose B, Benoit Y, et al: MicroRNA-193b-3p acts as a tumor
suppressor by targeting the MYB oncogene in T-cell acute
lymphoblastic leukemia. Leukemia. 29:798–806. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Scherr M, Elder A, Battmer K, Barzan D,
Bomken S, Ricke-Hoch M, Schröder A, Venturini L, Blair HJ, Vormoor
J, et al: Differential expression of miR-17~92 identifies BCL2 as a
therapeutic target in BCR-ABL-positive B-lineage acute
lymphoblastic leukemia. Leukemia. 28:554–565. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Mohnike KL, Kluba U, Mittler U, Aumann V,
Vorwerk P and Blum WF: Serum levels of insulin-like growth
factor-I, -II and insulin-like growth factor binding proteins −2
and −3 in children with acute lymphoblastic leukaemia. Eur J
Pediatr. 155:81–86. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yang S, Li W, Sun H, Wu B, Ji F, Sun T,
Chang H, Shen P, Wang Y and Zhou D: Resveratrol elicits
anti-colorectal cancer effect by activating miR-34c-KITLG in vitro
and in vivo. BMC Cancer. 15:9692015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhou C, Ding J and Wu Y: Resveratrol
induces apoptosis of bladder cancer cells via miR-21 regulation of
the Akt/Bcl-2 signaling pathway. Mol Med Rep. 9:1467–1473.
2014.PubMed/NCBI
|
|
35
|
Phuah NH and Nagoor NH: Regulation of
microRNAs by natural agents: New strategies in cancer therapies.
BioMed Res Int. 2014:8045102014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Li Z, Huang H, Chen P, He M, Li Y,
Arnovitz S, Jiang X, He C, Hyjek E, Zhang J, et al: miR-196b
directly targets both HOXA9/MEIS1 oncogenes and FAS tumour
suppressor in MLL-rearranged leukaemia. Nat Commun. 3:6882012.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Endo Y, Toy am a T, Takahashi S, Yoshimoto
N, Iwasa M, Asano T, Fujii Y and Yamashita H: miR-1290 and its
potential targets are associated with characteristics of estrogen
receptor α-positive breast cancer. Endocr Relat Cancer. 20:91–102.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Popovic R, Riesbeck LE, Velu CS, Chaubey
A, Zhang J, Achille NJ, Erfurth FE, Eaton K, Lu J, Grimes HL, et
al: Regulation of mir-196b by MLL and its overexpression by MLL
fusions contributes to immortalization. Blood. 113:3314–3322. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Wu J, Ji X, Zhu L, Jiang Q, Wen Z, Xu S,
Shao W, Cai J, Du Q, Zhu Y, et al: Up-regulation of microRNA-1290
impairs cytokinesis and affects the reprogramming of colon cancer
cells. Cancer Lett. 329:155–163. 2013. View Article : Google Scholar : PubMed/NCBI
|