Introduction
Approximately 60–70% of breast cancer express
estrogen receptor α (ERα) and (or) progesterone receptor and merit
the use of hormone therapies, such as the estrogen receptor
antagonist tamoxifen (1). Use of
tamoxifen has improved outcomes for patients with ER+
breast cancer, but in 40–50% of these patients the cancers recur,
usually as distant metastasis, and the majority of these patients
will die of the metastasis (2). One
possible explanation for the initial positive response to therapy
followed by drug-resistance is that current therapies eliminate the
bulk of the tumour, but they fail to eliminate cancer stem cells
(CSCs). Thus, studying molecular mechanism underlying the formation
of CSCs and the link between CSCs and tamoxifen-resistance will
help us to offer new molecular targets for effective therapies.
Emerging evidence suggests a strong link between
resistance to therapies and the induction of epithelial-mesenchymal
transition (EMT) in cancer (3).
Morphologically, EMT is characterized by the loss of tight
cell-cell junctions and accompanied by re-organization of the actin
cytoskeleton, resulting in spindle shaped mesenchymal-like cells,
which are capable of migrating and invading other tissues (4). Determining the mechanisms that connect
EMT and the development of drug resistance could be a key approach
for the development of novel therapeutic strategies to overcome
both drug resistance and potentially prevent metastasis
initiation.
MicroRNAs (miRs) are small non-coding RNAs that
modulate protein expression by binding to complementary or
partially complementary the 3-untranslated region (UTR) of target
mRNAs and thereby targeting the mRNA for degradation or
translational inhibition (5,6).
Recently, it has been reported that miRNA-375 was among the top
downregulated miRNAs in tamoxifen-resistant cells (7). Re-expression of miR-375 was sufficient
to sensitize tamoxifen-resistant cells to tamoxifen and partly
reversed EMT (7). Thus, restoration
of miR-375 might serve as a potential therapeutic approach for the
treatment of tamoxifen-resistant breast cancer.
In the present study, we showed that miR-375
inhibits CSC traits in breast cancer MCF-7 cells. Bioinformatics
analysis and experimental validation identified HOXB3 as a direct
target of miR-375. Overexpressing miR-375 degraded HOXB3 mRNA in
MCF-7 cells. Moreover, overexpression of HOXB3 induced formation of
CSC phenotypes, EMT and tamoxifen-resistance as well as enhanced
ability of migration and invasion in MCF-7 cells. Most ER-positive
breast cancer-related deaths occur because of resistance to
standard therapies and metastasis, restoring miR-375 or targeting
HOXB3 might serve as potential therapeutic approaches for the
treatment of tamoxifen-resistant breast cancer.
Materials and methods
Breast cancer MCF-7 cell line, HOXB3
expressing plasmids/empty vectors, pre-miR-375/control miR and
transfection
Human breast cancer cell line MCF-7 was obtained
from the American Type Culture Collection (ATCC; Manassas, VA,
USA). Briefly, cells were maintained in RPMI-1640 medium
supplemented with 10% fetal bovine serum (FBS; Gibco, Grand Island,
NY, USA) and penicillin/streptomycin at 37°C in a humidified
atmosphere with 5% CO2. HOXB3 expressing plasmids/empty
vectors (pcDNA3.1) were purchased from Tiangen Biotech, Co., Ltd.
(Beijing, China). pre-miR-375/control miR were purchased from
Ambion, Inc. (Austin, TX, USA). For transfection experiments, the
cells were cultured in serum-free medium without antibiotics at 60%
confluence for 24 h, and then transfected with transfection reagent
(Lipofectamine 2000; Invitrogen, Carlsbad, CA, USA) according to
the manufacturers instructions. After incubation for 6 h, the
medium was removed and replaced with normal culture medium for 48
h, unless otherwise specified.
Western blot analysis
Western blot analysis was performed as previously
described (8). Briefly, after
incubation with primary antibody anti-CD44 (1:500; Abcam,
Cambridge, MA, USA), anti-CD133 (1:500; Abcam), anti-HOXB3 (1:500;
Abcam), anti-MTDH (1:500; Abcam), anti-TWIST (1:500; Abcam) and
anti-β-actin (1:500; Abcam) overnight at 4°C, IRDye™-800 conjugated
anti-rabbit secondary antibodies (LI-COR Biosciences, Lincoln, NE,
USA) were used for 30 min at room temperature. The specific
proteins were visualized by Odyssey™ Infrared Imaging System (Gene
Co., Lincoln, NE, USA).
Sphere formation assay
Cells (103/ml) in serum-free RPMI-1640/1
mM Na-pyruvate were seeded on 0.5% agar precoated 6-well plates.
After 1 week, half the medium was changed every third day. Single
spheres were picked and counted.
Immunostaining assay for CD44 and
CD133 in breast cancer spheres
Single cell suspensions of MCF-7 cells transfected
as indicated were prepared and plated using ultra low adherent
wells of 6-well plate at 5,000 cells/well in sphere formation
medium, as described above. After 7 days of treatment, the breast
cancer spheres were collected by centrifugation, washed with 1X
phosphate-buffered saline (PBS) and fixed with 3.7% parformaldehyde
for immunofluorescence staining, as described previously by our
laboratory. Coverslips were counterstained with DAPI
(Invitrogen-Molecular Probes, Eugene, OR, USA) for visualization of
the nuclei. Microscopic analysis was performed with a confocal
laser scanning microscope (Leica Microsystems, Bensheim, Germany).
Fluorescence intensities were measured in a few viewing areas for
300 cells per coverslip and analyzed using ImageJ 1.37v software
(http://rsb.info.nih.gov/ij/index.html).
Immunofluorescence analyses for HOXB3
in MCF-7 cells
For immunofluorescence analyses, MCF-7 cells were
plated on glass coverslips in 6-well plates and transfected as
indicated. At 48 h after transfection, coverslips were stained with
HOXB3 (1:500; Abcam) Alexa Fluor 488 goat anti-rabbit IgG antibody
was used as secondary antibody (Invitrogen). Similarly to the
above, the coverslips were counterstained with DAPI
(Invitrogen-Molecular Probes) for visualization of the nuclei.
Microscopic analysis was performed with a confocal laser-scanning
microscope (Leica Microsystems). Fluorescence intensities were
measured in a few viewing areas for 300 cells per coverslip and
analyzed using ImageJ 1.37v software (http://rsb.info.nih.gov/ij/index.html).
Migration and invasion assay
Migration and invasion assay was performed as
described before (8).
Methods of bioinformatics
The analysis of potential microRNA target site using
the commonly used prediction algorithms, miRanda (http://www.microrna.org/).
Real-time PCR for microRNAs
Total RNA from cultured cells, with efficient
recovery of small RNAs, was isolated using the mirVana miRNA
isolation kit (Ambion). Detection of the mature form of miRNAs was
performed using the mirVana qRT-PCR miRNA detection kit and qRT-PCR
Primer Sets, according to the manufacturers instructions (Ambion).
The U6 small nuclear RNA was used as an internal control.
Reverse transcription-polymerase chain
reaction
RT-PCR was performed as described (9). Primers for CD44: forward,
5-GGATGGGCGGATGGAAGGAT-3 and reverse, 5-TCCGCTGGGCAATGAGGCTG-3.
Primers for CD133: forward, 5-CACTCTATACCAAAGCGTCAA-3 and reverse,
5-CACGATGCCACTTTCTCAC-3. Primers for GAPDH: forward,
5-CGGAGTCAACGGATTTGGTCGTAT-3 and reverse,
5-AGCCTTCTCCATGGTGGTGAAGAC-3.
MTT assay
The effect of the cell proliferation was assessed by
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT;
Sigma-Aldrich, St. Louis, MO, USA) assay and it was performed as
described (9). Absorbance was
directly proportional to the number of survival cells.
Statistical analysis
Data are presented as mean ± SEM. Students t-test
(two-tailed) was used to compare two groups (P<0.05 was
considered significant).
Results
miR-375 inhibits CSCs traits in breast
cancer MCF-7 cells
Re-expression of miR-375 can partly reverse EMT in
breast cancer (7).
Epithelial-mesenchymal transition (EMT) not only confers tumor
cells with a distinct advantage for metastatic dissemination, but
it also provides those cells with cancer stem cell-like
characteristics for proliferation and drug resistance (10–13).
To determine whether breast cancer cells with EMT phenotype induced
by miR-375 could have stem-like cell characteristics, we performed
sphere forming assay to assess the capacity of CSC or CSC-like cell
self renewal in MCF-7 cells. We tested whether pre-miR-375 could
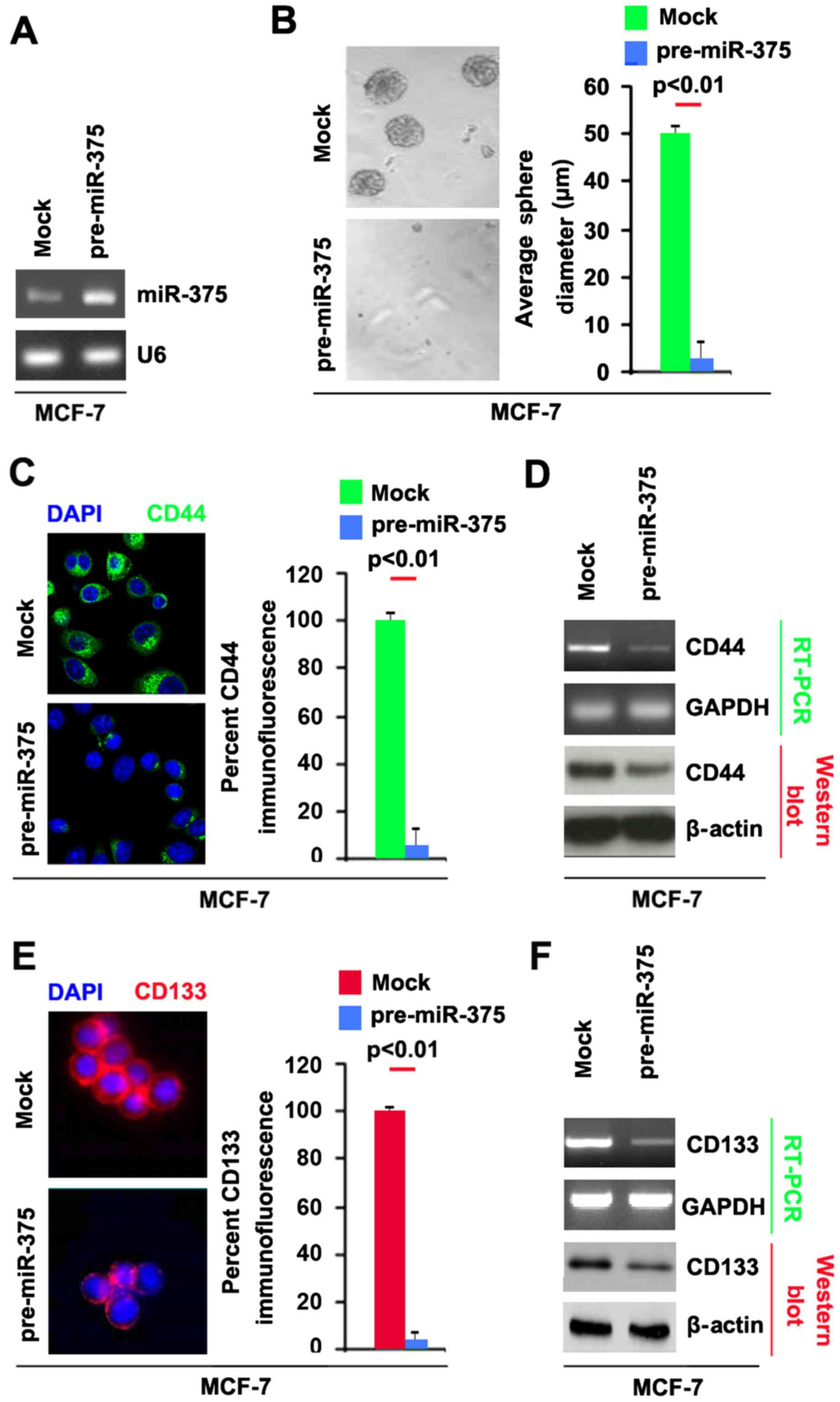
stably express miR-375 in MCF-7 cells. The results showed that
miR-375 could be significantly increased by pre-miR-375 in the
cells (Fig. 1A). Sphere forming
assay showed (Fig. 1B), miR-375
overexpressing cells formed much smaller spheres after 14 days of
culture as compared with control cells, indicating markedly
decreased CSCs traits by miR-375.
CD44 is positively associated with stem cell-like
characteristics in breast cancer (14). In order to assess whether CD44 can
be regulated by miR-375, we performed immunoflurescence to detect
CD44 protein in spheres formed by MCF-7 cells transfected with
pre-miR-375. We found that CD44 protein was significantly inhibited
by miR-375 in spheres formed by MCF-7 cells (Fig. 1C). To further confirm that CD44 can
be inhibited by miR-375, we performed RT-PCR and western blot
analysis to detect CD44 mRNA and protein (Fig. 1D). The results showed that CD44 mRNA
and protein were decreased in MCF-7 cells transfected with
pre-miR-375.
Moreover, CD133 has been proposed as a cancer stem
cell marker in breast cancer (15).
To identify whether miR-375 can affect CD133 expression, we
performed immunoflurescence to detect its expression in spheres
formed by MCF-7 cells transfected with pre-miR-375. We found that
CD133 protein was significantly inhibited by miR-375 in spheres
formed by MCF-7 cells (Fig. 1E). To
further confirm that CD133 can be inhibited by miR-375, we
performed RT-PCR and western blot analysis to detect CD133 mRNA and
protein. The results showed that CD133 mRNA and protein were
decreased in MCF-7 cells transfected with pre-miR-375 (Fig. 1F). Taken together, these data showed
that reintroduction of miR-375 in MCF-7 cells reduced the
stem cell-like population and greatly attenuated the ability of
stem cell-like breast cancer cells to retain stemness.
miR-375 degrades HOXB3 mRNA in MCF-7
cells
Having demonstrated that overexpressing miR-375
inhibited formation of stem cell-like population. Next, we studied
the mechanisms of miR-375 regulating CSCs. MicroRNAs (miRs) are a
class of small non-coding RNAs (~22 nucleotides) and negatively
regulate protein-coding gene expression by targeting mRNA
degradation or translation inhibition (16–18).
To find the target gene of miR-375, we used the prediction
algorithm, miRanda (http://www.microrna.org/microrna/home.do) to predict
its target gene. A dozen of target genes were found by the
algorithm, but we are interested in HOXB3, because aberrations in
HOX (a highly conserved subgroup of the homeobox superfamily) gene
expression have been reported in abnormal development and
malignancy, suggesting that altered expression of HOX genes could
be important for both oncogenesis or tumor suppression, depending
on context (19). Such studies note
an association of increased expression of a set of homeodomain
transcription factors, including HOXB3, with poor prognosis in
acute myeloid leukemia and breast cancer (20,21).
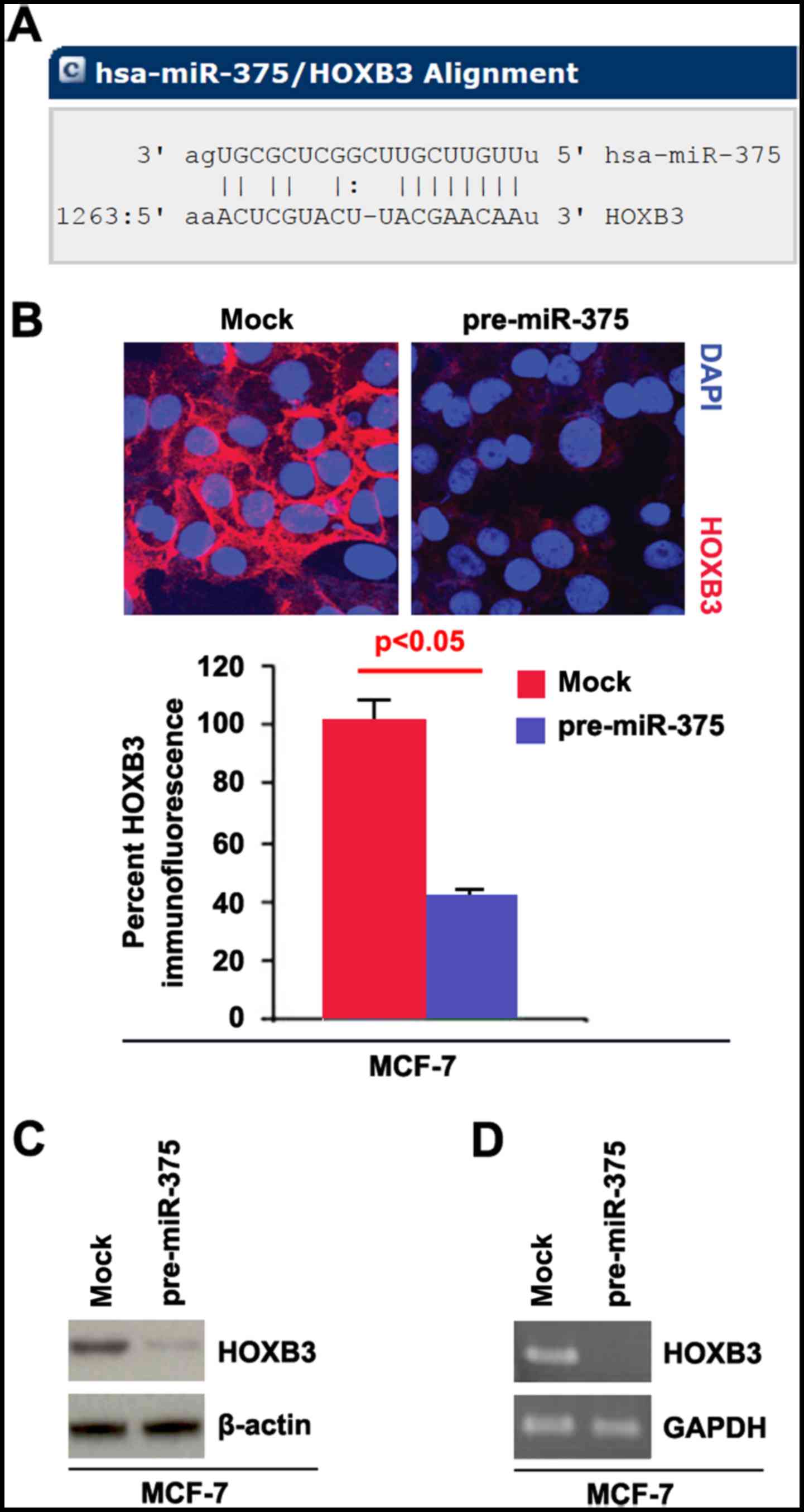
Target sites on 3UTR of HOXB3 are shown in Fig. 2A. We reasoned that miR-375 could
downregulate HOXB3 expression by targeting its 3UTR in breast
cancer.
In an attempt to identify the role of miR-375 in
regulating HOXB3 expression in breast cancer, we performed
immunofluorescence analyses in MCF-7 cells transfected with
pre-miR-375 and control miR. The results showed that HOXB3 protein
was evidently inhibited in the cells transfected with pre-miR-375
(Fig. 2B). We next performed RT-PCR
and western blot analysis to detect HOXB3 expression in MCF-7 cells
transfected with pre-miR-375 and control miR. The results showed
that HOXB3 protein (Fig. 2C) and
HOXB3 mRNA (Fig. 2D) were
significantly downregulated in the cells transfected with
pre-miR-375.
HOXB3 promotes CSCs phenotypes in
MCF-7 cells
In order to study the role of HOXB3 in breast
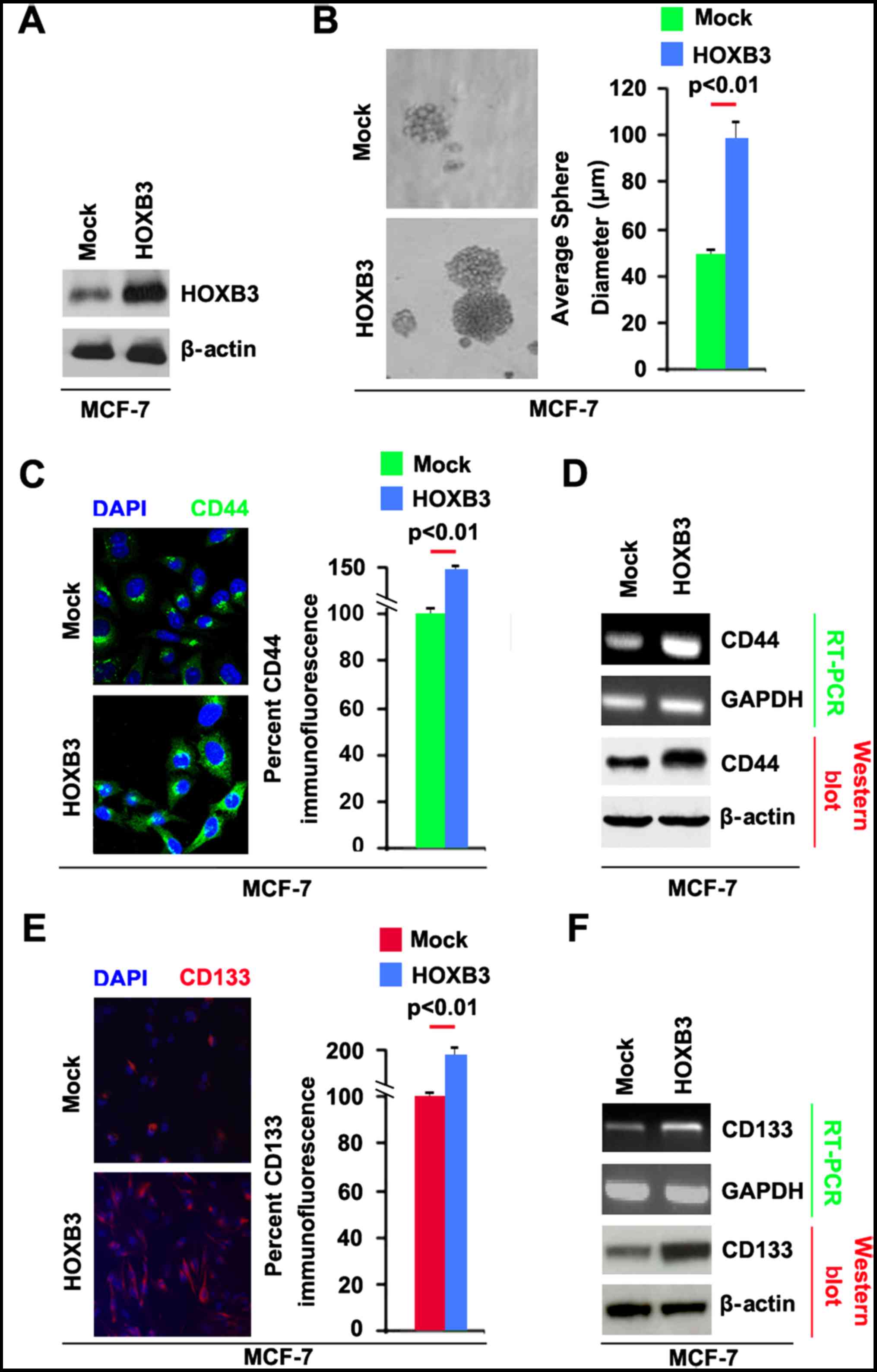
cancer, we transfected MCF-7 cells with HOXB3 expressing plasmids.
The results showed that HOXB3 protein could be significantly
increased by HOXB3 expressing plasmids in the cells (Fig. 3A). To determine whether HOXB3 can
regulate CSC traits in breast cancer, we performed sphere growth
assay and the result demonstrated that the capacity of formation of
CSCs or CSC-like cell self renewal was increased by HOXB3
expressing plasmids in MCF-7 cells (Fig. 3B).
To identify whether CD44 protein can be regulated by
HOXB3, we performed immunoflurescence to detect CD44 protein in
spheres formed by MCF-7 cells transfected with HOXB3 expressing
plasmids. We found that CD44 protein level was significantly
enhanced by HOXB3 in shperes formed by MCF-7 cells (Fig. 3C). To further confirm that CD44 can
be upregulated by HOXB3, we performed RT-PCR and western blot
analysis to detect CD44 mRNA and protein in MCF-7 cells. The
results showed that CD44 mRNA and protein were increased in MCF-7
cells transfected with HOXB3 expressing plasmids (Fig. 3D). To identify whether HOXB3 can
affect CD133 expression, we performed immunoflurescence to detect
CD133 protein in spheres formed by MCF-7 cells transfected with
HOXB3 expressing plasmids. We found that CD133 protein was
significantly increased by HOXB3 in spheres formed by MCF-7 cells
(Fig. 3E).
To further confirm that CD133 protein can be induced
by HOXB3, we performed RT-PCR and western blot analysis to detect
CD133 mRNA and protein. The results showed that CD133 mRNA and
protein were increased in MCF-7 cells transfected with HOXB3
expressing plasmids (Fig. 3F).
Taken together, these data showed that overexpression of HOXB3 in
MCF-7 cells promoted the formation of stem cell-like population and
greatly enhanced the ability of stem cell-like breast cancer cells
to retain stemness.
HOXB3 promotes EMT and tamoxifen
resistance in MCF-7
Re-expression of miR-375 can partly reverse EMT
(7) and degrade HOXB3 in breast
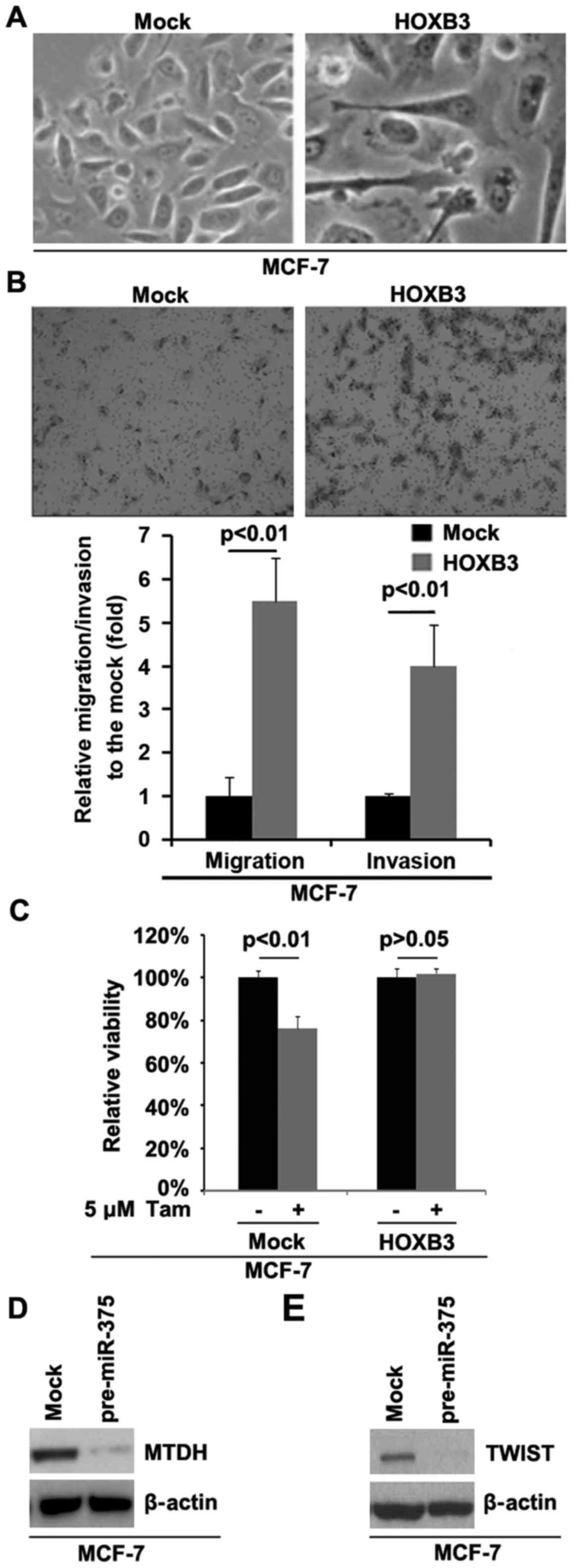
cancer. To identify whether HOXB3 can promote EMT in MCF-7 cells,
we transfected MCF-7 cells with HOXB3 expressing plasmids and then
its overexpression caused significant changes in MCF-7 cell
morphology (EMT, phenotype from a cobblestone-like to a
spindle-like morphology) (Fig. 4A).
Restoration of miRNA-375 expression sensitized tamoxifen resistant
cells to tamoxifen and inhibited invasion (7). In order to study whether HOXB3 can
affect migration and invasion in MCF-7 cells, we performed
migration and invasion assay. The results showed that HOXB3 can
significantly enhance migration and invasion in MCF-7 cells
(Fig. 4B). Moreover, the direct
targeting of HOXB3 by miRNA-375 led us to hypothesize that
upregulation of HOXB3 by lack of miRNA-375 could be involved in
tamoxifen-resistance in MCF-7 cells. For this purpose, we
overexpressed HOXB3 in tamoxifen-sensitive MCF-7 cells.
Overexpression of HOXB3 could transform tamoxifen-sensitive MCF-7
cells to tamoxifen-resistant cells (Fig. 4C), suggesting that its
over-expression is involved in the transformation from tamoxifen
sensitive to tamoxifen resistant cells. Metadherin (MTDH) is a
direct target of miR-375 (7). Our
results confirmed that MTDH protein was downregulated by miR-375
(Fig. 4D). Moreover, MTDH can
activate TWIST epigenetically and promote cancer stem-like cell
traits in breast cancer (22). We
performed western blot analysis to detect whether miR-375 can
regulate TWIST. The results demonstrated that TWIST was
significantly inhibited by miR-375 (Fig. 4E).
Discussion
Current breast cancer therapeutic strategies based
on tumor regression may target and kill differentiated tumor cells
which comprise the bulk of the tumor, but the therapeutic
strategies spared the rare CSC population. CSCs constitute a mostly
small subset of cancer cells that possess the ability to self-renew
and generate the diverse differentiated cell populations that
constitute the cancer mass (23–25).
Currently, a growing body of evidence indicates the idea that
cancers are diseases driven by subpopulation of self-renewing CSCs,
which have been identified and isolated from tumors of the breast,
gastric cancer, hematopoietic system, lung, colon, prostate, head
and neck, brain, endometrial and pancreatic cancer (24–28).
The CSCs possess the capacity for self-renewal and have the ability
to drive continued expansion of the population of malignant cells
with invasive and metastatic propensity. Tamoxifen-resistant MCF-7
cells possess CSCs characteristics (29). Recently, it was reported that
downregulation of miR-375 is an important cause for
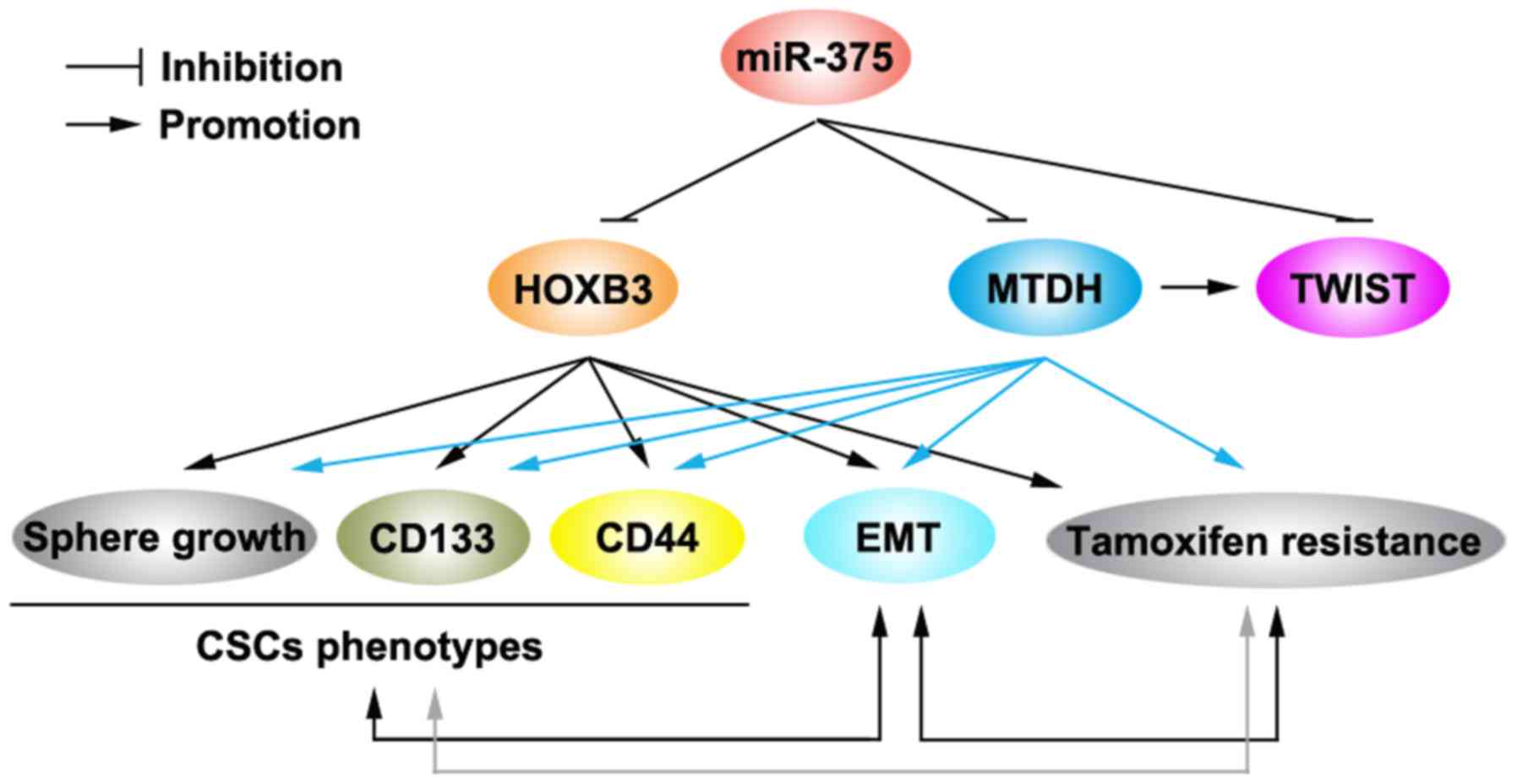
tamoxifen-resistance (Fig. 5)
(7). We showed that miR-375 can
inhibit formation of CSC phenotypes in MCF-7 cells. The results
indicated that restoration of miR-375 can inhibit
tamoxifen-resistance by regulating CSCs.
We found that HOXB3 is a target of miR-375 in MCF-7
cells. The HOX genes encode a family of highly conserved
transcription factors that normally regulate temporospatial
development of the extremities and organs. Aberrant expression of
these genes in different tissues has been associated with
tumorigenesis (30). HOXB13 genes
play an important role in tamoxifen-resistance (31). However, there is no report on the
role of HOXB3 in the development of tamoxifen-resistance. We showed
that HOXB3 can promote formation of CSC phenotypes and
tamoxifen-resistance (Fig. 5). EMT
is involved in formation of CSCs and tamoxifen-resistant breast
cancer cells (32,33). Our results showed that HOXB3 can
significantly promote EMT in MCF-7 cells.
Metadherin (MTDH) is a target of miRNA-375, which
was upregulated in tamoxifen-resistant cells (34). Elevated MTDH levels were inversely
correlated with miR-375 expression and positively correlated with
poorer disease-free survival in tamoxifen-treated patients
(34). MTDH plays a critical role
in mammary tumorigenesis by regulating oncogene-induced expansion,
EMT and activities of CSCs (Fig. 5)
(34). Our results confirmed that
miR-375 can inhibit MTDH protein in MCF-7 cells. Moreover,
epigenetic activation of TWIST by MTDH can promote cancer stem-like
cell traits in breast cancer (34).
Our data, together with previous studies (34), showed that TWIST can be inhibited by
miR-375 and suggested that the inhibition might be meditated by
MTDH.
Acknowledgements
The present study was supported by the grant from
the Focus on Research and Development Plan in Shandong Province
(2015GSF118132).
Glossary
Abbreviations
Abbreviations:
|
CSCs
|
cancer stem cells
|
|
EMT
|
epithelial-mesenchymal transition
|
References
|
1
|
Early Breast Cancer Trialists
Collaborative Group (EBCTCG), . Effects of chemotherapy and
hormonal therapy for early breast cancer on recurrence and 15-year
survival: An overview of the randomised trials. Lancet.
365:1687–1717. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Normanno N, Di Maio M, De Maio E, De Luca
A, de Matteis A, Giordano A and Perrone F: NCI-Naple Breast Cancer
Group: Mechanisms of endocrine resistance and novel therapeutic
strategies in breast cancer. Endocr Relat Cancer. 12:721–747. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Thiery JP, Acloque H, Huang RY and Nieto
MA: Epithelial-mesenchymal transitions in development and disease.
Cell. 139:871–890. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Polyak K and Weinberg RA: Transitions
between epithelial and mesenchymal states: Acquisition of malignant
and stem cell traits. Nat Rev Cancer. 9:265–273. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Baek D, Villén J, Shin C, Camargo FD, Gygi
SP and Bartel DP: The impact of microRNAs on protein output.
Nature. 455:64–71. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ward A, Balwierz A, Zhang JD, Küblbeck M,
Pawitan Y, Hielscher T, Wiemann S and Sahin Ö: Re-expression of
microRNA-375 reverses both tamoxifen resistance and accompanying
EMT-like properties in breast cancer. Oncogene. 32:1173–1182. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lu Y, Chopp M, Zheng X, Katakowski M,
Buller B and Jiang F: MiR-145 reduces ADAM17 expression and
inhibits in vitro migration and invasion of glioma cells. Oncol
Rep. 29:67–72. 2013.PubMed/NCBI
|
|
9
|
Tang L, Chen F, Pang EJ, Zhang ZQ, Jin BW
and Dong WF: MicroRNA-182 inhibits proliferation through targeting
oncogenic ANUBL1 in gastric cancer. Oncol Rep. 33:1707–1716.
2015.PubMed/NCBI
|
|
10
|
Kurrey NK, Jalgaonkar SP, Joglekar AV,
Ghanate AD, Chaskar PD, Doiphode RY and Bapat SA: Snail and slug
mediate radioresistance and chemoresistance by antagonizing
p53-mediated apoptosis and acquiring a stem-like phenotype in
ovarian cancer cells. Stem Cells. 27:2059–2068. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan
A, Zhou AY, Brooks M, Reinhard F, Zhang CC, Shipitsin M, et al: The
epithelial-mesenchymal transition generates cells with properties
of stem cells. Cell. 133:704–715. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Morel AP, Lièvre M, Thomas C, Hinkal G,
Ansieau S and Puisieux A: Generation of breast cancer stem cells
through epithelial-mesenchymal transition. PLoS One. 3:e28882008.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Santisteban M, Reiman JM, Asiedu MK,
Behrens MD, Nassar A, Kalli KR, Haluska P, Ingle JN, Hartmann LC,
Manjili MH, et al: Immune-induced epithelial to mesenchymal
transition in vivo generates breast cancer stem cells. Cancer Res.
69:2887–2895. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Park SY, Lee HE, Li H, Shipitsin M, Gelman
R and Polyak K: Heterogeneity for stem cell-related markers
according to tumor subtype and histologic stage in breast cancer.
Clin Cancer Res. 16:876e87. 2010. View Article : Google Scholar
|
|
15
|
Mansour SF and Atwa MM:
Clinicopathological significance of CD133 and ALDH1 cancer stem
cell marker expression in invasive ductal breast carcinoma. Asian
Pac J Cancer Prev. 16:7491–7496. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lee RC, Feinbaum RL and Ambros V: The C.
elegans heterochronic gene lin-4 encodes small RNAs with antisense
complementarity to lin-14. Cell. 75:843–854. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Pasquinelli AE, Reinhart BJ, Slack F,
Martindale MQ, Kuroda MI, Maller B, Hayward DC, Ball EE, Degnan B,
Müller P, et al: Conservation of the sequence and temporal
expression of let-7 heterochronic regulatory RNA. Nature.
408:86–89. 2000. View
Article : Google Scholar : PubMed/NCBI
|
|
18
|
Reinhart BJ, Slack FJ, Basson M,
Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR and Ruvkun G:
The 21-nucleotide let-7 RNA regulates developmental timing in
Caenorhabditis elegans. Nature. 403:901–906. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Shah N and Sukumar S: The Hox genes and
their roles in oncogenesis. Nat Rev Cancer. 10:361–371. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Eklund E: The role of Hox proteins in
leukemogenesis: Insights into key regulatory events in
hematopoiesis. Crit Rev Oncog. 16:65–76. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Palakurthy RK, Wajapeyee N, Santra MK,
Gazin C, Lin L, Gobeil S and Green MR: Epigenetic silencing of the
RASSF1A tumor suppressor gene through HOXB3-mediated induction of
DNMT3B expression. Mol Cell. 36:219–230. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Liang Y, Hu J, Li J, Liu Y, Yu J, Zhuang
X, Mu L, Kong X, Hong D, Yang Q, et al: Epigenetic activation of
TWIST1 by MTDH promotes cancer stem-like cell traits in breast
cancer. Cancer Res. 75:3672–3680. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Dirks P: Cancer stem cells: Invitation to
a second round. Nature. 466:40–41. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Gupta PB, Chaffer CL and Weinberg RA:
Cancer stem cells: Mirage or reality? Nat Med. 15:1010–1012. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Frank NY, Schatton T and Frank MH: The
therapeutic promise of the cancer stem cell concept. J Clin Invest.
120:41–50. 2010. View
Article : Google Scholar : PubMed/NCBI
|
|
26
|
Tang C, Ang BT and Pervaiz S: Cancer stem
cell: Target for anti-cancer therapy. FASEB J. 21:3777–3785. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Takaishi S, Okumura T, Tu S, Wang SS,
Shibata W, Vigneshwaran R, Gordon SA, Shimada Y and Wang TC:
Identification of gastric cancer stem cells using the cell surface
marker CD44. Stem Cells. 27:1006–1020. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Rutella S, Bonanno G, Procoli A, Mariotti
A, Corallo M, Prisco MG, Eramo A, Napoletano C, Gallo D, Perillo A,
et al: Cells with characteristics of cancer stem/progenitor cells
express the CD133 antigen in human endometrial tumors. Clin Cancer
Res. 15:4299–4311. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Liu H, Zhang HW, Sun XF, Guo XH, He YN,
Cui SD and Fan QX: Tamoxifen-resistant breast cancer cells possess
cancer stem-like cell properties. Chin Med J (Engl). 126:3030–3034.
2013.PubMed/NCBI
|
|
30
|
Wu X, Chen H, Parker B, Rubin E, Zhu T,
Lee JS, Argani P and Sukumar S: HOXB7, a homeodomain protein, is
overexpressed in breast cancer and confers epithelial-mesenchymal
transition. Cancer Res. 66:9527–9534. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Shah N, Jin K, Cruz LA, Park S, Sadik H,
Cho S, Goswami CP, Nakshatri H, Gupta R, Chang HY, et al: HOXB13
mediates tamoxifen resistance and invasiveness in human breast
cancer by suppressing ERα and inducing IL-6 expression. Cancer Res.
73:5449–5458. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Aktas B, Tewes M, Fehm T, Hauch S, Kimmig
R and Kasimir-Bauer S: Stem cell and epithelial-mesenchymal
transition markers are frequently overexpressed in circulating
tumor cells of metastatic breast cancer patients. Breast Cancer
Res. 11:R462009. View
Article : Google Scholar : PubMed/NCBI
|
|
33
|
Kim MR, Choi HK, Cho KB, Kim HS and Kang
KW: Involvement of Pin1 induction in epithelial-mesenchymal
transition of tamoxifen-resistant breast cancer cells. Cancer Sci.
100:1834–1841. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wan L, Lu X, Yuan S, Wei Y, Guo F, Shen M,
Yuan M, Chakrabarti R, Hua Y, Smith HA, et al: MTDH-SND1
interaction is crucial for expansion and activity of
tumor-initiating cells in diverse oncogene- and carcinogen-induced
mammary tumors. Cancer Cell. 26:92–105. 2014. View Article : Google Scholar : PubMed/NCBI
|