Introduction
Human bladder cancer is prevalent throughout the
world. According to statistics, human bladder cancer is the fourth
leading cause of malignancy in men and currently the tenth most
malignant tumor in women (1).
According to its pathological features, bladder cancer can be
divided into two major groups: urothelial carcinoma (UC) and
invasive bladder cancer. UC is the most widespread bladder cancer,
and it frequently recurs, but rarely progresses (2,3);
invasive bladder cancer easily progresses to distant metastases
(4). However, because the
pathophysiological mechanisms underlying bladder cancer are not
clear, the appropriate treatment to increase bladder cancer
survival rates is limited. Consequently, molecular mechanism
studies concerning the development mechanisms of bladder cancer are
urgent.
microRNAs (miRNAs) are a subgroup of endogenous
non-coding small RNAs that are 19–23 nucleotides in length and are
involved in post-transcriptional regulation by targeting the 3′UTR
of mRNA to affect biological processes (5–8). An
increasing number of studies have shown that miRNAs act as a group
of regulatory genes and participate in the development and
progression of various diseases, such as the progression of
tumorigenesis (6,9–11).
Many studies have revealed that various miRNAs, such as miR-126,
miR-143, miR-145, miR-23b, and miR-144-5p, are related to the
development of bladder cancer (12–16).
The mechanism and function of miR-24-3p have also been studied in
glioma (17), lung cancer (18), hepatocellular carcinoma (19), colorectal cancer (20), and breast cancer (21). However, the functional relevance of
miR-24-3p in bladder cancer is not fully understood.
DEDD, a member of the death effector
domain-containing protein family, participates in biological
processes, such as cell apoptosis, cell cycle, and cell mitosis
(22,23). Increasing evidence has shown that
the epithelial-mesenchymal transition (EMT), a crucial development
process, promotes cancer invasion and metastasis (24,25).
Studies have found that DEDD serves as a novel tumor repressor and
can suppress EMT and metastasis in breast and colon cancers
(26,27). Therefore, DEDD may act as a
prognostic marker and potential therapeutic target for the
treatment of cancer metastasis. However, the mechanism and function
of DEDD have not been reported in bladder cancer. To further study
the anti-metastatic roles of DEDD in bladder cancer, our study
investigate the expression level of DEDD in bladder cancer tissues
and the interaction relationship between DEDD and miR-24-3p in
bladder cancer.
Here, we confirmed that miR-24-3p was highly
expressed and that DEDD was expressed at a low level in bladder
cancer tissues. miR-24-3p promoted the abilities of proliferation,
migration and invasion; inhibited apoptosis; participated in
autophagy of bladder cancer cells by LC3, DEDD, and p62 in
vitro; and promoted the growth of bladder tumor in vivo.
Furthermore, miR-24-3p suppressed DEDD gene transcription and
accelerated proliferation and invasion through DEDD in bladder
cancer cells. Therefore, miR-24-3p could be a pivotal potential
therapeutic target for the treatment of bladder cancer.
Materials and methods
Cell lines and transfection
Human ureter epithelium (HCV29) cells, the human
bladder cancer cell line T24, human bladder transitional cell
carcinoma (UM-UC-3) cells, human bladder cancer (HBC) cells, and
malignant bladder carcinoma (BLU87) cells were purchased from
College of Life Science, Hunan Normal University, China. All of the
cells were cultured in Dulbecco's modified Eagle's medium (DMEM;
Gibco-Invitrogen, Carlsbad, CA, USA) containing 10% fetal bovine
serum (FBS; Invitrogen, Carlsbad, CA, USA), penicillin (100 U/ml;
Invitrogen), and streptomycin (100 µg/ml; Invitrogen). All of the
cultures were maintained at 37°C with 5% CO2. For
treatment, according to the manufacturer's protocol, T24 and HBC
cells (2×105 cells/well) were seeded in 6-well plates
and were transfected with 200 µl of mature miR-24-3p mock, mimic,
scramble, or inhibitor (GenePharma Co., Ltd., Shanghai, China)
using Lipofectamine™ 3000 (Invitrogen, Carlsbad, CA, USA) for 72 h.
Similarly, T24 and HBC cells were transfected with scramble and
vector, miR-24-3p and pcMV6, and miR-24-3p and DEDD with
Lipofectamine 3000, respectively.
Clinical specimens
Our study obtained ethics committee approval from
the First Affiliated Hospital of Henan University of Science and
Technology. We collected the bladder cancer tissues and adjacent
non-cancerous tissues samples from the First Affiliated Hospital of
Henan University of Science and Technology between 2014 and 2016.
All of the tissue samples were snap-frozen at −80°C. Each patient
provided written informed consent.
Quantitative real-time reverse
transcription PCR (qRT-PCR)
According to the manufacturer's instructions, total
RNA was extracted from bladder cancer tissues, matched adjacent
noncancerous tissues and treated T24 and HBC cells using the RNeasy
Plus Mini kit. cDNAs were synthesized using the RevertAid First
Strand cDNA Synthesis kit (Thermo Fisher). The samples were
normalized to U6 according to their miRNA and GAPDH for mRNA. As
previously described (28), the
mRNA expression levels were analyzed using the SYBR-Green PCR
Master Mix kit (Takara). For the detection of mRNA, the primer
sequences for GAPDH were: 5′-TGT TCG TCA TGG GTG TGA AC-3′ (the
forward primer) and 5′-ATG GCA TGG ACT GTG GTC AT-3′ (the reverse
primer); the primer sequences for DEDD were: 5′-TCC CCA GCC CTC TAA
AAC AG-3′ (the forward primer) and 5′-CCG CAG TCT GAT GTC ACA TG-3′
(the reverse primer); the primer sequences for PAK1 were: 5′- TTG
GGA ATG GAT GGC TCT GT-3′ (the forward primer) and 5′-ATG TCA ACC
TTG GGC CCA TA-3′ (the reverse primer). For the detection of miRNA,
the primer sequence for miR-24-3p was 5′-CCC ATT CAG CAG GAA CAG
AAA-3′ and that for U6 was 5′-ACG CAA ATT CGT GAA GCG TT-3′.
Western blot analysis
Treated T24 and HBC cells were lysed using RIPA
buffer containing a protease inhibitor cocktail (P8340; Sigma).
Equivalent protein samples were separated on 8% SDS/PAGE gels and
were then transferred to PVDF membranes (Millipore, Billerica, MA,
USA). The PVDF membranes were incubated in 5% skim milk (BD
Biosciences) for 2 h with a primary antibody at 4°C overnight,
followed by incubation with horseradish peroxidase-conjugated
secondary antibodies for 1 h at room temperature. The results were
obtained using an enhanced chemiluminescence detection system
(Amersham Biosciences, Piscataway, NJ, USA). In this study, the
used primary antibodies were anti-LC3 (1:200; MBL International
Co., Woburn, MA, USA), anti-DEDD (1:200; ab56480, Abcam), and
anti-P62 (1:5000, Progen, GP62-C); the anti-GAPDH antibody (1:4000,
Beverly, MA, USA) was used as an internal control.
Luciferase reporter assays
Treated T24 and HBC cells (5×104
cells/well) were cultured in 24-well plates and were co-transfected
with miR-24-3p and the wild-type pGL3-promoter-DEDD reporter vector
or mutant pGL3-promoter-DEDD reporter vector as well as a renilla
plasmid (RL-SV40) using Lipofectamine 3000 (Invitrogen). The
renilla plasmid was used as an internal control. According to the
manufacturer's instructions, the luciferase activities were
measured by the Dual-Luciferase reporter assay system (Promega,
Madison, WI, USA).
Colony forming unit (CFU) assay
For the colony forming unit assay, T24 and HBC cells
(5×103 cells/well) transfected with miR-24-3p mock,
mimics, scramble, or inhibitors were cultured into a 10-cm culture
dish with complete DMEM medium for 12 days. Next, the colonies were
fixed using methanol and dyed using Giemsa dye solution. The colony
forming units were observed and photographed.
MTT assay
T24 and HBC cells (3×103 cells/well)
transfected with miR-24-3p mock, mimics, scramble, or inhibitors
were seeded in 96-well plates for 0, 12, 24, 48, and 72 h. Next, 20
µl of a 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium
bromide (MTT, 5 mg/ml) solution was added into each well. After 4
h, formazan was dissolved in a dimethyl sulfoxide (DMSO) solution.
A microplate reader was used to measure the absorbance at 570
nm.
Flow cytometric analysis of the cell
apoptosis
The treated T24 and HBC cells were resuspended in
PBS and were stained with FITC-Annexin V and propidium iodide (PI).
The flow cytometry results were analyzed using FlowJo software. The
cells were segmented into four types, viable cells, dead cells,
early-stage apoptotic cells, and late-stage apoptotic cells.
Migration and invasion assays
According to the manufacturer's instructions, the
migration abilities of the treated T24 and HBC cells were carried
out in 24-well Transwell chambers with 8-µm pore cell culture
inserts (Costar). The treated cells (5×105 cells/well)
in serum-free media were seeded into the upper chamber, and DMEM
medium with 10% FBS was seeded into the lower chambers for 24 h in
an incubator at 37°C with 5% CO2. Thereafter, the
migratory cells were fixed, stained and counted under a microscope.
For the invasion assay, 10 µl of 1:5 diluted matrigel (BD
Biosciences) was pre-paved into Transwell inserts before the
experiment at 37°C for 2 h.
Tumor formation in nude mice
The Institutional Committee for Animal Research
approved the animal experiments of this study, and we performed
tumor formation in nude mice according to the Institutional Animal
Care and Use Committee. The flanks of 5-week-old BALB/c athymic
nude mice were subcutaneously implanted with T24 cells
(1×107 cells in 100 µl) transfected with miR-24-3p
mimics, mock, inhibitor or scramble for 0, 8, 16, 24, 32, 40, and
48 days. The tumor volume and tumor weight were measured.
Statistical analysis
The data were analyzed by Student's t-test and
analysis of variance (ANOVA) using Prism6 (GraphPad Software, Inc.,
San Diego, CA, USA) and SPSS 15.0 software (SPSS, Chicago, IL,
USA). All of the results are presented as the means ± SD. A P-value
<0.05 was considered to indicate a statistically significant
difference.
Results
miR-24-3p is overexpressed, and DEDD
expression is low in bladder cancer
We collected bladder cancer tissues and paired
non-carcinoma tissues from 26 patients. The mRNA expression levels
of miR-24-3p and DEDD were measured by qRT-PCR. The results
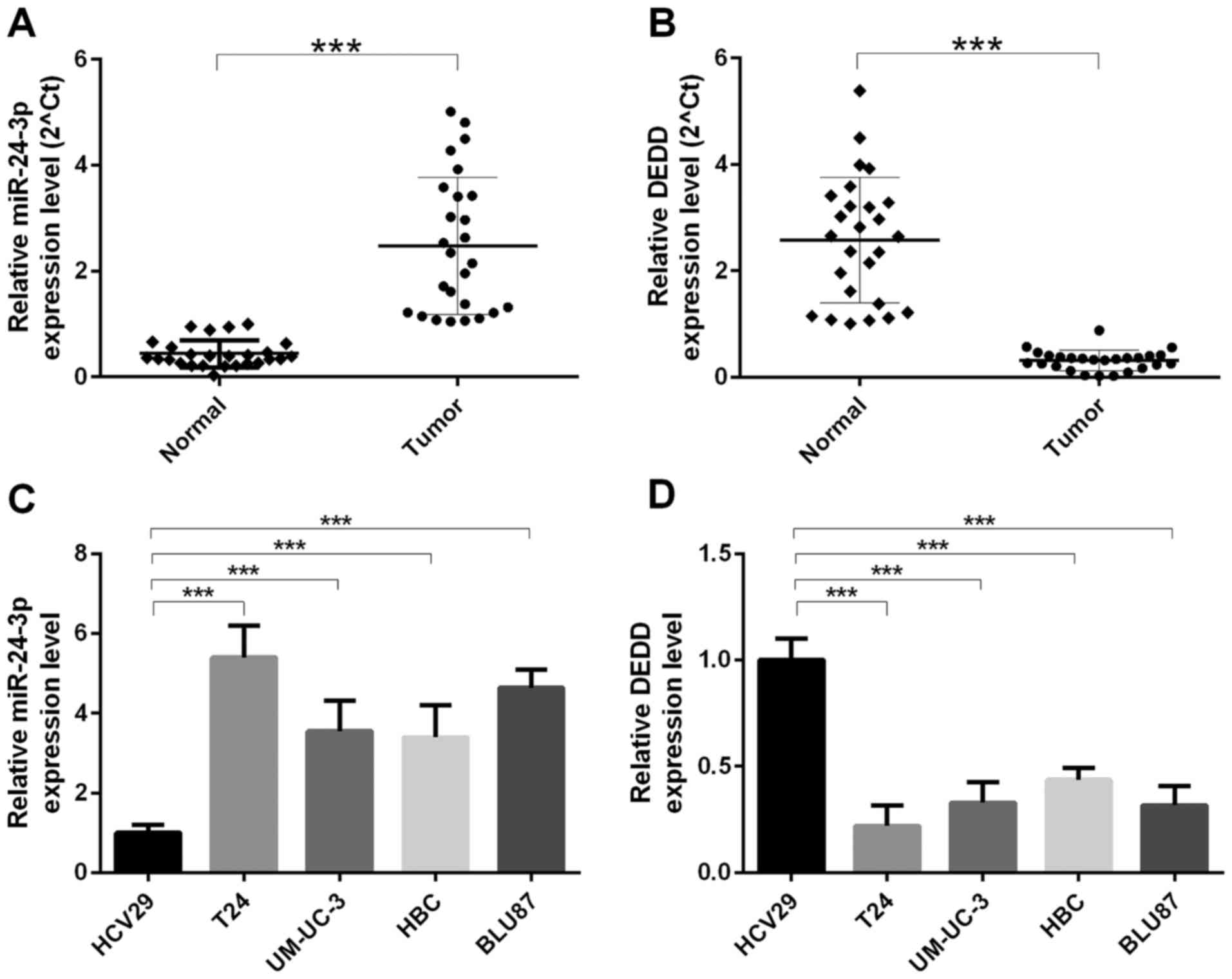
indicated that the expression level of miR-24-3p was increased in
bladder cancer tissues (n=26) compared with that in para-carcinoma
tissues (n=26) (P<0.0001) (Fig.
1A); the mRNA expression level of DEDD was decreased in bladder
cancer tissues (n=26) compared with that in para-carcinoma tissues
(n=26) (P<0.0001) (Fig. 1B). We
also found that the mRNA expression level of miR-24-3p was
increased in human ureter epithelium (HCV29) cells compared with
that in bladder cancer cells (T24, UM-UC-3, HBC, and BLU87)
(P<0.0001). The highest level of miR-24-3p was found in T24
cells (P<0.0001) (Fig. 1C); the
mRNA expression level of DEDD was decreased in human ureter
epithelium (HCV29) cells compared with that in bladder cancer cells
(T24, UM-UC-3, HBC, and BLU87) (P<0.0001). The lowest level of
DEDD was found in T24 cells (P<0.0001) (Fig. 1D).
miR-24-3p promotes cell proliferation
and invasion in bladder cancer cells
The impact of miR-24-3p on the proliferation and
invasion abilities of bladder cancer cells was detected in T24 and
HBC cells transfected with miR-24-3p mimics, mock (control),
inhibitor or scramble (control). First, the mRNA expression level
of miR-24-3p was measured by qRT-PCR. Our results indicated that
the transfection effects of miR-24-3p mimics and inhibitor were
high (P<0.05) (Fig. 2A). The
bladder cell proliferation ability was detected by the MTT assay,
and the results indicated that miR-24-3p promoted the proliferation
of T24 cells (P<0.05) (Fig. 2B)
and HBC cells (P<0.05) (Fig.
2C). Similarly, the colony forming unit assay was performed to
determine the bladder cell proliferation. Our results found that
the proliferation ability was significantly increased in T24 and
HBC cells transfected with miR-24-3p mimics relative to mock and
that the proliferation ability was significantly decreased in T24
and HBC cells transfected with miR-24-3p inhibitor relative to
scramble (P<0.05) (Fig. 2D and
E). Furthermore, we found that miR-24-3p promoted the invasion
ability of T24 and HBC cells (Fig. 2F
and G).
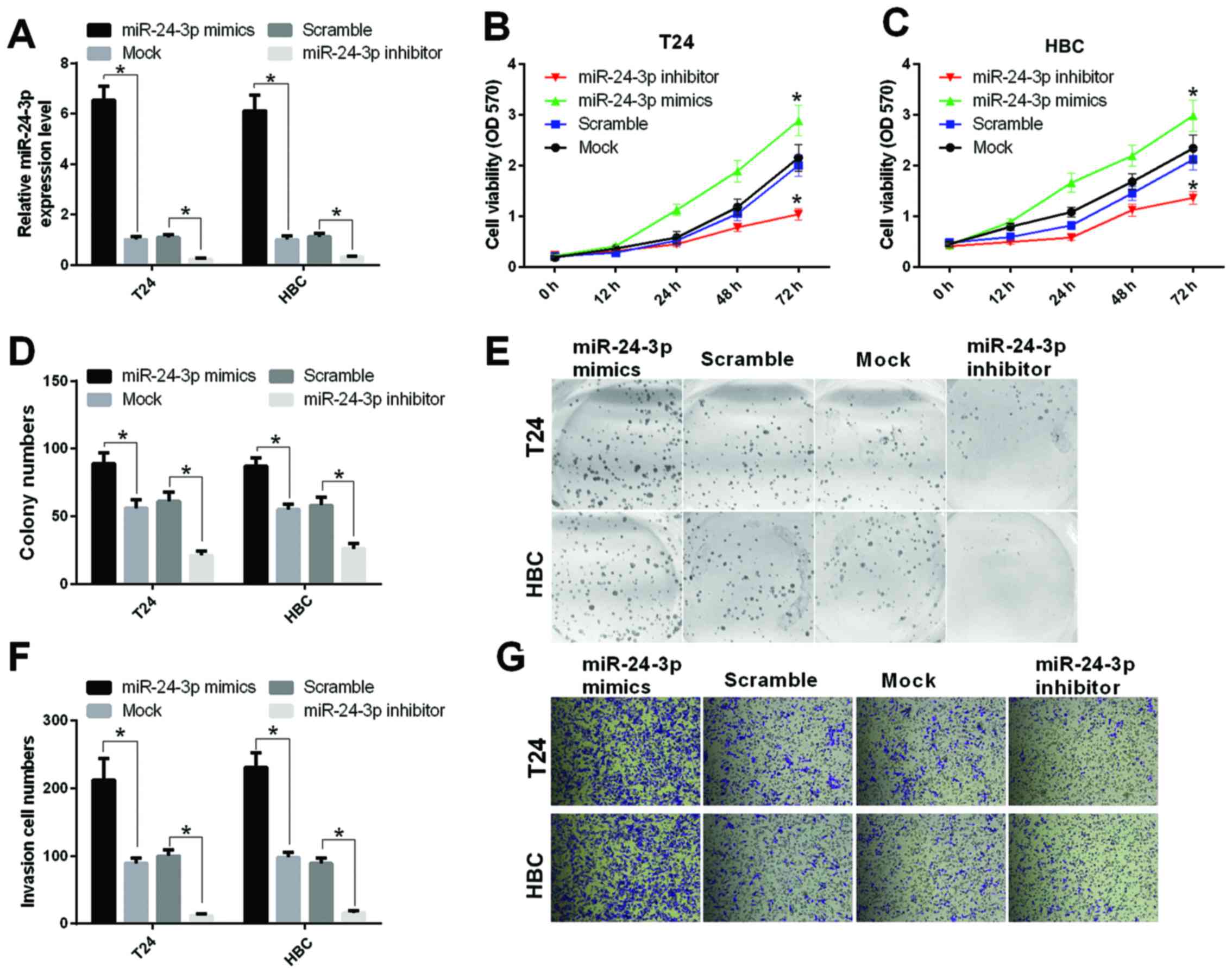 | Figure 2.miR-24-3p promotes the abilities of
cell proliferation and invasion in bladder cancer cells. (A) The
mRNA expression level of miR-24-3p was measured by qRT-PCR in T24
and HBC cells transfected with miR-24-3p mimics, mock (control),
inhibitor or scramble (control), respectively (*P<0.05). (B) The
MTT assay was performed to detect proliferation of T24 cells
transfected with miR-24-3p mimics, mock, inhibitor or scramble,
respectively, at 0, 12, 24, 48, and 72 h (*P<0.05). (C) The MTT
assay was used to measure the proliferation of Hbc cells
transfected with miR-24-3p mimics, mock, inhibitor or scramble,
respectively (*P<0.05). (D) The colony numbers were counted
(*P<0.05). (E) Cell proliferation was detected by colony forming
assay in T24 and HBC cells transfected with miR-24-3p mimics, mock,
inhibitor or scramble. (F) The invasion cell numbers were counted
(*P<0.05). (G) Cell invasion was determined by the Transwell
assay in T24 and HBC cells transfected with miR-24-3p mimics, mock,
inhibitor or scramble, respectively. |
miR-24-3p promotes cell migration,
inhibits apoptosis, and participates in autophagy
We further studied the cell migration and apoptosis
that were affected by miR-24-3p in bladder cancer cells. First, the
migration ability of T24 and HBC cells transfected with miR-24-3p
mimics, mock, inhibitor or scramble were measured by the migration
assay. The results indicated that the migration of T24 and HBC
cells transfected with miR-24-3p mimics were significantly
increased compared with mock (P<0.05); the cell migration
capacity of T24 and HBC cells transfected with the miR-24-3p
inhibitor was significantly decreased compared with scramble
(P<0.05) (Fig. 3A and B).
Second, cell apoptosis was measured by Annexin V-FITC/PI staining.
The results showed that apoptosis T24 and HBC cells transfected
with miR-24-3p mimics was significantly decreased compared with
that of the mock (P<0.05); apoptosis of T24 and HBC transfected
with miR-24-3p inhibitor was significantly increased compared with
that of scramble (P<0.05) (Fig. 3C
and D). Furthermore, the western blot results showed that
miR-24-3p upregulated the protein expression levels of LC3 and p62
and downregulated the protein expression level of DEDD in T24 and
HBC cells (Fig. 3E). These results
indicated that miR-24-3p participated in autophagy by repressing
DEDD and then increasing LC3 and p62 in bladder cancer cells.
 | Figure 3.miR-24-3p promotes cell migration,
inhibits apoptosis, and participates in autophagy by LC3, DEDD, and
p62 in bladder cancer cells. (A) The migration cell numbers were
counted (*P<0.05). (B) Cell migration was detected by the
Transwell assay in T24 and HBC cells transfected with miR-24-3p
mimics, mock, inhibitor or scramble. (C) Apoptotic cells were
analyzed (*P<0.05). (D) Cell apoptosis was measured by Annexin
V-FITC/PI staining in T24 and HBC cells transfected with miR-24-3p
mimics, mock, inhibitor or scramble. (E) Western blotting detected
the protein expression levels of LC3, DEDD, and p62 in T24 and HBC
cells transfected with miR-24-3p mimics, mock, inhibitor or
scramble. Actin was used as a protein-loading control. |
miR-24-3p suppresses DEDD gene
transcription in bladder cancer
To further explore the mechanism of miR-24-3p in
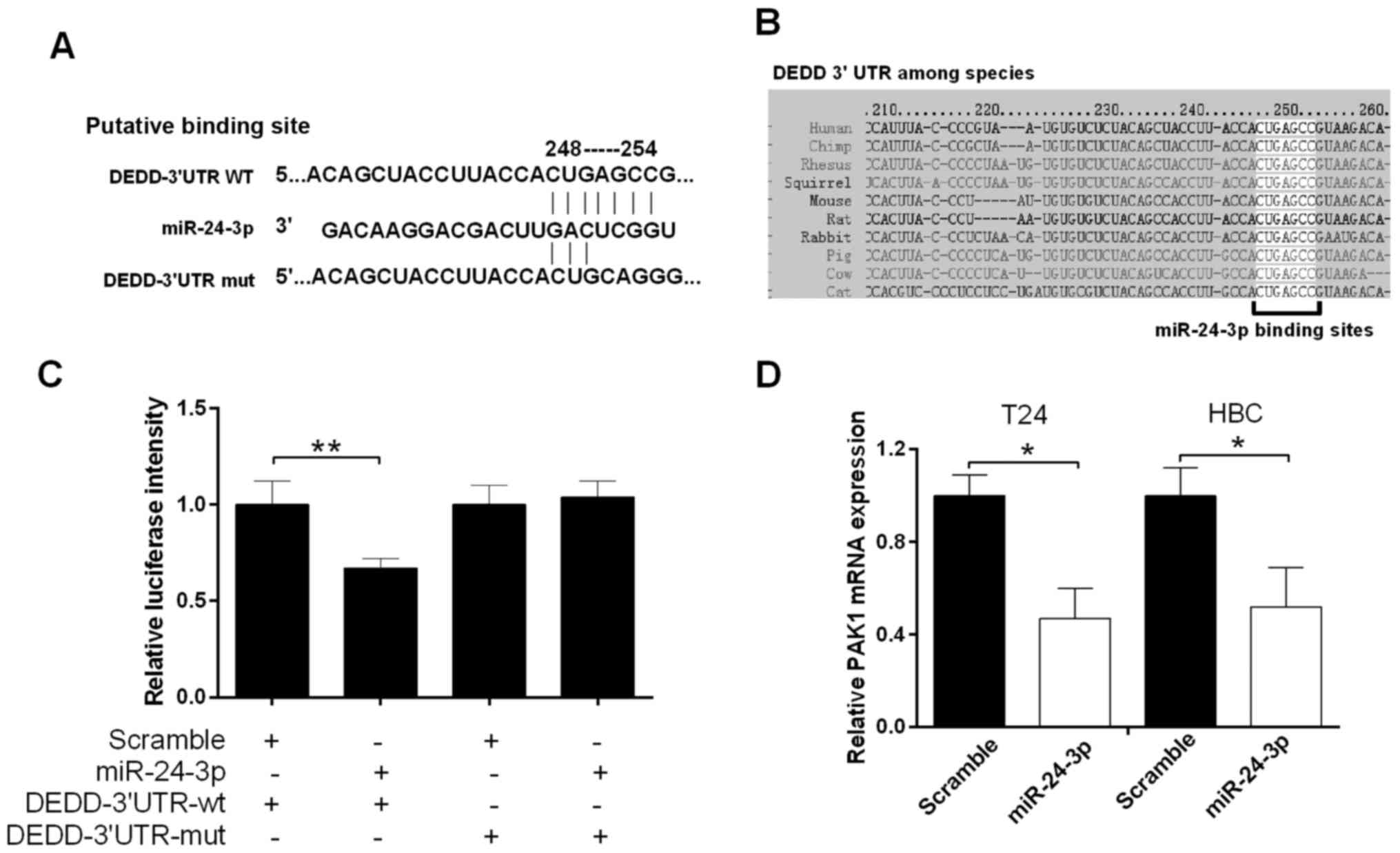
bladder cancer, we studied the relationship between miR-24-3p and
DEDD. According to TargetScan, the miR-24-3p target sites in the
sequence of DEDD were found (Fig.
4A). As shown in Fig. 4B, the
target sites between miR-24-3p and the DEDD 3′UTR among human and
other species were also analyzed. Therefore, we hypothesized that
miR-24-3p may directly bind to DEDD. The luciferase reporter gene
assay was used to measure the luciferase activity of the DEDD
3′UTR. The results showed a significant decrease in fluorescence
activity after cotransfection of miR-24-3p and the wild-type DEDD
3′UTR vector (P<0.01), but a not significant change in the
mutant DEDD 3′UTR vector (Fig. 4C).
The qRT-PCR results revealed that miR-24-3p inhibited the mRNA
expression level of DEDD T24 and HBC cells (P<0.05) (Fig. 4D).
miR-24-3p promotes the growth of
bladder tumor in vivo
To assess the effect of miR-24-3p on tumorigenesis
in vivo, T24 cells transfected with miR-24-3p mimics, mock,
inhibitor or scramble were implanted subcutaneously into nude mice.
The tumor volume was calculated, and the tumor weight was measured
at 0, 8, 16, 24, 32, 40, and 48 days. The results showed that mice
injected with cells transfected with miR-24-3p mimics were larger
than control mice (P<0.05) (Fig.
5A). Mice injected with cells transfected with miR-24-3p mimics
were heavier than control mice (P<0.05) (Fig. 5B). The tumors are shown in Fig. 5C, and RNA was extracted from the
tumors. The qRT-PCR results indicated that the mRNA expression
level of miR-24-3p was decreased significantly in mice injected
with cells transfected with miR-24-3p mimics compared with mock
(P<0.05); the mRNA expression level of miR-24-3p was increased
significantly in mice injected with cells transfected with the
miR-24-3p inhibitor compared with scramble (P<0.05) (Fig. 5D).
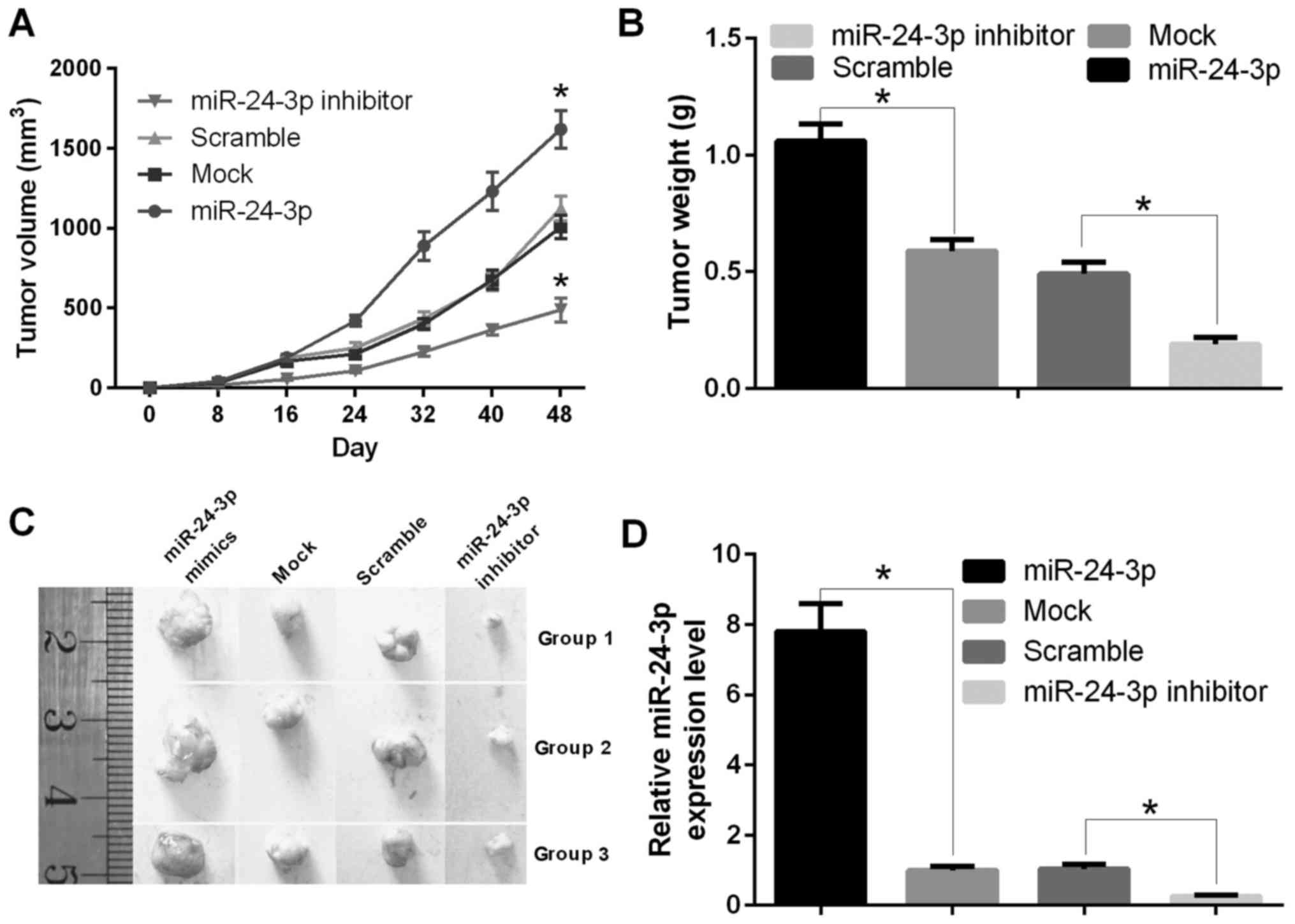 | Figure 5.miR-24-3p promotes the growth of
bladder tumors in vivo. (A) The tumor volume was counted in
nude mice with T24 cells transfected with miR-24-3p mimics, mock,
inhibitor or scramble at 0, 8, 16, 24, 32, 40, or 48 days
(*P<0.05). (B) The tumor weight was measured in nude mice with
T24 cells transfected with miR-24-3p mimics, mock, inhibitor or
scramble (*P<0.05). (C) miR-24-3p increased the tumor size in
nude mice. Athymic mice were treated, and the tumors were removed
and photographed. (D) The treated tumors in nude mice were removed,
and RNA was extracted; the mRNA expression level of miR-24-3p was
detected by qRT-PCR (*P<0.05). |
miR-24-3p accelerates the
proliferation and invasion through DEDD in bladder cancer
cells
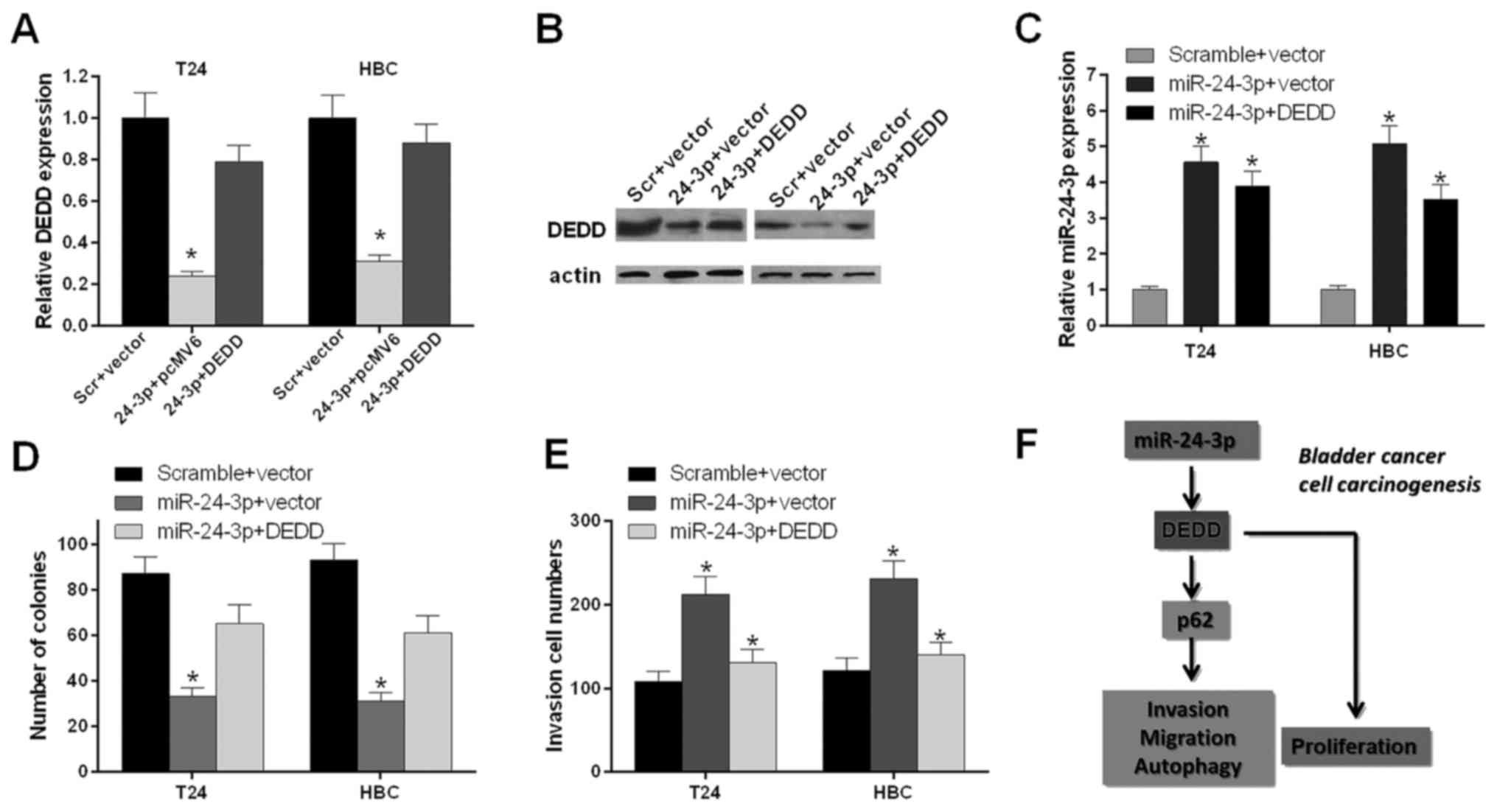
We further investigated whether miR-24-3p expression
influenced the bladder cancer cell proliferation and invasion
through DEDD. T24 and HBC cells were transfected with scramble and
vector, miR-24-3p and pcMV6, and miR-24-3p and DEDD, respectively.
The qRT-PCR results indicated that the mRNA expression level of
DEDD was decreased significantly in T24 and HBC cells transfected
with miR-24-3p and pcMV6 compared with scramble and vector
(P<0.05); the mRNA expression level of DEDD was increased
significantly in T24 and HBC cells transfected with miR-24-3p and
DEDD compared with those transfected with miR-24-3p and pcMV6
(P<0.05) (Fig. 6A). The western
blot results also indicated that miR-24-3p inhibited the protein
expression level of DEDD (Fig.
6B).
The qRT-PCR results showed that the mRNA expression
level of miR-24-3p was increased significantly in T24 and HBC cells
transfected with miR-24-3p and pcMV6 compared with that in scramble
and vector (P<0.05); the mRNA expression level of miR-24-3p was
decreased significantly in T24 and HBC cells transfected with
miR-24-3p and DEDD compared with miR-24-3p and pcMV6 (P<0.05)
(Fig. 6C). The bladder cell
proliferation ability was further detected by the colony forming
unit assay. We found that miR-24-3p inhibited the proliferation
ability of T24 and HBC cells and that DEDD promoted the
proliferation ability of T24 and HBC cells mediated by miR-24-3p
(P<0.05) (Fig. 6D). We also
found that miR-24-3p promoted the invasion ability of T24 and HBC
cells and that DEDD inhibited the invasion ability of T24 and HBC
cells mediated by miR-24-3p (P<0.05) (Fig. 6E). Therefore, as shown in Fig. 6F, miR-24-3p accelerated cell
proliferation, migration, invasion and autophagy and inhibited
apoptosis by inhibiting DEDD and activating p62 in bladder
cancer.
Discussion
miRNAs serve as a class of small non-coding RNAs
that regulate mRNAs according to the 3′UTR. Several studies have
demonstrated that miRNAs are involved in multiple biological
functions, including cell proliferation, migration, metastasis, and
inflammation, as well as tumor angiogenesis, by targeting mRNAs in
cancer (29,30). Various studies have indicated that
miRNAs participate in the occurrence and development of diversified
diseases (6,9,11). A
handful of studies has found that various miRNAs are involved in
the development of bladder cancer (31). For example, miR-126 and miR-182 are
related to urinary bladder cancer (12); miR-125b inhibits the development of
bladder cancer through E2F3 (32);
miR-143 serves as a tumor inhibitor of bladder cancer (13); miR-200 regulates EMT in bladder
cancer cells and reverses the resistance to epidermal growth factor
(EGF) receptor (33).
miR-24-3p has been studied in many cancers. For
example, miR-24-3p promotes cell proliferation in glioma cells via
MXI1 (17); the diagnosis and
prognosis of miR-24-3p have been investigated in HBV-related
hepatocellular carcinoma (19);
miR-24-3p inhibits VP16-DDP resistance in small cell lung cancer
via ATG4A(18). However, the
functional relevance of miR-24-3p in bladder cancer is still not
known. In our study, we indicated that the expression level of
miR-24-3p was increased in bladder cancer tissues compared with
para-carcinoma tissues. We also found that miR-24-3p promotes cell
proliferation, migration and invasion and inhibited apoptosis in
bladder cancer cells. In addition, we found that miR-24-3p promotes
the growth of bladder tumors in vivo.
DEDD, an important effector molecule for cell death
signaling receptors, plays critical roles in cell apoptosis, the
cell cycle, and cell mitosis (22,23). A
recent study also reported that DEDD can weaken the EMT and
inhibits tumor growth and metastasis (26). Studies have found that DEDD inhibits
transforming growth factor-β1 signaling (34). DEDD has also been correlated with
apoptosis (35,36) and reverses EMT by activating
selective autophagy (26,27). In our study, we found that miR-24-3p
suppressed DEDD gene transcription. miR-24-3p accelerated
proliferation and invasion through DEDD in bladder cancer
cells.
Autophagy, also called type 2 non-apoptotic cell
death, is a prominent mechanism of self-destruction, distinguished
by a catabolic process beginning with the formation of
autophagosomes and degradation of cytoplasmic material in the
autophagy-lysosome by lysosomal enzymes to affect health and
disease (37–40). Various studies have suggested that
LC3 can serve as an autophagosome marker, and p62 has been shown to
be related to autophagy (41–43).
Our study showed that miR-24-3p participated in autophagy through
DEDD, LC3 and p62 in bladder cancer cells.
In summary, we found that miR-24-3p was highly
expressed and that DEDD was expressed at a low level in bladder
cancer tissues. miR-24-3p promoted proliferation, migration and
invasion and inhibited apoptosis in bladder cancer cells. miR-24-3p
also participated in the autophagy of bladder cancer cells through
LC3, DEDD, and p62. We found that miR-24-3p promoted the growth of
bladder tumors in vivo. Furthermore, miR-24-3p suppressed
DEDD gene transcription. miR-24-3p accelerated proliferation and
invasion through DEDD in bladder cancer cells. Therefore, our study
indicated that miR-24-3p promoted bladder cancer progression by
inhibiting DEDD. Thus, miR-24-3p could be a pivotal potential
therapeutic target for the treatment of bladder cancer.
Acknowledgements
The study was supported by the Key Project of
Education Department of Henan Science and Technology
(13A320243).
References
|
1
|
Ying L, Chen Q, Wang Y, Zhou Z, Huang Y
and Qiu F: Upregulated MALAT-1 contributes to bladder cancer cell
migration by inducing epithelial-to-mesenchymal transition. Mol
Biosyst. 8:2289–2294. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bo J, Yang G, Huo K, Jiang H, Zhang L, Liu
D and Huang Y: microRNA-203 suppresses bladder cancer development
by repressing bcl-w expression. FEBS J. 278:786–792. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Egerod FL, Bartels A, Fristrup N, Borre M,
Ørntoft TF, Oleksiewicz MB, Brünner N and Dyrskjøt L: High
frequency of tumor cells with nuclear Egr-1 protein expression in
human bladder cancer is associated with disease progression. BMC
Cancer. 9:3852009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Said N, Sanchez-Carbayo M, Smith SC and
Theodorescu D: RhoGDI2 suppresses lung metastasis in mice by
reducing tumor versican expression and macrophage infiltration. J
Clin Invest. 122:1503–1518. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
5
|
Iorio MV and Croce CM: MicroRNAs in
cancer: Small molecules with a huge impact. J Clin Oncol.
27:5848–5856. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Farazi TA, Hoell JI, Morozov P and Tuschl
T: MicroRNAs in human cancer. Adv Exp Med Biol. 774:1–20. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Djuranovic S, Nahvi A and Green R: A
parsimonious model for gene regulation by miRNAs. Science.
331:550–553. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kasinski AL and Slack FJ: Epigenetics and
genetics. MicroRNAs en route to the clinic: Progress in validating
and targeting microRNAs for cancer therapy. Nat Rev Cancer.
11:849–864. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Baer C, Claus R and Plass C: Genome-wide
epigenetic regulation of miRNAs in cancer. Cancer Res. 73:473–477.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zaman MS, Maher DM, Khan S, Jaggi M and
Chauhan SC: Current status and implications of microRNAs in ovarian
cancer diagnosis and therapy. J Ovarian Res. 5:442012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Di Leva G and Croce CM: The role of
microRNAs in the tumorigenesis of ovarian cancer. Front Oncol.
3:1532013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Hanke M, Hoefig K, Merz H, Feller AC,
Kausch I, Jocham D, Warnecke JM and Sczakiel G: A robust
methodology to study urine microRNA as tumor marker: microRNA-126
and microRNA-182 are related to urinary bladder cancer. Urol Oncol.
28:655–661. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lin T, Dong W, Huang J, Pan Q, Fan X,
Zhang C and Huang L: MicroRNA-143 as a tumor suppressor for bladder
cancer. J Urol. 181:1372–1380. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Noguchi S, Yasui Y, Iwasaki J, Kumazaki M,
Yamada N, Naito S and Akao Y: Replacement treatment with
microRNA-143 and −145 induces synergistic inhibition of the growth
of human bladder cancer cells by regulating PI3K/Akt and MAPK
signaling pathways. Cancer Lett. 328:353–361. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Majid S, Dar AA, Saini S, Deng G, Chang I,
Greene K, Tanaka Y, Dahiya R and Yamamura S: MicroRNA-23b functions
as a tumor suppressor by regulating Zeb1 in bladder cancer. PLoS
One. 8:e676862013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Matsushita R, Seki N, Chiyomaru T,
Inoguchi S, Ishihara T, Goto Y, Nishikawa R, Mataki H, Tatarano S,
Itesako T, et al: Tumour-suppressive microRNA-144-5p directly
targets CCNE1/2 as potential prognostic markers in bladder cancer.
Br J Cancer. 113:282–289. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Xu W, Liu M, Peng X, Zhou P, Zhou J, Xu K,
Xu H and Jiang S: miR-24-3p and miR-27a-3p promote cell
proliferation in glioma cells via cooperative regulation of MXI1.
Int J Oncol. 42:757–766. 2013.PubMed/NCBI
|
|
18
|
Pan B, Chen Y, Song H, Xu Y, Wang R and
Chen L: Mir-24-3p downregulation contributes to VP16-DDP resistance
in small-cell lung cancer by targeting ATG4A. Oncotarget.
6:317–331. 2015.PubMed/NCBI
|
|
19
|
Meng F-L, Wang W and Jia W-D: Diagnostic
and prognostic significance of serum miR-24-3p in HBV-related
hepatocellular carcinoma. Med Oncol. 31:1772014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Gao Y, Liu Y, Du L, Li J, Qu A, Zhang X,
Wang L and Wang C: Down-regulation of miR-24-3p in colorectal
cancer is associated with malignant behavior. Med Oncol.
32:3622015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Lu K, Wang J, Song Y, Zhao S, Liu H, Tang
D, Pan B, Zhao H and Zhang Q: miRNA-24-3p promotes cell
proliferation and inhibits apoptosis in human breast cancer by
targeting p27Kip1. Oncol Rep. 34:995–1002. 2015.PubMed/NCBI
|
|
22
|
Hua F, Xue J, Lü X and Hu Z: DEDD
decreases Smad3 activity, promotes tumor cell apoptosis and
inhibits proliferation. Yao Xue Xue Bao. 48:680–685. 2013.(In
Chinese). PubMed/NCBI
|
|
23
|
He L, Zhong G and Zhu B: microRNA-15b
expressed in lung cancer-infiltrated CD8-positive memory T cell
inhibit cell apoptosis by repressing DEDD. Eur J Cancer (Suppl).
S147. 2013.
|
|
24
|
Carstens JL, Lovisa S and Kalluri R:
Microenvironment-dependent cues trigger miRNA-regulated feedback
loop to facilitate the EMT/MET switch. J Clin Invest.
124:1458–1460. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
25
|
De Craene B and Berx G: Regulatory
networks defining EMT during cancer initiation and progression. Nat
Rev Cancer. 13:97–110. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lv Q, Hua F and Hu Z-W: DEDD, a novel
tumor repressor, reverses epithelial-mesenchymal transition by
activating selective autophagy. Autophagy. 8:1675–1676. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lv Q, Wang W, Xue J, Hua F, Mu R, Lin H,
Yan J, Lv X, Chen X and Hu ZW: DEDD interacts with PI3KC3 to
activate autophagy and attenuate epithelial-mesenchymal transition
in human breast cancer. Cancer Res. 72:3238–3250. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Jiang L, Lai YK, Zhang J, Wang H, Lin MC,
He ML and Kung HF: Targeting S100P inhibits colon cancer growth and
metastasis by Lentivirus-mediated RNA interference and proteomic
analysis. Mol Med. 17:709–716. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wang J, Paris PL, Chen J, Ngo V, Yao H,
Frazier ML, Killary AM, Liu CG, Liang H, Mathy C, et al: Next
generation sequencing of pancreatic cyst fluid microRNAs from low
grade-benign and high grade-invasive lesions. Cancer Lett.
356:404–409. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Stahlhut C and Slack FJ: MicroRNAs and the
cancer phenotype: Profiling, signatures and clinical implications.
Genome Med. 5:1112013. View
Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ichimi T, Enokida H, Okuno Y, Kunimoto R,
Chiyomaru T, Kawamoto K, Kawahara K, Toki K, Kawakami K, Nishiyama
K, et al: Identification of novel microRNA targets based on
microRNA signatures in bladder cancer. Int J Cancer. 125:345–352.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Huang L, Luo J, Cai Q, Pan Q, Zeng H, Guo
Z, Dong W, Huang J and Lin T: MicroRNA-125b suppresses the
development of bladder cancer by targeting E2F3. Int J Cancer.
128:1758–1769. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Adam L, Zhong M, Choi W, Qi W, Nicoloso M,
Arora A, Calin G, Wang H, Siefker-Radtke A, McConkey D, et al:
miR-200 expression regulates epithelial-to-mesenchymal transition
in bladder cancer cells and reverses resistance to epidermal growth
factor receptor therapy. Clin Cancer Res. 15:5060–5072. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Xue J-F, Hua F, Lv Q, Lin H, Wang ZY, Yan
J, Liu JW, Lv XX, Yang HZ and Hu ZW: DEDD negatively regulates
transforming growth factor-β1 signaling by interacting with Smad3.
FEBS Lett. 584:3028–3034. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Lee JC, Schickling O, Stegh AH, Oshima RG,
Dinsdale D, Cohen GM and Peter ME: DEDD regulates degradation of
intermediate filaments during apoptosis. J Cell Biol.
158:1051–1066. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Schutte B, Henfling M and Ramaekers FC:
DEDD association with cytokeratin filaments correlates with
sensitivity to apoptosis. Apoptosis. 11:1561–1572. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Klionsky DJ: Autophagy: From phenomenology
to molecular understanding in less than a decade. Nat Rev Mol Cell
Biol. 8:931–937. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
38
|
Huett A, Goel G and Xavier RJ: A systems
biology viewpoint on autophagy in health and disease. Curr Opin
Gastroenterol. 26:302–309. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Li ZY, Yang Y, Ming M and Liu B:
Mitochondrial ROS generation for regulation of autophagic pathways
in cancer. Biochem Biophys Res Commun. 414:5–8. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Liu JJ, Lin M, Yu JY, Liu B and Bao JK:
Targeting apoptotic and autophagic pathways for cancer
therapeutics. Cancer Lett. 300:105–114. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Kuma A, Matsui M and Mizushima N: LC3, an
autophagosome marker, can be incorporated into protein aggregates
independent of autophagy: Caution in the interpretation of LC3
localization. Autophagy. 3:323–328. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Pankiv S, Clausen TH, Lamark T, Brech A,
Bruun JA, Outzen H, Øvervatn A, Bjørkøy G and Johansen T:
p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of
ubiquitinated protein aggregates by autophagy. J Biol Chem.
282:24131–24145. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Ichimura Y, Kumanomidou T, Sou YS,
Mizushima T, Ezaki J, Ueno T, Kominami E, Yamane T, Tanaka K and
Komatsu M: Structural basis for sorting mechanism of p62 in
selective autophagy. J Biol Chem. 283:22847–22857. 2008. View Article : Google Scholar : PubMed/NCBI
|