Introduction
Acute kidney injury (AKI) is defined as a set of
clinical-based syndromes characterized by rapid deterioration of
glomerular filtration rate (1). The
pathogenesis rate of AKI is approximately 30% in intensive care
unit (ICU) patients with complications of clinical nephrology with
mortality rate of up to 50% (2,3). Apart
from being a strong independent risk factor in developing
progressive chronic kidney disease (CKD) and end-stage renal
disease (ESRD), AKI also determines non-renal outcomes, for
instance as seen in cardiovascular disease (4–8). One
of the primary causes of AKI is ischemia-reperfusion (I/R) injury
of the kidney which occurs by means of severely restricting blood
supply to the kidney, particularly towards renal medullar. At this
point, hypotension and hypoxia-based injuries are more prone on
epithelial cells located at the S3 segment of the outer medullary
of proximal tubule (9). In animal
models of I/R-induced AKI, reperfusion causes cellular
inflammation, oxidative stress, endothelial dysfunction and
necro-apoptotic processes, paradoxically exacerbating cell damage
(10). Furthermore, the kidney
tubular epithelial cells are paramount in tubulointerstitial
fibrosis, as a result of oxidative stress and excessive protein
exposure. Subsequent overexpression and accumulation of
extracellular matrix proteins (ECM) mainly accounts for it
(11). With growing body of
evidence, it was further established that variety of forms of AKI
are associated with CKD (12). In
fact, up to 50% of clinically defined AKI cases displayed certain
degree of fibrosis.
Chronic AKI can be pathologically defined as
extensive collection of ECM in glomeruli and tubulointerstitial
that serves as the major cause of renal failure worldwide. ECM is
actively involved in cellular signaling, and thus plays an
essential role in renal damage and repair during I/R by modulating
scaring, inflammatory processes, proliferation and
transdifferentiation of fibroblasts. Due to the fact that
I/R-induced AKI involves disruption of certain signal activation
and transduction pathways, it is vital for us to fully understand
the mechanism especially in terms of working and signaling
molecules (13,14). At the moment, a suitable solution
for this disorder has not yet been made available.
Epithelial-mesenchymal transition (EMT) is a complex
reprogramming process that provides epithelial cells with a
mesenchymal phenotype. EMT takes place via numerous different
pathways and plays an essential role during organogenesis, tissue
repair and fibrosis, in addition to tumor invasion and metastasis
(15). Studies have shown that the
EMT can take place in adult kidney during chronic injury, which
plays very important roles in renal fibrosis (16,17).
Foremost causes for the AKI are ischemia, hypoxia and
nephrotoxicity. The mechanisms involved in kidney injury and repair
are complex. The kidney is particularly susceptible to ischemia and
toxins, resulting in vasoconstriction, endothelial damage, and
activation of inflammatory processes. This susceptibility arises in
part from the vascular-tubular relationships in the outer medulla
of the kidney, where the partial pressure of oxygen is low, even at
baseline, making them more vulnerable to a decreased renal blood
flow (18). A general strategy for
treating renal fibrosis is focusing on the signal pathways that
causes tubular EMT, or even on myofibroblasts transformation
(19).
Recently, another group of powerful regulators which
are given high regards from disease initiation and/or progression
viewpoint are the microRNAs (miRNAs). They are essentially small
noncoding RNA transcripts with approximate length of 22
nucleotides. Their high complementary sequences make them bind to
3′-untranslated region of mRNA targets and subsequently lead to
repression of protein synthesis (20). Another unique feature of these
regulators are the fact they could modify the expression of
multiple genes at any point of time and this serves as a major
regulator of whole disease-specific signaling cascades and pathways
instead of single gene. This feature specifically highlights the
immense potential of miRNAs. As such, any alterations in the levels
of miRNAs could possibly be the underlying mechanism of
dysregulated expression of protein which also includes kidney
disease progression. They are even being studied as internal
biomarkers for polycystic disease (21), renal cancer (22) and fibrosis (23). Regardless, only few studies were
made available in describing the role played by miRNAs in chronic
AKI induced by I/R. In addition, the alteration of miRNA
dysregulation and mRNA expression especially in chronic AKI induced
by I/R are not fully elucidated. Despite this, the presence of
miRNAs and corresponding functions were studied in a limited extent
especially pertaining to chronic AKI induced by I/R. Same pattern
holds true regarding network integrating dysregulation of miRNA as
well as changed mRNA expression which takes place in chronic AKI by
I/R.
So far, most published miRNA expression studies
utilized whole kidney tissues in I/R-induced AKI mouse without
separating the tubular cells that have a central role in
tubulointerstitial fibrosis after AKI, following excessive protein
exposure. Kidney medulla and cortex are extensively described as
having differential patterns of kidney-related miRNAs (24). The expression profile of miRNA
specific to kidney samples/specimens is always challenged with
heterogenicity of cells which are reflected in the analysis that
most of time drowns the specific markers of renal tubular
epithelia.
Our work attempted to understand the network
properties in regulating both mRNAs and miRNAs that maintain the
progression of ECM after AKI induced by lethal ischemia (45-min,
unilateral) and following reperfusion injury. This study focussed
on elucidating groups of miRNAs that have direct relations with ECM
after lethal ischemia and following reperfusion injury. Here the
combination of few methods which include RT-qPCR, microarray and
laser capture microdissection (LCM) to screen transcription
molecule patterns originating from concentrated cell groups
harvested from murine kidney with I/R-induced AKI, as an effort to
describe networking pathways related to pathologically-defined
fibrotic issues. By using the array technology we obtained a
thorough selection of miRNAs related to tubular epithelial cells
those captured by LCM and the differential profiles of mRNAs in the
kidney tissues, as verified by qPCR with subsequent analysis of
bioinformatics.
Materials and methods
Animal studies
C57BL/6J male mice were obtained from Shanghai SLAC
Laboratory Animal Co. Ltd. (licensed no.: SCXK Hu 2007–0005) and
were taken care as per the conditions described by Guide for Care
and Use of Laboratory Animals. All mice were kept in animal housing
facility under temperature of 21±1°C with 50–80% relative humidity
with 12-h light-dark cycle every alternate day except during
surgery. They were acclimatized for 2 weeks prior to experiments
and their food was commercial rodent chow with water supply ad
libitum. Ethical approval was obtained from Xinhua Hospital
Animal Care and Use Committees.
Renal ischemia-reperfusion injury
A total of 40 C57BL/6J mice (weight range: 20–24 g)
were subjected to renal unilateral I/R injury for 45 min as
previously described (25). Among
them, 8 mice underwent sham surgery. In brief, all the experiments
were performed at room temperature (24±0.5°C), as described by the
standard operating procedures. By using heating pad (HK-3, DOPS)
the intra-abdominal temperature was kept within acceptable range.
The animals were anesthetized via intraperitoneal (i.p.) injection
of 45 mg/kg pentobarbital. Lethal (45 min) ischemia injury was
obtained via clamping of left renal artery. Preparation for
sham-operated control mice were carried out with no clamping on the
renal pedicle.
Sacrifice and organ collection
Animals were sacrificed upon completion of the
experimental treatment by adopting cervical dislocation method at
3, 24, 72 and 128 h following reperfusion. Sham mice were
sacrificed by cervical dislocation method 24 h later (n=8/time
point). Both kidneys were taken out upon deliberate trans-cardiac
perfusion with 20 ml pre-cooled (4°C) physiological salt solution.
Both kidneys from randomly chosen mice undergoing I/R were excised
and stored accordingly for further use in microarray analysis of
microRNA. Other renal tissue samples were kept at −80°C via
snap-frozen method in liquid nitrogen.
Histology of renal tissue injury and
immunohistochemistry
Fixed renal tissue samples at 4% buffered
formaldehyde were dehydrated and embedded in paraffin wax (FFPE)
for histology and immunohistochemistry experiments. They were
morphologically evaluated by using tissue microarray (TMA) as
previously described (26).
Subsequent immuno-staining required TMA sections to
be incubated in 1% BSA in TBS (pH 7.4) buffer for 15 min, followed
by primary antibody [rabbit anti-mouse α-SMA immunoglobulin-G (IgG)
(1:100; R&D Systems] for 16 h and in secondary antibody [goat
anti-rabbit IgG EnVision-peroxidase polymer kit (Dako)] for 40 min,
all at room temperature. Immunostained slides were then digitalized
using a Pannoramic Scan, and the results were analyzed via the
Pannoramic Viewer software (3DHISTECH).
Laser capture microdissection
The kidney samples were subjected to LCM with the
Leica LMD7000 System as suggested by the manufacturer. In brief,
serial cut sections of 10 µm were mounted on polyethelene membrane
slides (Leica Membrane Slides), and were fixed in 100% ice-cold
ethanol for 15 min and kept for approximately 2 h at −80°C. The
slides were then washed with DEPC treated water and stained with
hematoxylin solution (MHS128; Sigma-Aldrich, St. Louis, MO, USA)
for one min. Then, the slides were dehydrated for 204 sec with 100%
ethanol and xylene for 5 min and later air-dried. The slides were
positioned onto Leica LMD7000 Laser Microdissection Instrument
(Leica LMD7000, DE). As described by manufacturer's recommended
protocol, kidney tubular epithelial cell populations
(~2×105) were selected and captured using UV laser
cutting. Immediately, LCM cap was placed in a micro-centrifuge tube
that contained 400 µl lysis/binding buffer (Ambion, Austin, TX,
USA), and stored for RNA isolation at −80°C.
Total RNA isolation and quality
control
Total RNA was extracted from tissue sections by
means of mirVana™ RNA Isolation kit (Applied Biosystems, Foster
City, CA, USA) as per provided protocol. Quantification of RNA was
performed with NanoDrop ND-2000 (Thermo Scientific) and its
integrity was evaluated via Agilent Bioanalyzer 2100 (Agilent
Technologies, Santa Clara, CA, USA). The quality was determined by
RNA integrity number (RIN). On a scale of 1–10, a RIN score was
generated for each sample. The quality is considered to be
excellent for RIN ≥7-10, good for RIN ≥5 and poor for RIN
<5.
Genome-wide miRNA profiling
analysis
Agilent Mouse miRNA V21.0 (8×60 K) microarrays
(Agilent Technologies) were utilized to match the expression
profiles of LCM selected epithelial cells taken from 6 kidney
tissues (n=3 left kidney at 24 h following reperfusion, n=3 right
kidney of the same mouse at 24 h following reperfusion). The
microarray consists of probes with 1881 mouse mature miRNAs from
Sanger miRBase V21.0. Cy3 incorporation was performed on a total
RNA of 100 ng from each of the kidney samples. Scanning of
microarray slides were by the Agilent Scanner G2505C (Agilent
Technologies). The subsequent processing of labeling and
hybridization were accomplished at Shanghai Oebiotech Company as
per the protocols in the Agilent miRNA system. In brief, total RNA
were dephosphorylated, denatured and then labeled with Cy3. After
purification, RNAs were hybridized onto the microarray. Upon
washing, the arrays tentatively were scanned with the Agilent
Scanner G2505C (Agilent Technologies). Next, Gene-spring software
(version 12.5; Agilent Technologies) was engaged to complete the
primary analysis with the raw data. Initially, the raw data were
normalized with quantile algorithm. In conditions where the probes
detects at least 1 condition out of 2 conditions have flags and
were chosen for further data analysis. miRNAs expressed
differentially were then recognized through fold change and P-value
was generated using t-test. The threshold for up- and
down-regulated genes was set at a fold change of ≥2.0 and a P-value
≤0.05, respectively. Target genes of differentially expressed
miRNAs were the intersection predicted with three databases
(TargetScan, microRNAorg, Pita). GO analysis and KEGG analysis were
functional to govern the parts of these interested target genes
played in GO terms or the pathways. Expression pattern of miRNAs
among samples was distinguished by means of Hierarchical
Clustering.
MicroRNA-mRNA co-expression network
analysis
Putative functions mediated by mRNA targets were
identified by the miRNA of interest. We first predicted
expression-based mRNA targets of miRNA. Differentially expressed
miRNA-related target genes of interest were the intersection
predicted with three databases (TargetScan, microRNAorg and Pita).
GO and KEGG analysis were performed to detect the function of these
target genes played in GO terms or the pathways. Predicted mRNAs
were validated by microarray profiling of differential mRNA
expression in kidney tissues, and mRNA targets that showed
consistent expression patterns were chosen for further study.
Predicted mRNAs were validated by microarray profiling of
differential mRNA expression in kidney tissues, and mRNA targets
that show consistent expression patterns were chosen for further
study.
RT-qPCR analysis of miRNAs and
mRNA
Total RNA of 2 µg was reverse transcribed with
Bulge-Loop miRNA-specific reverse transcription-primers (RiboBio,
Guangzhou, China). Real-time PCR reactions were carried out with
Platinum SYBR Green qPCR SuperMix-UDG chemicals (Invitrogen,
Carlsbad, CA, USA) and Bulge-Loop primers (RiboBio) on PRISM 7900HT
(Applied Biosystems, Carlsbad, CA, USA) with small nuclear RNA U6
as the normalization control. Initially, a total of 5 ng RNA
samples were reverse-transcribed (RT) to complementary DNAs (cDNAs)
using a miRNA-specific, stem-loop RT primer (for miR-21, miR-483,
miR-5115, miR-30e, miR-128, miR-181c, miR-203, miR-204, miR-30c,
the primers are shown in Table I)
as described by the manufacturer. The PCR end products were
augmented from the cDNA samples using the TaqMan Small RNA Assay
along with the TaqMan Universal PCR Master Mix2.
 | Table I.The primers for AKI mice. |
Table I.
The primers for AKI mice.
| miRNA | Primer | Sequence (5′-3′) |
|---|
| mmu-mir-204 | RNA |
UUCCCUUUGUCAUCCUAUGCCU |
| MIMAT0000237 | RT stem-loop |
GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACAGGCAT |
|
| Forward |
GCGCTTCCCTTTGTCATCCT |
|
| Reverse |
CAGTGCAGGGTCCGAGGTA |
| mmu-mir-203 | RNA |
AGUGGUUCUUGACAGUUCAACA |
| MIMAT0004547 | RT stem-loop |
GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACTGTTGA |
|
| Forward |
GCGCAGTGGTTCTTGACAGT |
|
| Reverse |
CAGTGCAGGGTCCGAGGTA |
| mmu-mir-181c | RNA |
AACAUUCAACCUGUCGGUGAGU |
| MIMAT0000674 | RT stem-loop |
GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACACTCAC |
|
| Forward |
GCGCAACATTCAACCTGTCG |
|
| Reverse |
CAGTGCAGGGTCCGAGGTA |
| mmu-mir-128 | RNA |
CGGGGCCGUAGCACUGUCUGA |
| MIMAT0016982 | RT stem-loop |
GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACTCAGAC |
|
| Forward |
GCGCCGGGGCCGTAGCACT |
|
| Reverse |
CAGTGCAGGGTCCGAGGTA |
| mmu-mir-30c | RNA |
UGUAAACAUCCUACACUCUCAGC |
| MIMAT0000514 | RT stem-loop |
GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACGCTGAG |
|
| Forward |
GCGCTGTAAACATCCTACACT |
|
| Reverse |
CAGTGCAGGGTCCGAGGTA |
| mmu-mir-30e | RNA |
UGUAAACAUCCUUGACUGGAAG |
| MIMAT0000248 | RT stem-loop |
GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACCTTCCA |
|
| Forward |
GCGCTGTAAACATCCTTGAC |
|
| Reverse |
CAGTGCAGGGTCCGAGGTA |
| mmu-U6 | RT |
GGGCCATGCTAAATCTTCTC |
| NR_003027 | Forward |
ATGGGTCGAAGTCGTAGCC |
|
| Reverse |
TTCTCGGCGTCTTCTTTCTCG |
Western blotting
The expression of TGF-β, SP1, E-cadherin, vimentin,
α-SMA and Collagen I were detected by means of standard Western
blotting. Cells were lysed in RIPA buffer and 20 µg of proteins
were separated on 8–10% SDS/PAGE gel before transferring onto PVDF
membranes (Millipore, Billerica, MA, USA). Upon incubation with
necessary antibodies, the membranes were then visualized using the
ECL system.
Cell culture and cell
transfection
HK2 cells were cultured in DMEM supplemented with
100 mg/ml of streptomycin, 100 U/ml penicillin and 10% fetal bovine
serum (FBS). They were cultivated at 37°C in a humidified incubator
of 5% CO2. They were seeded in 24-well plates and
incubated overnight, before transiently being transfected with
Lipofectamine 2000 (Invitrogen) reverse transfection protocol as
per manufacturer's protocol. miR-204 inhibitor (10 nM; both
Qiagen), acted as negative control, unspecific AllStars negative
control RNA from Qiagen or LNA control from Exiqon was used and all
the primers are shown in Table
II.
 | Table II.The primers for HK-2. |
Table II.
The primers for HK-2.
| miRNA | Primer | Sequence
(5′-3′) |
|---|
| hsa-mir-204 | RNA |
UUCCCUUUGUCAUCCUAUGCCU |
| MIMAT0000265 | RT stem-loop |
GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACAGGCAT |
|
| Forward |
GCGCTTCCCTTTGTCATCCT |
|
| Reverse |
CAGTGCAGGGTCCGAGGTA |
| SP1 | Forward |
TGGCAGCAGTACCAATGGC |
| NM_001251825 | Reverse |
CCAGGTAGTCCTGTCAGAACTT |
| GAPDH | Forward |
GGAGCGAGATCCCTCCAAAAT |
| NM_001256799 | Reverse |
GGCTGTTGTCATACTTCTCATGG |
| α-SMA | Forward |
GTGTTGCCCCTGAAGAGCAT |
| NM_001613 | Reverse |
GCTGGGACATTGAAAGTCTCA |
| Collagen I | Forward |
GAGGGCCAAGACGAAGACATC |
| NM_000088 | Reverse |
CAGATCACGTCATCGCACAAC |
| E-cadherin | Forward |
CGAGAGCTACACGTTCACGG |
| NM_004360 | Reverse |
GGGTGTCGAGGGAAAAATAGG |
| Vimentin | Forward |
GCCCTAGACGAACTGGGTC |
| NM_001017921 | Reverse |
GGCTGCAACTGCCTAATGAG |
Luciferase reporter assay
HK2 cells were seeded on 24-well plates and
co-transfected with 100 ng per well of the resulting luciferase
UTR-report vector, 20 ng per well of miR-204 precursor molecules or
control precursor (Applied Biosystems) and 2 ng per well of pRLCMV
vector (internal control, Promega) using Lipofectamine 2000
(Invitrogen) as per manufacturer's protocol. The cells were lysed
after 24 h and their corresponding luciferase activity was assessed
with the DualLuciferase Assay Reporter System (Promega).
Statistical analysis
Statistical analysis was accomplished with SigmaStat
software (Jandel Scientific Software, San Rafael, CA, USA). One-way
analysis of variance, followed by the Student-Newman-Keuls was
performed for comparison between groups. P<0.05 was considered
statistically significant in all cases.
Results
I/R injury results in significant
renal tubule interstitial tissue chronic fibrotic changes
In order to characterize whether I/R injury could
cause renal tubule interstitial tissue chronic fibrotic changes, we
employed a well-designed mouse settings of 45 min unilateral renal
I/R injury. The murine models were sacrificed at 24 h, 72 h and 7
days upon reperfusion exhibited renal tubular cell damage evaluated
by H&E (Fig. 1A). Extensive
tubule-interstitial damage was noted especially in S3 segments
located at tubules of proximal origin and also in the
corticomedullary as well as outer medulla junctions. In addition,
events of necrosis and degenerative shifts were profound in
individual tubules with lumen closure via cellular debris and
desquamated cells. Mild infiltration of chronic inflammation was
seen after 24 h of post-treatment where less quantity of necrotic
cells were noted in the tubules, especially in their lumens.
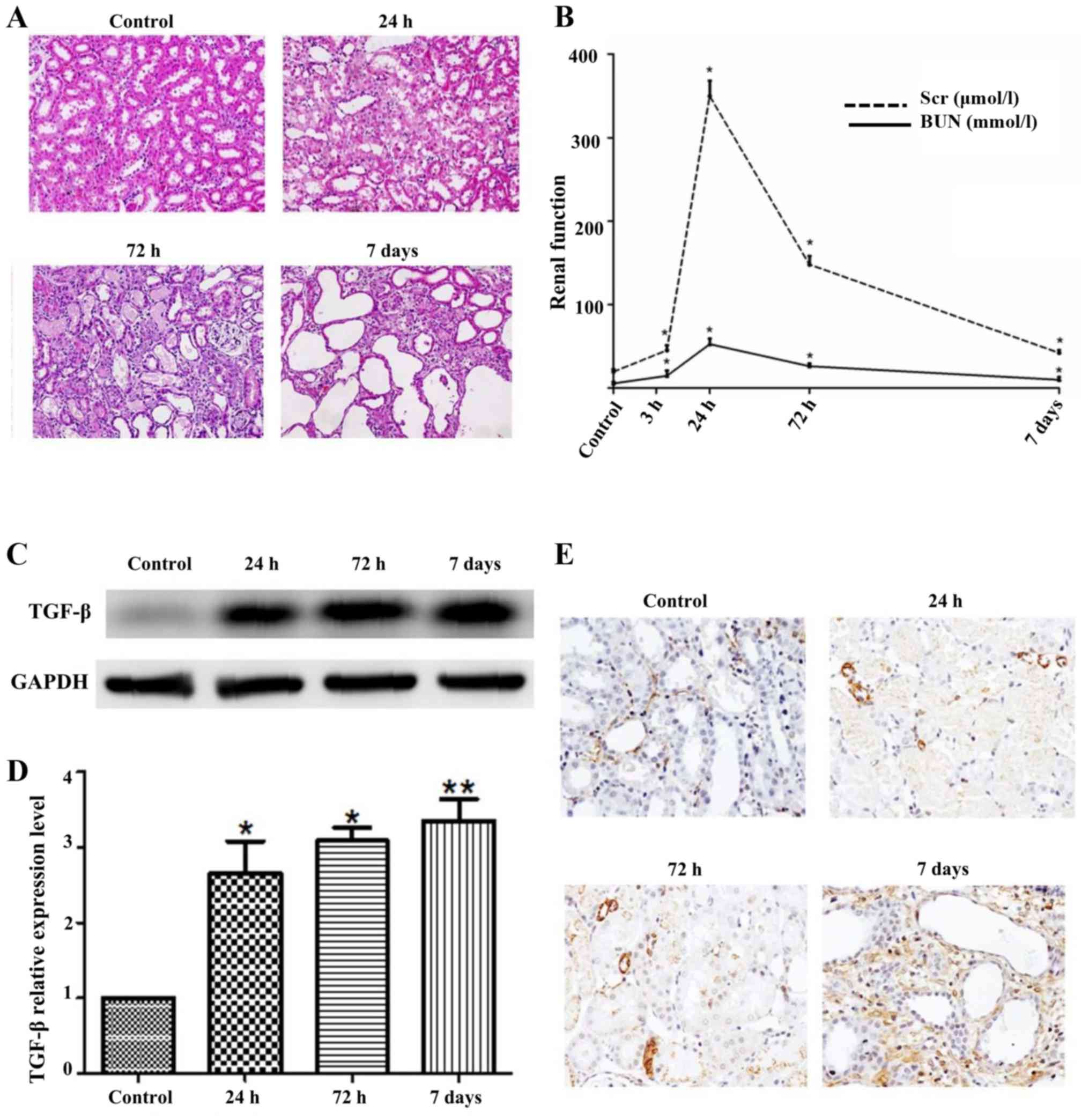 | Figure 1.I/R injury results in significant
renal tubule interstitial tissue chronic fibrotic changes. (A)
H&E of mouse kidney tissue after I/R 24 h, 72 h, 7 days,
control with sham operation; (B) Renal function after I/R 3 h, 24
h, 72 h, 7 days, control with sham operation; (C) western blotting
of TGF-β after I/R, control with sham operation; (D) the relative
expression levels of TGF-β; (E) IHC of α-SMA after I/R, control
with sham operation. Data are shown as the mean ± SD, *P<0.05
vs. control, **P<0.01 vs. control. Student's t-test was
performed to evaluate the statistical difference. |
In addition, renal function indicated that Scr level
was induced at 24 h, but gradually reduced after 24 h, while the
level of BUN changed slightly with time (Fig. 1B). Western blot results revealed
that the level of TGF-β gradually increased in time dependent
manner (Fig. 1C). Moreover, the
mRNA expression of TGF-β was seen to be consistent with the WB
outcome (Fig. 1D).
Immunohistochemical staining of alpha smooth muscle actin (α-SMA),
was an important marker of renal tubule interstitial tissue chronic
fibrotic change, and showed stronger staining for α-SMA at 7 days
than the control (Fig. 1E). Above
all, these data demonstrated that I/R injury could cause
significant renal tubule interstitial tissue chronic fibrotic
changes.
I/R injury induces distinct
downregulation of miR-204 in the kidney tissue
To explore the molecular mechanism how I/R injury
altered the miRNA expression, we assessed the miRNA profile by
means of a novel micro-bead based technology Agilent Mouse miRNA
Microarray Scanner formats (8×60 K) which contain probes for 1881
mouse mature miRNAs from the Sanger miRBase V21.0. Upon excluding
miRNAs that have minute fluorescent indication (<100 MFI), a
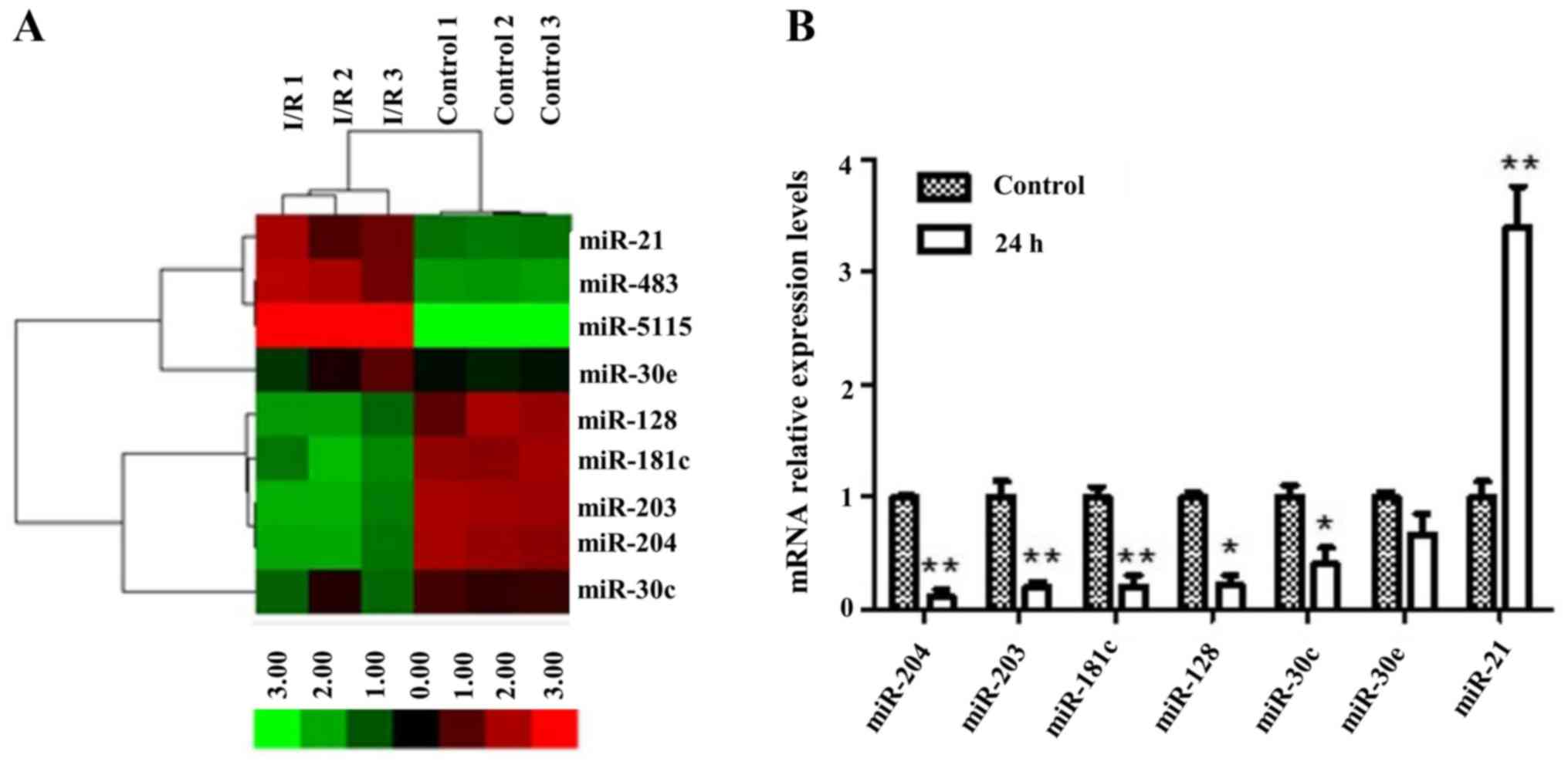
total of 9 miRNAs (namely miR-483, miR-21, miR-5115, miR-30e,
miR-128, miR-181c, miR-203, miR-204, miR-30c) of the tubule
epithelial cell population were selected and captured by LCM. These
miRNAs had significantly different expression relative to that of
right kidney without damage by I/R. These studies revealed that the
miR-204 levels were decreasing in I/R injury group as compared to
that of control group. Profound changes in levels of miRNA upon I/R
injury in all groups are depicted in Fig. 2A.
Validation of array data was performed by means of
Real-time quantitative PCR (RT-qPCR) to detect levels of miR-204
when injury occurs. Fig. 2B shows
5- to 6-fold lower expression of miR-204 under I/R-caused damage as
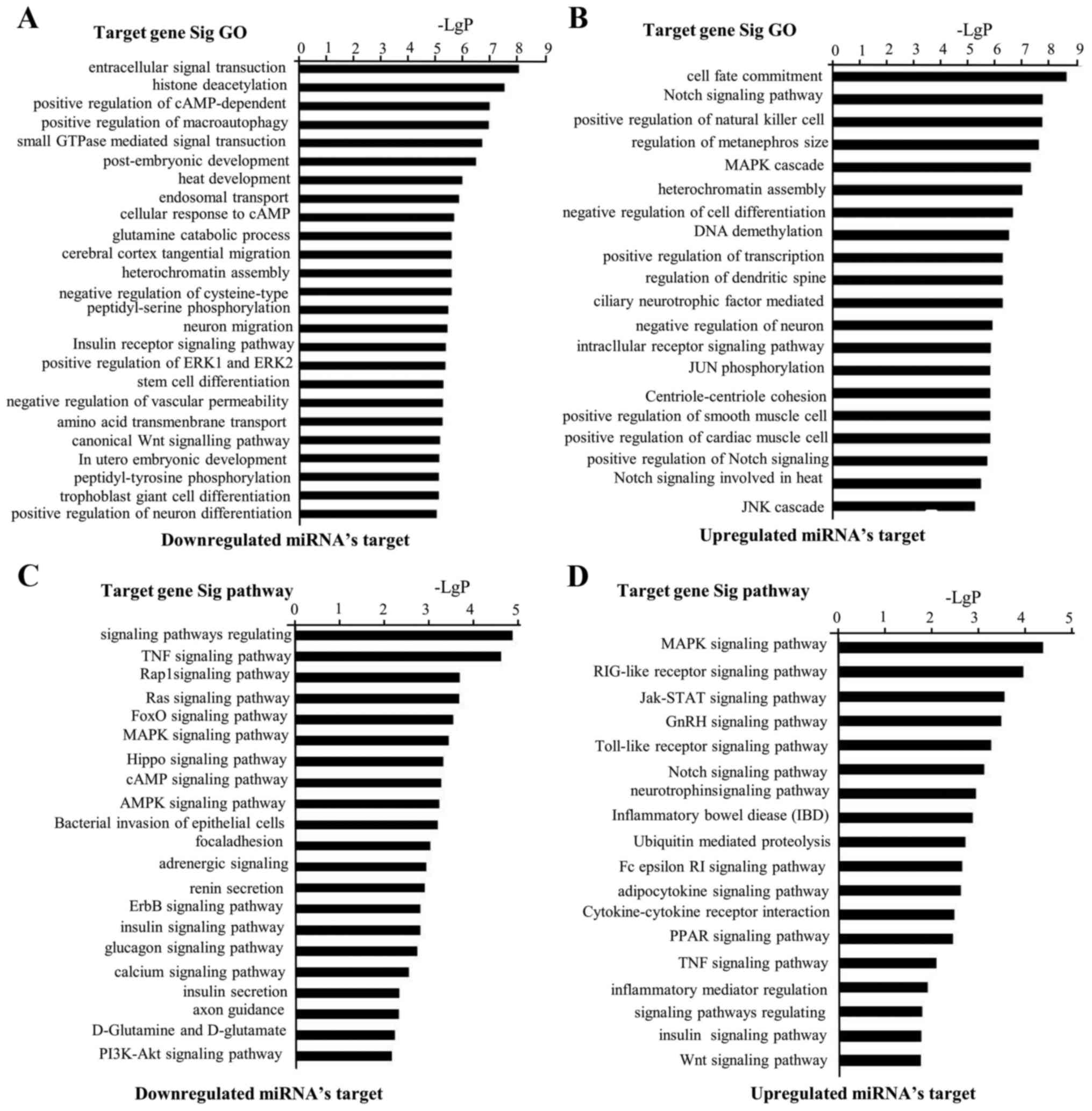
compared to control group. The analysis of Gene Ontology was
performed to elucidate distinct biological processes and molecular
functions in relation with I/R injury groups and control
groups.
The most enriched term for each group can be seen in
Fig. 3A and B listing all
significant GO categories with a P<0.05. Noteworthy, we observed
the GOs which showed target gene of downregulation. miRNAs were
potentially relevant in intracellular signal transduction,
positively modulated cAMP-dependent protein kinase activity, small
GTPase mediated signal generation, augmentation of ERK1 and ERK2
cascade and canonical Wnt signaling pathway. Enriched GO terms
associated with target gene of upregulated miRNAs were involved in
the functions of cell fate commitment, Notch signaling pathway,
augmentation of cytotoxicity due to natural killer cells,
attenuation of cell differentiation and augmentation of smooth
muscle cell apoptotic process. In Fig.
3C and D, the most enriched pathway for each group is shown.
Pathway analysis showed that target gene of downregulation miRNAs
were implicated in the following pathways: Signaling pathways
regulating stem cell pluripotency, TNF signaling pathways, Rap1
signaling pathways, Ras signaling pathways, Hippo signaling
pathways, ErbB signaling pathways and PI3K-Akt signaling pathways.
Moreover, analysis of pathways demonstrated that target gene of
upregulation miRNAs were implicated in following pathways: MAPK
signaling pathways, RIG-1-like receptor signaling pathways,
Jak-STAT signaling pathways, Notch signaling pathways,
Cytokine-cytokine receptor interactions, PPAR signaling pathways,
TNF signaling pathways and Wnt signaling pathways.
miR-204 expression was downregulated
while SP1 expression was upregulated by I/R injury
To search for potential protein-coding RNAs and
miRNAs involved in kidney from I/R-induced AKI mice, we globally
analyzed the protein-coding RNA and miRNA expression profiles of
normal group and I/R injury group. Co-expression networks of
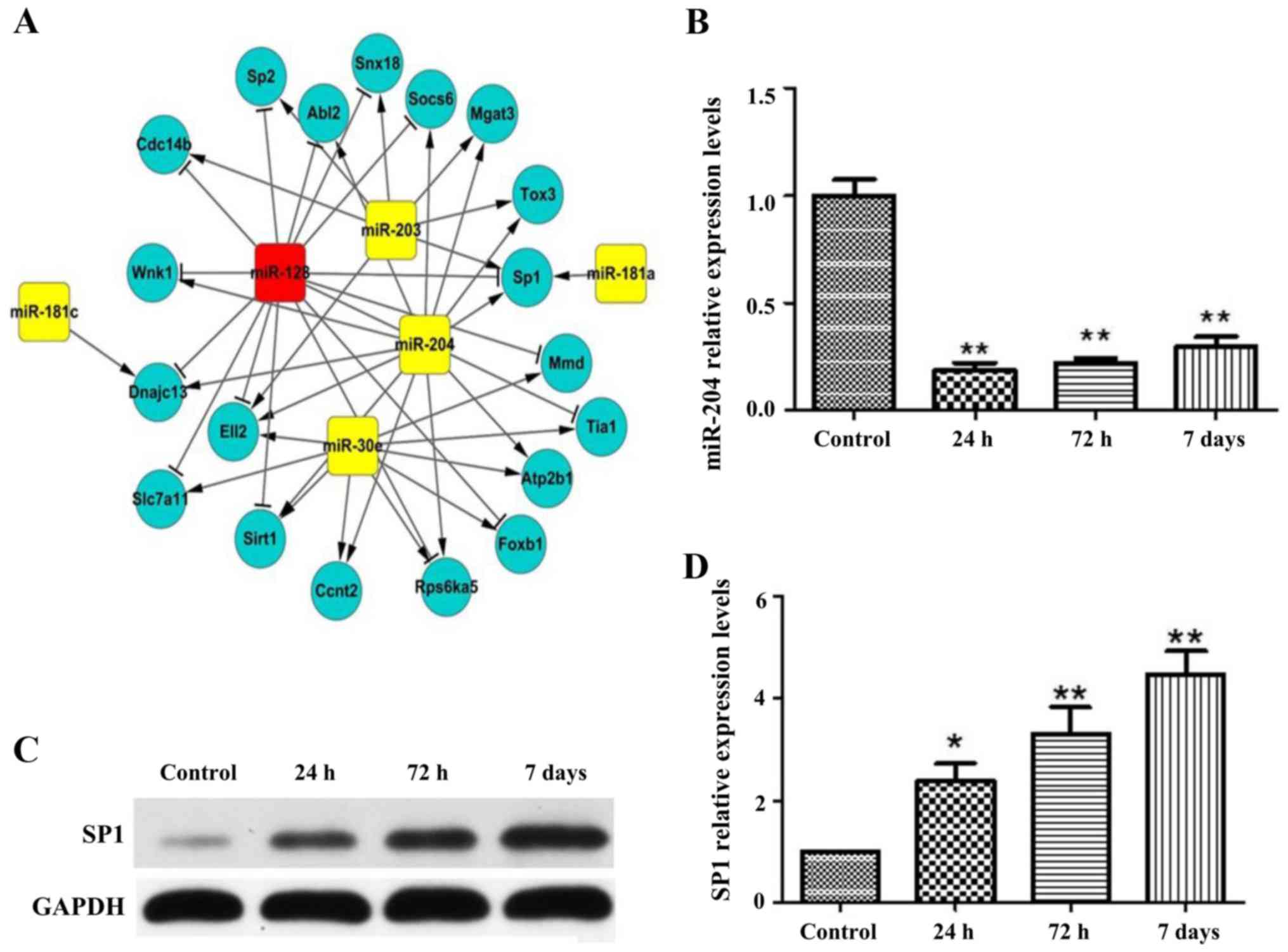
protein-coding RNAs and miRNAs were constructed (Fig. 3A). This is due to co-expression of
modules that may correspond to functional and biological pathways
and that of protein-coding RNAs can also be searched from RefSeq in
National Center for Biotechnology Information (NCBI), we
concentrated further on the co-expression models that have high
proportion of protein coding RNAs in the CRC co-expression
networks. miR-204 in CRC was identified via such modality. In this
co-expression network, miR-204 is connected to 13 protein-coding
transcription molecules which are highly relevant for genes
involving I/R by modulating scaring, inflammatory processes,
proliferation and transdifferentiation of fibroblasts (Fig. 4A). The expression pattern of miR-204
in Fig. 3A was validated using
RT-qPCR analysis and the expression of miR-204 was suppressed at 24
h, 72 h and 7 days (Fig. 4B).
Correspondingly, SP1 protein and mRNA level was induced at I/R
injury 24 h, 72 h and 7 days with western blot and RT-qPCR analysis
(Fig. 4C and D).
miR-204 suppresses hypoxia-induced
SP1/EMT signaling pathway by directly targeting SP1
We hypothesized that miRNA/mRNA network would
operate I/R injury under hypoxia. The expression of miR-204 was
markedly decreased by hypoxia while SP1 mRNA level was induced
significantly under hypoxia (Fig.
5A). Furthermore, the increased levels of SP1 induced by
hypoxia could be partially reversed by overexpression of miR-204
with miR-204 mimic (Fig. 5A). The
mRNA expression of vimentin, α-SMA and Collagen I, acting as
mesenchymal cell markers, were induced under hypoxic condition
(Fig. 5A). As expected, the
interruption approach using miR-204 mimic also reversed the
hypoxia-induced vimentin, α-SMA and Collagen I mRNA level (Fig. 5A). E-cadherin, as expression of
epithelial cell markers, displayed the opposite expression patterns
to mesenchymal cell markers (Fig.
5A). Consistent with RT-qPCR data, western blot also proved
that hypoxia-induced SP1 protein level could be partially reversed
by overexpression of miR-204 (Fig. 5B
and C). Similarly, hypoxia promoted the expression levels of
vimentin, α-SMA and Collagen, which could be partly terminated by
miR-204 (Fig. 5B and C).
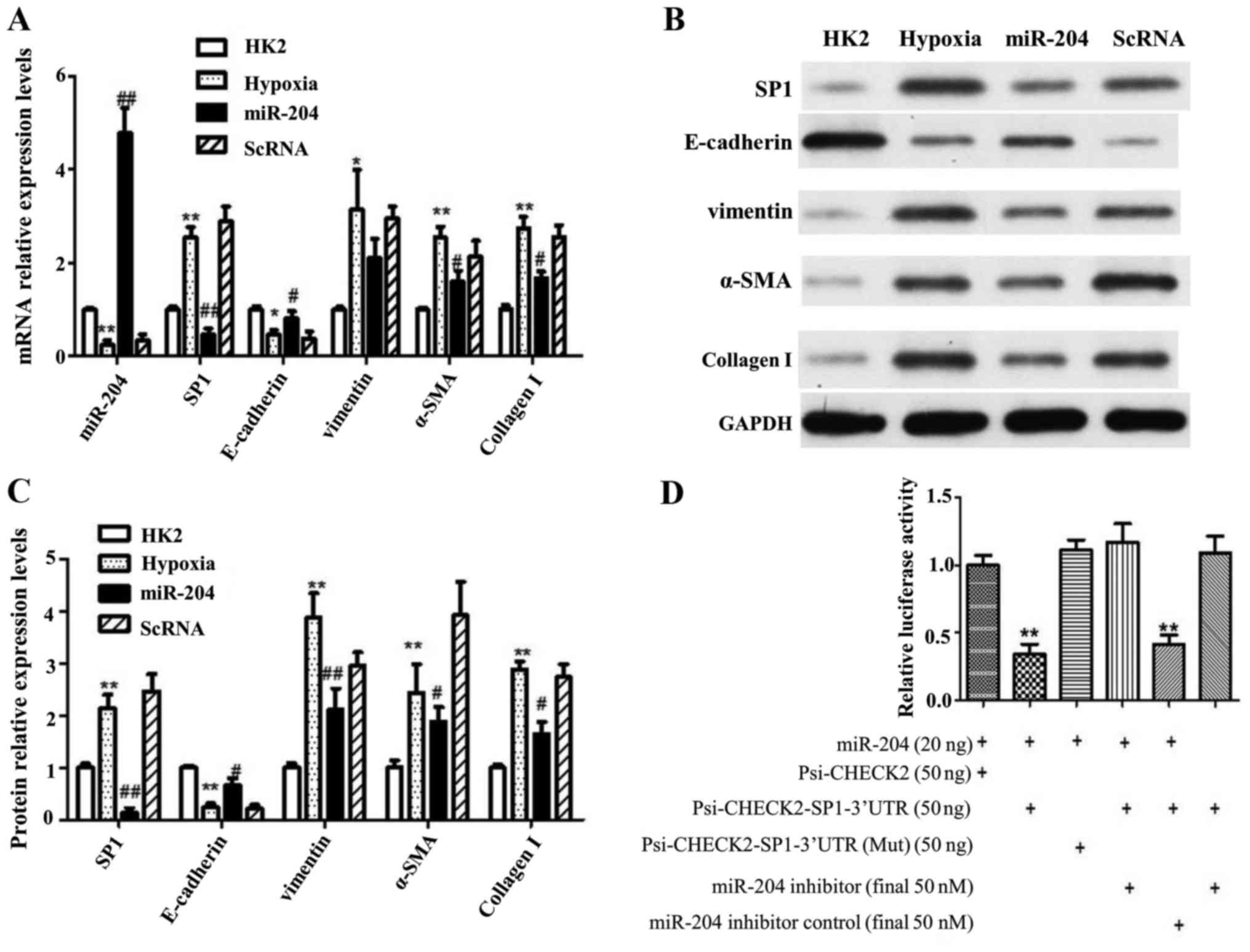 | Figure 5.miR-204 suppresses hypoxia-induced
SP1/EMT signaling pathway by directly targeting SP1. (A) relative
expression levels of mRNA, as a control, HK2 was normally treated,
hypoxia was treated with hypoxia in HK2 cells as I/R model, miR-204
was treated with both hypoxia and overexpression miR-204, and ScRNA
was treated with both hypoxia and random sequence; (B) western
blotting of EMT proteins and SP1; (C) the relative expression
levels of western blotting; (D) luciferase activity decreased
significantly in HEK293 cells co-transfected with miR-204 wild-type
3′UTR of SP1, but mutant of 3′UTR of SP1 did not. Data are shown as
the mean ± SD, *P<0.05 vs. control, **P<0.01 vs. control,
#P<0.05 vs. model, ##P<0.01 vs. model.
Student's t-test was performed to evaluate the statistical
difference. |
Using TargetScan software, SP1 gene was further
validated as candidate target of miR-204 since it carries
corresponding miR-204 target sites in the 3′-UTR. As a way to
verify this phenomenon, a luciferase-based reporter vector
containing oligo-nucleotides that fully complements the 3′-UTR of
wild-type SP1, or its relevant mutants was constructed. Using HK2
cell lines, non-functional control miR-NC RNAs or pre-hsa-miR-204
RNAs co-transfection were performed. It was further noted that
wild-type SP1 3′-UTR and miR-204 target sequences had distinct
reduction in its relative luciferase activity, especially upon
presence of miR-204 and not the mutant (Fig. 5D). Additionally, miR-204 inhibitor
could also induce the relative luciferase activity of SP1 3′-UTR
but not mutant sequence. Taken together, the data from luciferase
reporter assay presented that SP1 is both a direct and an exact
target of miR-204.
Discussion
Numerous studies have depicted that simultaneous
clamping especially with non-traumatic microaneurysm clamps, of the
renal pedicle of both kidneys could induce acute kidney injury
(AKI) (5). In our current study, we
first globally screened microRNAs whose levels are affected during
AKI using a microarray; then we validated these screened-out
targets by qPCR on a one-by-one basis. Our goal is to identify
specific microRNA candidates that might play essential roles in AKI
so that we can further study in detail these validated microRNA
molecules in cell culture to animal models of AKI. It is a known
observation, which we also have taken notice of, that there are
fluctuations of even base-line RNA levels among individual mouse;
and considering also the high-throughput nature of microarrays, we
aimed to minimize RNA level fluctuations in each individual mouse,
therefore, in our study, AKI were induced instead by unilaterally
clamping using non-traumatic microaneurysm clamps on the renal
pedicle of the left kidney and paired comparison were done on
microRNA levels between the left and right kidneys within the same
mouse. Reflecting on our interpretations of opposing expression
levels in both left and right kidney tissue, it is plausible to
roughly assess the different levels of miRNAs playing a crucial
role on the mechanism of extracellular matrix proteins (ECM)
through the progress of kidney injury and tissue repair process
following I/R.
Laser capture microdissection (LCM) has been proven
useful in separating cell populations from tissue sections and the
standard genomic methods have served as an added tool and further
improved the analysis of complex tissues. The limitation, however,
is that only small amount of material with compromised quality
could be recovered from LCM for genomic usage. To overcome this
issue, tissue preparation protocol was optimized and specific miRNA
expression profile approach was performed. Furthermore, miRNAs are
highly stable and difficult to degrade in instances such as in
kidney tissues and clinical plasma (27). Several studies have reported that
differences in miRNA expression did not overlap each other in the
post-I/R tissues (28). As a
result, we attempted extra work to detect I/R injury-altered
miRNAs.
The goal of this study was to describe the
expression patterns of miRNAs in kidney tissues upon induction with
I/R. It was obviously noted that miR-21, miR-483, miR-5115,
miR-204e, miR-128, miR-181c, miR-203, miR-204, miR-204c were highly
regulated as compared to sham. This was indeed a brand new protocol
in identifying 29 core mRNA target genes related to miR-204 ECM
upon kidney injury. miRNA expression over extended period of time
is crucial to detect key miRNA that are relevant to different
levels of kidney injury which include repair and damage processes
(29). Our study demonstrated that
renal tubules interstitial tissue chronic fibrotic changes were
significant after I/R injury at 24 h. We shifted our focus of
analysis on 24 h as the time point encircling miRNA microarray for
the reason that we presented a complete outline for prediction of
the relationship between the progression of the chronic fibrotic
changes of renal tubule interstitial tissue and miRNAs, and in
detail, the molecule mechanisms of inhibition and interruption of
chronic AKI.
Our results demonstrated that the miR-204 expression
was increased during the recovery and maintenance phase in
I/R-induced AKI models. It is known that miR-204 belongs to
miR-204/211 family, which includes miR-204 and miR-211. All of them
have similar sequence in their 5′ terminus known as ‘seed
sequence’. This miRNA is profound in kidney and play vital roles
for podocyte homeostasis and pro-nephros development. It has been
previously shown that hepatic miR-204 was spotted in CCl4-induced
liver fibrosis. The downregulation of miR-204 was described to
stimulate epithelial-mesenchymal transition (EMT) of epithelial
thyroid cells especially in anaplastic thyroid pancreatitis. In
this study, a drastic decline of miR-204 expression was noted
especially in fibrotic kidney.
Moreover, Jiang et al (30) reported that miR-204 was
downregulated by TGF-β1 in NRK-52E cells though there was partial
blockage by Smad7 transfection. As such, implication by Smad
signaling activation has direct effect on downregulation of the
miR-204. Taken together, Jiang et al (30) results indicate that the miR-204
downregulation could be a vital facilitator for tubular-cell
extracellular matrix production induced by TGF-β1 in renal fibrosis
in a mouse model. This study however depicted that level of miR-204
corresponds to severity of AKI though the correlation is not yet
fully understood as it can also indicate protective behaviour. As
previously described (6), miR-204
had control over apoptosis and necrosis of renal tubular epithelial
cells as well as promoting cell proliferation. As such, clearly
this molecule has a huge role to play as kidney protector in AKI.
In addition, release of miR-204 also takes place in response to
inflammation, stress and also protects the kidneys from delayed
ischemia preconditioning.
It should be noted that the whole body responds to
I/R injury and microRNA circulation could perhaps be initiated in
another place. Even if injured kidneys are speculated to be the
primary source of microRNAs in circulation, an extensive amount of
work are required to recognize the exact cell type which produce
them. In our study, our data concluded that miR-204 is a specific
I/R induced chronic AKI suppressor gene, thereby antagonizing the
SP1/EMT-signaling pathway.
Less information is available regarding the
integration of miRNA dysregulation networks which are prevalent in
chronic AKI. In that sense, perhaps an integrative network
consisting of both mRNA and miRNAs my hold the clue to detect key
factors of chronic AKI induced by I/R. Our current work identified
an integrative regulatory network of transcript levels and altered
miRNAs that govern chronic AKI induced by I/R. The current study
was designed to be a pilot with limitation in terms of mechanistic
experiments that would give us a clearer data regarding microRNAs
individually. As such, we truly consider that profiling of
microRNAs systematically is essential to study their molecular
properties. To conclude, this study provided distinct indication
that miR-204 are linked with hazard of chronic AKI by I/R. Studies
using large cohort would be paramount to ascertain the molecular
mechanisms involved in pathophysiology of chronic AKI. Accurate
measures to detect and classify the miRNAs with respect to AKI
would help in terms of healing mediations that can be harnessed, in
near future.
Acknowledgements
We are thankful to all authors for their efforts and
assistance in the Department of Nephrology, Xin Hua Hospital
Affiliated to Shanghai Jiao Tong University School of Medicine.
This work was supported by the grants of The National Natural
Science Foundation (nos. 81200496, 81270823), Shanghai Science and
Technology Committee (no. 12DJ1400200).
References
|
1
|
Bellomo R, Kellum JA and Ronco C: Acute
kidney injury. Lancet. 380:756–766. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Waikar SS, Curhan GC, Wald R, McCarthy EP
and Chertow GM: Declining mortality in patients with acute renal
failure, 1988 to 2002. J Am Soc Nephrol. 17:1143–1150. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lafrance JP and Miller DR: Acute kidney
injury associates with increased long-term mortality. J Am Soc
Nephrol. 21:345–352. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ishani A, Xue JL, Himmelfarb J, Eggers PW,
Kimmel PL, Molitoris BA and Collins AJ: Acute kidney injury
increases risk of ESRD among elderly. J Am Soc Nephrol. 20:223–228.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Heung M and Chawla LS: Predicting
progression to chronic kidney disease after recovery from acute
kidney injury. Curr Opin Nephrol Hypertens. 21:628–634. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Belayev LY and Palevsky PM: The link
between acute kidney injury and chronic kidney disease. Curr Opin
Nephrol Hypertens. 23:149–154. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Heung M and Chawla LS: Acute kidney
injury: Gateway to chronic kidney disease. Nephron Clin Pract.
127:30–34. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Chawla LS and Kimmel PL: Acute kidney
injury and chronic kidney disease: An integrated clinical syndrome.
Kidney Int. 82:516–524. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Basile DP: Rarefaction of peritubular
capillaries following ischemic acute renal failure: A potential
factor predisposing to progressive nephropathy. Curr Opin Nephrol
Hypertens. 13:1–7. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Eefting F, Rensing B, Wigman J, Pannekoek
WJ, Liu WM, Cramer MJ, Lips DJ and Doevendans PA: Role of apoptosis
in reperfusion injury. Cardiovasc Res. 61:414–426. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Manninen A: Epithelial polarity -
generating and integrating signals from the ECM with integrins. Exp
Cell Res. 334:337–349. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Stallons LJ, Whitaker RM and Schnellmann
RG: Suppressed mitochondrial biogenesis in folic acid-induced acute
kidney injury and early fibrosis. Toxicol Lett. 224:326–332. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Villa P, Bigini P, Mennini T, Agnello D,
Laragione T, Cagnotto A, Viviani B, Marinovich M, Cerami A, Coleman
TR, et al: Erythropoietin selectively attenuates cytokine
production and inflammation in cerebral ischemia by targeting
neuronal apoptosis. J Exp Med. 198:971–975. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kobayashi M, Sugiyama H, Wang DH, Toda N,
Maeshima Y, Yamasaki Y, Masuoka N, Yamada M, Kira S and Makino H:
Catalase deficiency renders remnant kidneys more susceptible to
oxidant tissue injury and renal fibrosis in mice. Kidney Int.
68:1018–1031. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Thiery JP, Acloque H, Huang RYJ and Nieto
MA: Epithelial-mesenchymal transitions in development and disease.
Cell. 139:871–890. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Vongwiwatana A, Tasanarong A, Rayner DC,
Melk A and Halloran PF: Epithelial to mesenchymal transition during
late deterioration of human kidney transplants: The role of tubular
cells in fibrogenesis. Am J Transplant. 5:1367–1374. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Rastaldi MP: Epithelial-mesenchymal
transition and its implications for the development of renal
tubulointerstitial fibrosis. J Nephrol. 19:407–412. 2006.PubMed/NCBI
|
|
18
|
Bonventre JV: Pathophysiology of AKI:
Injury and normal and abnormal repair. Contrib Nephrol. 165:9–17.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Jiang YS, Jiang T, Huang B, Chen PS and
Ouyang J: Epithelial-mesenchymal transition of renal tubules:
Divergent processes of repairing in acute or chronic injury? Med
Hypotheses. 81:73–75. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Bagga S, Bracht J, Hunter S, Massirer K,
Holtz J, Eachus R and Pasquinelli AE: Regulation by let-7 and lin-4
miRNAs results in target mRNA degradation. Cell. 122:553–563. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chandrasekaran K, Karolina DS, Sepramaniam
S, Armugam A, Wintour EM, Bertram JF and Jeyaseelan K: Role of
microRNAs in kidney homeostasis and disease. Kidney Int.
81:617–627. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Pandey P, Brors B, Srivastava PK, Bott A,
Boehn SNE, Groene HJ and Gretz N: Microarray-based approach
identifies microRNAs and their target functional patterns in
polycystic kidney disease. BMC Genomics. 9:624–629. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kato M, Arce L, Wang M, Putta S, Lanting L
and Natarajan R: A microRNA circuit mediates transforming growth
factor-β1 autoregulation in renal glomerular mesangial cells.
Kidney Int. 80:358–368. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Tian Z, Greene AS, Pietrusz JL, Matus IR
and Liang M: MicroRNA-target pairs in the rat kidney identified by
microRNA microarray, proteomic, and bioinformatic analysis. Genome
Res. 18:404–411. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Vaidya VS, Ozer JS, Dieterle F, Collings
FB, Ramirez V, Troth S, Muniappa N, Thudium D, Gerhold D, Holder
DJ, et al: Kidney injury molecule-1 outperforms traditional
biomarkers of kidney injury in preclinical biomarker qualification
studies. Nat Biotechnol. 28:478–485. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Krenacs T, Ficsor L, Varga SV, Angeli V
and Molnar B: Digital microscopy for boosting database integration
and analysis in TMA studiesTissue Microarrays: Methods and
Protocols. Ronald Simon: Humana press publisher; New York: 664. pp.
163–175. 2010, doi: 10.1007/978-1-60761-806-5_16. View Article : Google Scholar
|
|
27
|
Mitchell PS, Parkin RK, Kroh EM, Fritz BR,
Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O'Briant
KC, Allen A, et al: Circulating microRNAs as stable blood-based
markers for cancer detection. Proc Natl Acad Sci USA.
105:10513–10518. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Godwin JG, Ge X, Stephan K, Jurisch A,
Tullius SG and Iacomini J: Identification of a microRNA signature
of renal ischemia reperfusion injury. Proc Natl Acad Sci USA.
107:14339–14344. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
El Sabbahy M and Vaidya VS: Ischemic
kidney injury and mechanisms of tissue repair. Wiley Interdiscip
Rev Syst Biol Med. 3:606–618. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
30
|
Jiang L, Qiu W, Zhou Y, Wen P, Fang L, Cao
H, Zen K, He W, Zhang C, Dai C, et al: A microRNA-30e/mitochondrial
uncoupling protein 2 axis mediates TGF-β1-induced tubular
epithelial cell extracellular matrix production and kidney
fibrosis. Kidney Int. 84:285–296. 2013. View Article : Google Scholar : PubMed/NCBI
|