Introduction
Acute promyelocytic leukemia (APL) is a distinct
subtype of acute myeloid leukemia (AML). APL is characterized by
the selective expansion of immature myeloid precursor cells that
are blocked at the promyelocytic stage. Almost 98% of patients with
APL have transformed promyelocytes carrying the t(15;17)(q22;q12)
translocation that generates the promyelocytic leukemia-retinoic
acid receptor α (PML-RARα) fusion gene product (1). PML-RARα has been proposed to act as a
dominant negative inhibitor of endogenous PML and RARα functions,
causing resistance to apoptosis and blocking myeloid
differentiation (2).
Several studies revealed that transgenic and
knock-in animal models expressing PML-RARα in early myeloid cells
developed APL (3–5). However, when PML-RARα was expressed in
late myeloid cells, APL did not develop (6). Moreover, one study suggested that
PML-RARα may require cleavage to become fully oncogenic (7).
Neutrophil elastase (NE), an early myeloid-specific
serine protease, cleaves the PML-RARα protein of bcr-1 into two
parts. Mice deficient in NE develop APL at a greatly reduced rate
when crossed with PML-RARα knock-in mice (8). We recently found that overexpression
of the PML protein lacking the nuclear localization signal
[PML(NLS−)] can inhibit apoptosis and promote the growth
of HL60 cells (9). We also found
that NLS-RARα promotes proliferation and inhibits differentiation
of HL60 cells (10). Together,
these observations suggest that activation of PML-RARα by
NE-mediated cleavage plays a key role in the pathogenesis of
APL.
PML is a RING finger protein that belongs to the
tripartite motif protein family (11). There are several variants of human
PML generated by alternative splicing with variable C-terminal
lengths, and sizes ranging from 47 to 160 kDa (12). However, all isoforms have the same
N-terminus containing the NLS, B-Boxes, and an α-helical coiled
region. PML is an essential component of highly dynamic nuclear
structures known as PML oncogenic domains or nuclear bodies (NBs)
(13). PML and PML NBs have been
implicated in regulating growth inhibition, senescence and
apoptosis (14), and a myriad of
PML activities have been linked to its function in NB formation.
Deregulation of PML can be oncogenic and has been observed in
multiple human cancers (15). PML
can be SUMOylated on three different lysine residues and contains a
SIM domain. Lys65 in the RING finger domain, Lys160 in the B1 Box,
and Lys490 in the NLS contribute three major sentrinization sites
(16–18).
In normal human cells, PML has a nuclear
localization. A minority of PML types (such as PML VIb) show a
cytoplasmic localization (19). The
best understood system for the transport of macromolecules between
the cytoplasm and the nucleus is the classical nuclear import
pathway. In this pathway, a protein containing a basic NLS is
imported by a heterodimeric receptor consisting of β-karyopherin.
This receptor mediates interactions between the nuclear pore
complex and the adaptor protein importin α, which binds directly to
the classical NLS (20). This
indicates that the NLS sequence is critical for both PML
SUMOylation and PML NB formation, and that a deficiency of the NLS
sequence influences PML nuclear localization.
In the present study, PML and PML(NLS−)
expression was investigated in normal human cells and APL blasts.
An immunofluorescence co-localization assay and
co-immunoprecipitation were used to demonstrate that PML was
localized in the nucleus by its NLS sequence interacting with
importin α. However, aberrant PML(NLS−) disrupted the
activity of the NLS in the cytoplasm and eliminated the interaction
with importin α. These results indicated that PML(NLS−)
may be a potential biomarker for APL.
Materials and methods
Cell lines and primary APL blasts
HL-60, NB4 and HEK293 cell lines were purchased from
the Shanghai Institutes for Biological Sciences (Shanghai, China).
HEK293 cells were grown at 37°C with 5% CO2 in
Dulbecco's modified Eagle's medium supplemented with 10% fetal
bovine serum (FBS; Gibco, Grand Island, NY, USA) and 5 mg/ml of
gentamicin (Invitrogen, Carlsbad, CA, USA). HL-60 and NB4 cells
were cultured in RPMI-1640 (Gibco) supplemented with 10% FBS, 100
U/ml penicillin and 100 µg/ml streptomycin.
APL blast cells were obtained from human bone marrow
aspirates. Leukemia cells were isolated using Ficoll-Hypaque
(Sigma-Aldrich, St. Louis, MO, USA) density gradient
centrifugation.
Patient samples
Bone marrow and peripheral blood samples were
obtained with informed consent, in accordance with the Declaration
of Helsinki, from new untreated APL patients or patients with
non-APL hematological disorders (the controls), presenting at the
Department of Hematology, The First Affiliated Hospital of
Chongqing Medical University. Blasts were >70% in all APL
samples. Hemograms, peripheral blood smears, and bone marrow
examinations were performed in all APL and control cases using
Jenner-Giemsa and cytochemistry for routine diagnostic morphology.
The results were compared with those obtained by immunofluorescence
staining. Details of the patients examined in the present study are
presented in Table I.
 | Table I.Clinical details of patients. |
Table I.
Clinical details of patients.
| Patient no. | Gender | Age (years) | FAB | Blast (%) | WBC
(x109) |
|---|
| 001 | F | 16 | M3 | 85 | 30.5 |
| 002 | F | 25 | M3 | 79 | 3.2 |
| 003 | M | 30 | M3 | 72 | 2.5 |
| 004 | M | 29 | M2 | 75 | 17.8 |
| 005 | F | 33 | M2 | 70 | 22.9 |
| 006 | M | 22 | M4 | 66 | 10.5 |
| 007 | M | 35 | M4 | 69 | 25.7 |
Analyses of transfected cells
PML and PML(NLS−) eukaryotic plasmids
encoding C-terminal HA-tagged proteins [PCMV-HA-PML and
PCMV-HA-PML(NLS−), respectively], and the importin α
expression vector encoding C-terminal Myc-tagged proteins
(PCMV-MYC-imp α), were constructed for indirect immunofluorescence
and the in vivo binding assay. Briefly, HEK293 cells seeded
on 75 cm2 dishes or plated on coverslips, were
transfected with tagged constructs using Lipofectamine (Invitrogen)
according to the manufacturer's instructions. Western blotting and
immunofluorescence localization assays were performed by
conventional methods to investigate the expression of PML and
PML(NLS−). Co-immunoprecipitation was used to identify
the interaction of PML/PML(NLS−) with importin α. NB4
cells (2×106) were electroporated with 7 µg of
supercoiled plasmid DNA containing the NE cDNA pcDNA3.1. Cells were
harvested at various time points after electroporation, and cell
lysates were prepared for co-immunoprecipitation.
Western blotting
Cells were washed with ice-cold phosphate-buffered
saline (PBS) and lysed in cell lysis buffer [0.5% Triton X-100, 150
mM NaCl, 20 mM Tris (pH 7.5), 5 mM EDTA and 1 mM
phenylmethanesulfonyl fluoride] on ice for 20 min. Lysates were
pelleted by centrifugation at 14,000 × g for 10 min at 4°C. Total
protein concentrations in the supernatant were determined using a
Bio-Rad protein assay (Bio-Rad, Hercules, CA, USA). Whole cell
lysates were separated by 10% sodium dodecyl sulfate-polyacrylamide
gel electrophoresis and transferred to polyvinylidene difluoride
membranes (Millipore, Darmstadt, Germany). Membranes were blocked
with 5% milk in PBS for 1 h and incubated with primary antibodies
at 4°C overnight. A polyclonal rabbit anti-PML antibody (ab72137;
Abcam, Cambridge, UK) and a polyclonal rabbit anti-actin antibody
(Santa Cruz Biotechnology, Santa Cruz, CA, USA) were used as
primary antibodies. Immunocomplexes were detected with appropriate
horseradish peroxidase-conjugated secondary antibodies and detected
by enhanced chemiluminescence (GE Healthcare, Marlborough, MA,
USA).
Immunofluorescence microscopy
Transfected or untransfected HEK293 cells were
cultured on glass slides and fixed in 4% paraformaldehyde in PBS at
4°C for 20 min. Cytospins of non-adherent cells were performed with
40,000 cells at 400 rpm for 10 min, followed by air-drying and
fixation as previously described (21). After blocking with 1% bovine serum
albumin for 30 min, slides were incubated for 1 h at room
temperature with a rabbit polyclonal anti-PML antibody (Abcam) or a
mouse monoclonal anti-importin α antibody (Sigma-Aldrich), diluted
at 1:200 in Tris-buffered saline with Tween-20 (TBST). Fluorescein
isothiocyanate-conjugated goat anti-rabbit IgG and
tetramethylrhodamine isothiocyanate-conjugated goat anti-mouse IgG
(both from CW Biotech, Beijing, China) were used as the secondary
antibodies. Cells were mounted in mounting media (Sigma-Aldrich)
containing 1 µg/ml 4′,6-diamidino-2-phenylindole (DAPI) (Beyotime,
Shanghai, China) and visualized by standard immunofluorescence
microscopy (Olympus BX51; Olympus, Tokyo, Japan).
Co-immunoprecipitation
For in vivo binding assays, cells were seeded
in 75 cm2 plates. Collected cells were lysed in ice-cold
cell lysis buffer (20 mM Tris, 150 mM NaCl, 1 mM EDTA, 100 µg/ml
phenylmethanesulfonyl fluoride and 1% NP40). One milligram of
protein was incubated with an anti-PML or anti-importin α antibody
at 4°C overnight. Normal mouse or rabbit IgG (Santa Cruz
Biotechnology) was used as a negative control. Protein
A/G-Sepharose beads were added for 5 h at 4°C to bind the immune
complex from the incubation solution. Precipitates were washed four
times with lysis buffer, dissociated by boiling with the sample
buffer, and subjected to sodium dodecyl sulfate-polyacrylamide gel
electrophoresis. The remaining immunoblotting procedures are
described in the western blotting section in the ‘Materials and
methods’. Co-immunoprecipitated proteins were visualized using
anti-PML, anti-importin α, anti-HA tag or anti-cMyc tag antibodies
(Abcam).
Results
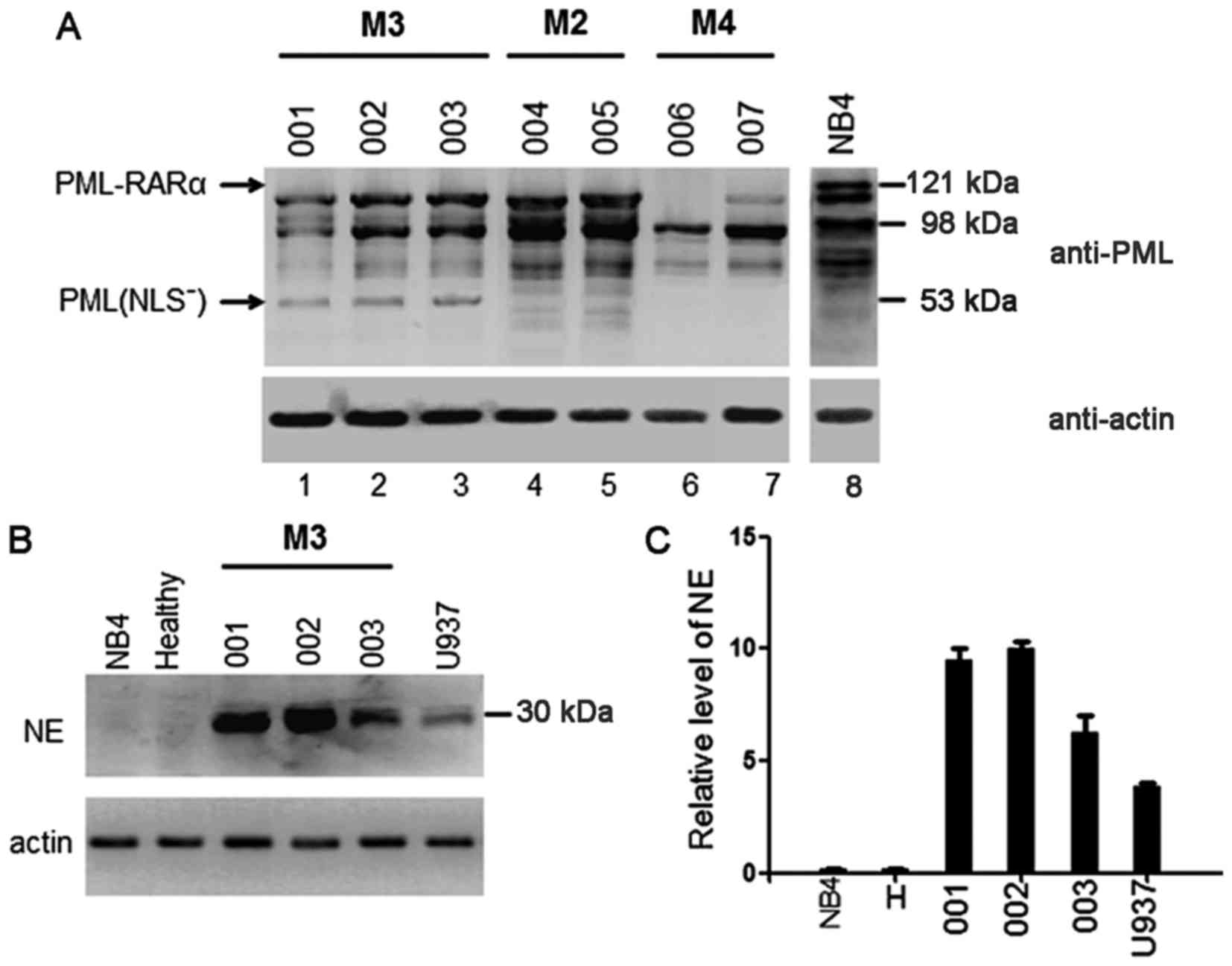
APL cells derived from human M3
patients contain the mutant PML(NLS−) protein
To verify whether PML-RARα was cleaved in
vivo, primary leukemia cells were obtained from the bone marrow
of three patients with M3 AML (APL), two with M2, and two with M4
as negative controls. APL cells from the M3 patients harbored a
bcr-1 PML-RARα fusion mRNA, which predicts a protein of 120 kDa.
Western blotting performed on lysates of primary AML cells showed
that none of the M3 samples expressed the full-length 120 kDa
PML-RARα protein. Instead, each contained multiple protein
fragments recognized by the anti-PML antibody, and a mutant PML
protein of ~53 kDa (Fig. 1A, lanes
1–3). Samples derived from M2 and M4 AML cells contained no
detectable PML-RARα protein as expected, although multiple proteins
were recognized by the anti-PML antibody (Fig. 1A, lanes 4–7).
NB4 cells that contained the bcr-1 isoform of
PML-RARα (22,23) expressed the predicted 120 kDa
PML-RARα protein (Fig. 1A, lane 8),
but did not express NE. Samples derived from healthy cells also
contained no detectable NE protein (Fig. 1B). Western blotting performed on all
three peripheral blood neutrophil extracts showed that, similar to
U937 cells, M3 cells expressed the 30 kDa NE protein (Fig. 1B). The quantity of NE protein in the
APL samples was much higher than that in the U937 cells (Fig. 1C). In contrast, extracts from
healthy and NB4 cells did not contain measurable quantities of NE
protein (Fig. 1C).
The NLS is important for the nuclear
localization of PML
To determine whether the absence of the NLS sequence
influenced the localization of PML, immunofluorescence microscopy
was used to determine the subcellular localization of wild-type PML
and PML(NLS−) (Fig. 2A and
C). Transfected cells were easily identified due to the intense
signals created by both overexpressed proteins. In western blot
analyses of transfected HEK293 cells, the HA-tagged PML or
PML(NLS−) proteins appeared as a group of bands at 70
and 53 kDa, respectively (Fig. 2B).
This was consistent with their calculated weights. When wild-type
PML was transfected transiently into HEK293 cells, the PML protein
was detected as a few large nuclear dots (Fig. 2C). When PML(NLS−) was
transfected into HEK293 cells, it was detected as large cytoplasmic
aggregates (Fig. 2C). This pattern
was consistent with the reported cytoplasmic aggregation of
full-length PML lacking its NLS (24). When cells were transfected with
NLS-RARα (containing the PML NLS plus the RARα portion of
PML-RARα), this protein was detected in a homogeneous nuclear
pattern, similar to that of RARα in RARα-transfected cells
(Fig. 2D).
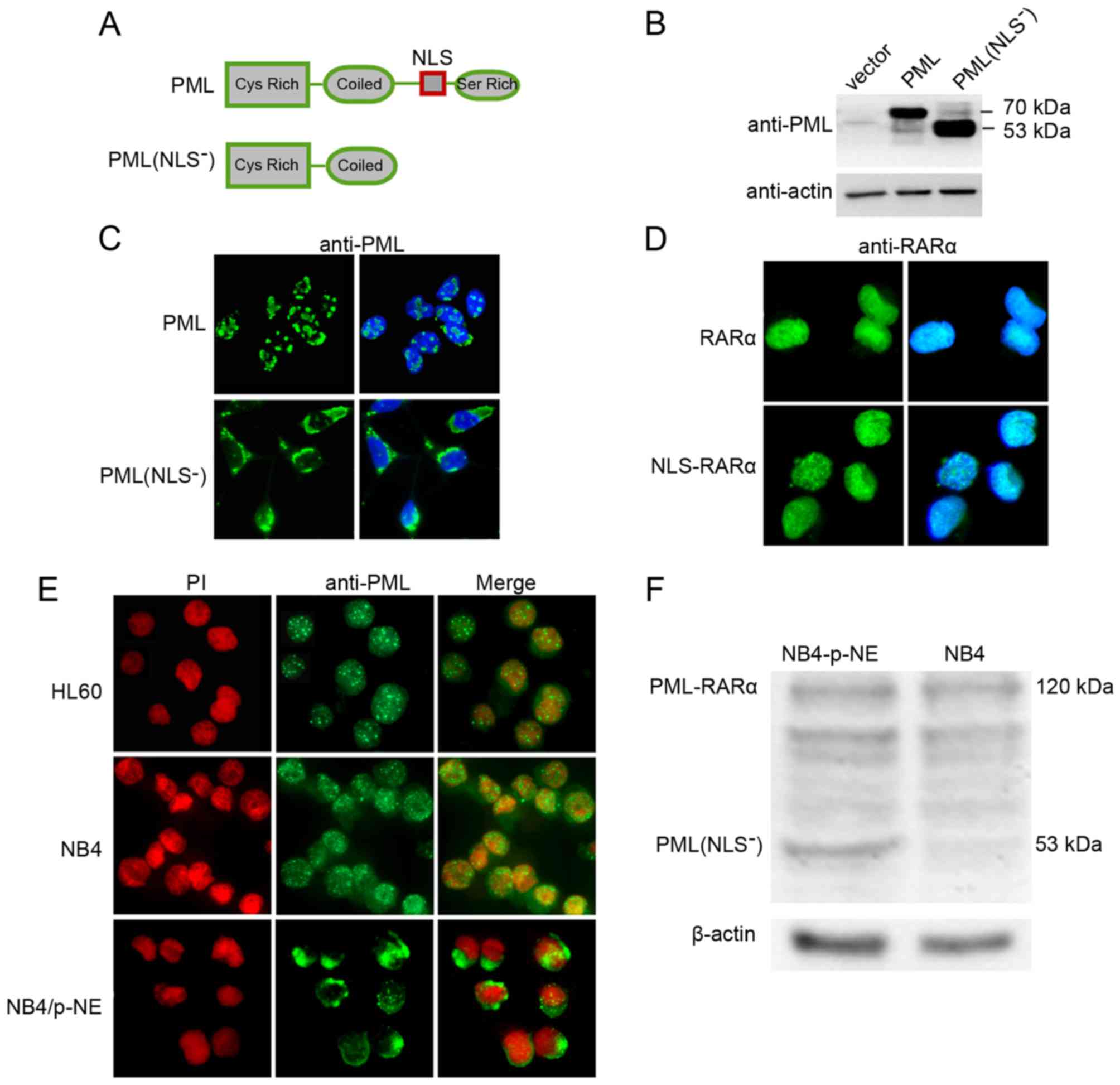 | Figure 2.Analyses of PML localization in
transfected HEK293 cells and leukemia cell lines. (A) Schematic
illustration of PML and its deletion mutant, PML(NLS−).
The positions of the NLS and its main domains are indicated. (B)
Western blot analysis of transfected HEK293 cells for PML. Vector,
pCMV-HA vector transfected control; PML, PML expression vector;
PML(NLS−), PML(NLS−) expression vector. (C
and D) Immunofluorescence analyses of PML, PML(NLS−),
RARα and NLS-RARα in transfected HEK293 cells. HEK293 cells were
transfected with the PML, PML(NLS−), RARα and NLS-RARα
expression vectors. Immunofluorescence staining (green) was
performed using anti-PML polyclonal and anti-RARα polyclonal
antibodies, followed by a fluorescein isothiocyanate-conjugated
secondary antibody. Nuclei were visualized by DAPI staining. (E)
Immunofluorescence microscopy of HL60, NB4 and NB4/p-NE cells 24 h
after electroporation with the NE expression construct. All cells
were stained with an anti-PML antibody (green fluorescence). Nuclei
were stained with propidium iodide (red fluorescence). (F) Western
blot analysis for PML and actin in protein extracts from NB4 and
NB4/p-NE cells. |
The intracellular expression of PML and PML-RARα was
also examined in APL cell lines. HL60, a human myeloid cell line
from peripheral blood leukocytes of a patient with APL (25), does not harbor t(15;17) and
expresses only normal PML. The APL-derived cell line, NB4, is a
valuable system to study the expression of PML-RARα since both the
normal and rearranged PML genes are present. Immunofluorescence
microscopy of HL60 cells revealed a punctate nuclear pattern of PML
expression (Fig. 2E). In NB4 cells,
a micro-punctate pattern consisting of hundreds of very small dots,
predominately located in the nucleus, was observed (Fig. 2E). This demonstrated the expected
nuclear location of PML-RARα. In contrast, when NB4 cells were
transfected with the NE expression vector, the PML-RARα pattern was
almost lost and replaced with a typical PML(NLS−)
profile as previously shown in NE transfected HEK293 cells. In NB4
cells, at 24 h post-transfection with NE, immunofluorescence
microscopy showed that the PML-relevant protein was mainly
cytoplasmic, although some nuclear dots were also detected
(Fig. 2E). Likewise, western blot
analysis revealed that PML(NLS−) existed in the
NE-transfected cells but not in the parental NB4 cells (Fig. 2F).
PML and importin α interact in vivo
and co-localize in the PML NBs
Our previous study revealed that PML was located in
the nucleus and that the NLS was responsible for efficient nuclear
targeting. In the present study, HEK293 cells were co-transfected
with either PCMV-HA-PML and PCMV-Myc-importin α, or
PCMV-HA-PML(NLS−) and PCMV-Myc-importin α. HA-PML, but
not ML(NLS−) was, co-immunoprecipitated with
Myc-importin α (Fig. 3A). When an
anti-HA antibody was used to immunoprecipitate HA-PML and
HA-PML(NLS−), importin α was detected using an anti-Myc
antibody. Nonetheless, we were unable to detect an interaction
between the HA-PML(NLS−) protein and Myc-importin α
(Fig. 3B). This demonstrated that
the NLS of PML was required for the binding of PML to importin
α.
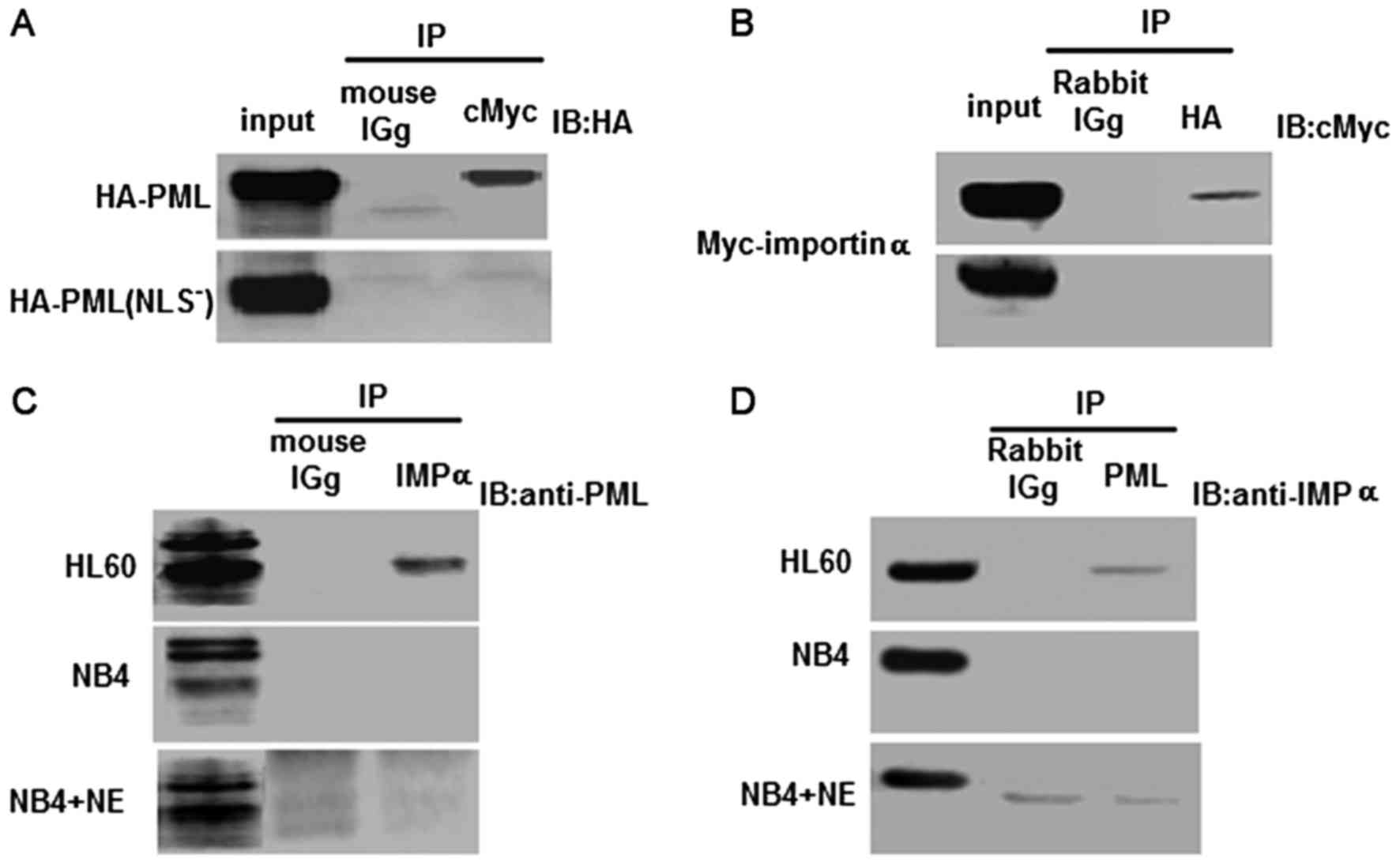 | Figure 3.Analyses of the interaction between
PML and importin α in transfected HEK293 cells and leukemia cell
lines. (A and B) Co-immunoprecipitation was employed to detect the
interaction between PML/PML(NLS−) and importin α in
transfected HEK293 cells. Upper blot, co-transfection with the PML
and importin α expression vectors. Lower blot, co-transfection with
the PML(NLS−) and importin α expression vectors. (A) IP,
Anti-cMyc antibody; IB, anti-HA antibody mouse IgG was used as the
negative control. (B) IP, Anti-HA antibody; IB, anti-cMyc antibody.
Rabbit IgG was used as the negative control. (C and D) Reciprocal
immunoprecipitation analyses were used to evaluate the physical
interaction between PML and importin α in HL60, NB4 and NB4/−NE
cell lines. (C) IP, Anti-importin α antibody; IB, anti-PML
antibody. Mouse IgG was used as the negative control. (D) IP,
Anti-PML antibody; IB, anti-importin α antibody. Rabbit IgG was
used as the negative control. |
To examine whether the same NLS alteration could
abrogate the interaction between PML and importin α in NB4 cells,
the NE expression vector was electroporated into NB4 cells for an
in vivo binding assay. Importantly, a specific interaction
was observed between endogenous PML and importin α in HL60 cells,
but not in NB4 and NB4/p-NE cells (Fig.
3C and D). These results clearly demonstrated that PML and
importin α interacted in vivo at physiological levels, and
that there was insufficient PML to detect an interaction with
importin α in NB4 and NB4/p-NE cells.
HEK293 cells, co-transfected with HA-PML and
Myc-importin α, were fixed 24 h later and subjected to
immunofluorescence staining. Importin α co-localized with PML in
the PML NBs, although staining of importin α was also diffusely
found within the nucleus and was evident in the cytoplasm (Fig. 4A). In contrast, PML(NLS−)
was mainly localized in the cytoplasm, without co-localization with
importin α in the nucleus (Fig.
4A). To verify that co-localization with importin α was not due
to the overexpression of PML, the experiment was repeated in HL60
and NB4 cells to determine whether co-localization also occurred at
physiological levels of PML. As shown in Fig. 4C, importin α co-localized with
endogenous PML in HL60 cells, but not in NB4 cells.
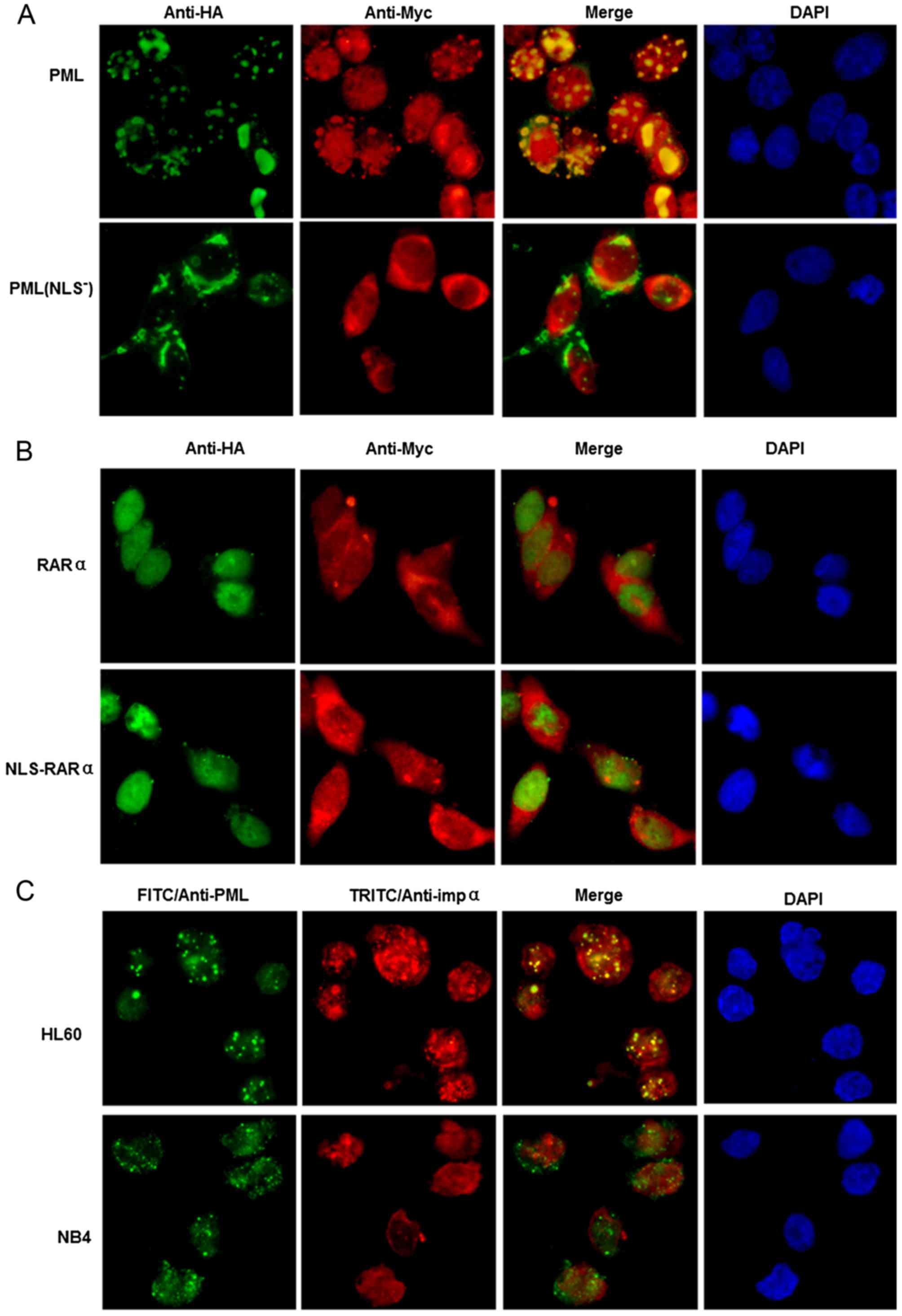 | Figure 4.Analyses of PML and importin α
co-localization in transiently transfected HEK293 cells and APL
cell lines. (A nd B) Localization of PML, PML(NLS−),
NLS-RARα and RARα, and their co-localization with importin α, in
transfected HEK293 cells. (A) Top panel, co-transfection with
PCMV-HA-PML and PCMV-Myc-importin α. Bottom panel, co-transfection
with PCMV-HA-PML(NLS−) and PCMV-Myc-importin α. (B) Top
panel, co-transfection with PCMV-HA-RARα and PCMV-Myc-importin α.
Bottom panel, co-transfection with PCMV-HA-NLS-RARα and
PCMV-Myc-importin α. Nuclei were visualized by
4′,6-diamidino-2-phenylindole staining. An anti-HA antibody was
used for HA-tagged proteins (green fluorescence). An anti-Myc
antibody was used for Myc-tagged importin α (red fluorescence). (C)
Co-localization of PML and importin α in HL60 and NB4 cells. Cells
were analyzed by immunofluorescence staining with anti-PML
polyclonal (green fluorescence) and anti-importin α monoclonal (red
fluorescence) antibodies. Nuclei were visualized by
4′,6-diamidino-2-phenylindole staining. Co-localization of PML and
importin α is illustrated by yellow fluorescence after overlaying
the images. |
We also examined whether an interaction existed
between NLS-RARα and importin α. Based on the immunofluorescence
assay, NLS-RARα was localized in the nucleus but not co-localized
with importin α. There was no noticeable difference in the
localization of NLS-RARα and RARα, and there was no determinable
interaction between NLS-RARα and importin α or RARα (Fig. 4B).
Evaluation of PML for the diagnosis of
APL
Although immunofluorescence of NB4 cells transfected
with NE demonstrated the expected ‘dispersed’ cytoplasmic pattern
with PML-specific antibodies, the localization of PML and its
interaction with importin α were unknown in primary APL cells. In
healthy blood neutrophils, PML showed a clear punctate nuclear
pattern of immunostaining (green fluorescence, Fig. 5A and B) that represented its nuclear
localization. Importin α immunostaining (red fluorescence, Fig. 5B) also showed a discrete, punctate
pattern that was very well co-localized with PML in healthy cells
(merged image of yellow fluorescence, Fig. 5B). This result demonstrated
excellent co-localization of PML with importin α in healthy cells.
However, in primary APL cells, PML exhibited a mostly cytoplasmic
localization (Fig. 5A and B).
Similarly, importin α had a discrete cytoplasmic localization
without co-localization with PML (Fig.
5B). In contrast to primary APL cells, primary M2, M4, M6, and
CML leukemia cells had a punctate nuclear pattern of PML expression
(Fig. 5A), similar to that in
healthy blood neutrophils.
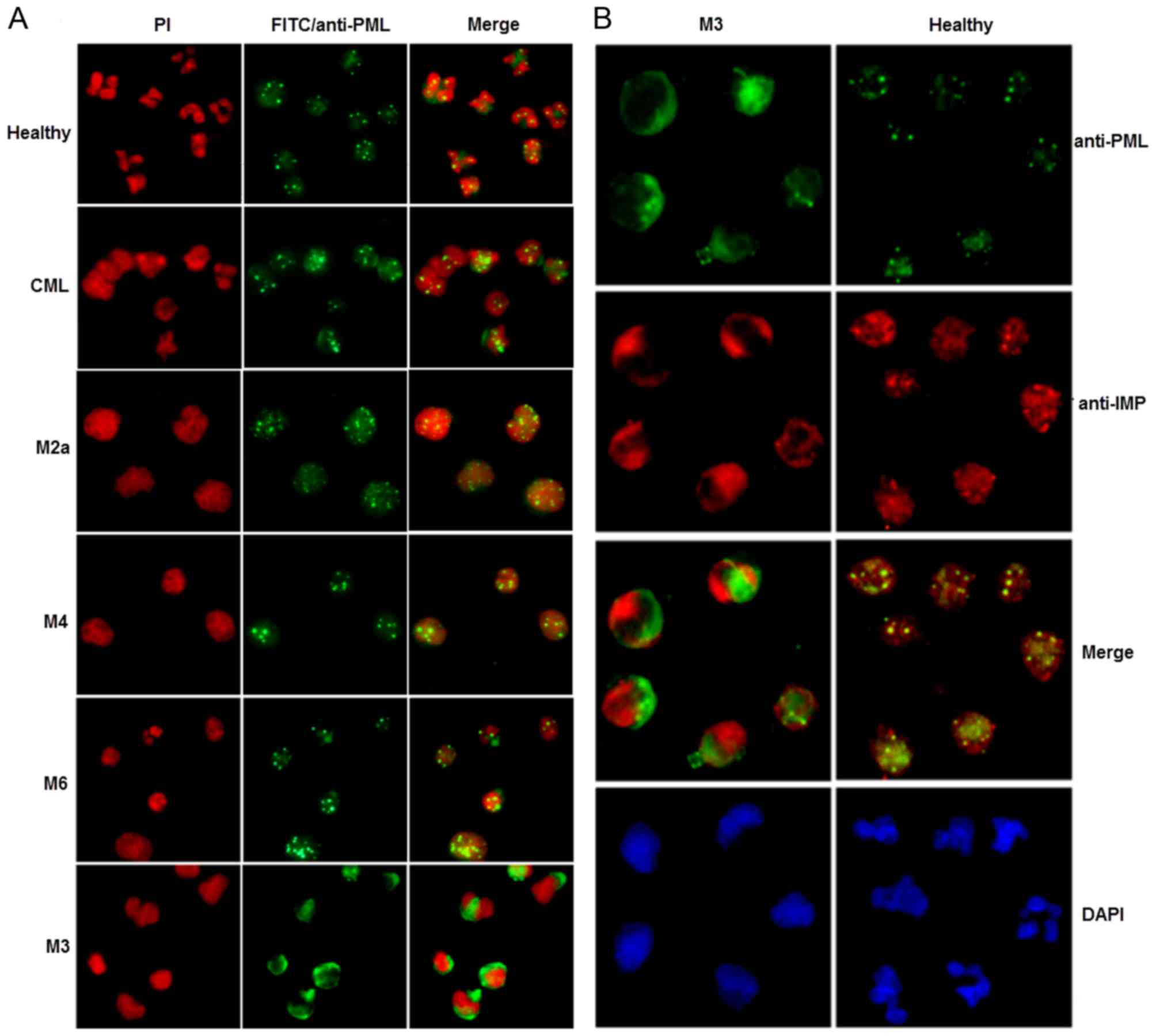 | Figure 5.Subcellular localization of PML, and
co-localization of PML with importin α, in primary human APL cells.
(A) Subcellular localization of PML in primary M3, M2a, M4 and M6
AML cells. CML, primary CML cells. Healthy, peripheral blood
neutrophils from healthy individuals. Cells were visualized by
immunofluorescence microscopy. Propidium iodide was used to stain
the nuclei. (B) Co-localization of PML with importin α in primary
APL cells. Immunofluorescence with rabbit anti-PML, detected with
anti-rabbit IgG-fluorescein isothiocyanate (green fluorescence) and
mouse anti-importin α, detected with anti-mouse
IgG-tetramethylrhodamineisothiocyanate (red fluorescence) was used.
Co-localization of PML and importin α is illustrated by yellow
fluorescence after overlaying the images. M3, primary M3 AML cells.
Healthy, peripheral blood neutrophils from healthy patients. Nuclei
were visualized by 4′,6-diamidino-2-phenylindole staining. |
The median age of the APL patients examined in the
present study was 30 years (range 14–60) and the study included 11
men and 16 women. Using morphology alone for diagnosis, 23 of 27
APL cases (85.2%) were correctly identified (Table II). Four cases (14.8%) were
misdiagnosed as either M2 (2 cases) or M4 (2 cases). Of the 22
non-APL AMLs, 13 (59.1%) were categorized correctly by morphology.
However, the possibility of APL could not be ruled out conclusively
in 9 (40.9%) cases due to the presence of atypical cells
reminiscent of promyelocytes. These 9 cases were finally
categorized as M4 (6 cases) or M5 (3 cases).
 | Table II.Comparison of various diagnostic
tests for APL. |
Table II.
Comparison of various diagnostic
tests for APL.
|
| APL (N=27) | Non-APL (N=22) |
|
|
|
|
|---|
|
|
|
|
|
|
|
|
|---|
| Test | Correctly
identified (%) | False negative
(%) | Correctly
identified (%) | False negative
(%) | Sensitivity
(%) | Specificity
(%) | PPV (%) | NPV (%) |
|---|
| Morphology | 23 (85.2) | 4 (14.8) | 13 (59.1) | 9 (40.9) | 85.1 | 59.1 | 71.9 | 76.5 |
| IF (PML) | 25 (92.6) | 2 (7.4) | 17 (77.3) | 5 (22.7) | 92.6 | 77.3 | 82.8 | 85.0 |
Twenty-five of the 27 APL cases (92.6%) showed an
acytoplasmic or microgranular nuclear pattern of immunofluorescence
using an anti-PML antibody. Two cases (7.4%) showed a punctate
pattern (false-negative). Seventeen of 22 non-APL AML controls
(77.3%) showed a punctate pattern and 5 (22.7%) showed a
microgranular pattern (false-positive) (Table II). Overall, immunofluorescence was
significantly superior to morphology alone for the diagnosis of APL
(P<0.01).
Discussion
In the present study, we showed that a truncated PML
protein, PML(NLS−), that contained the N-terminal domain
of PML without the C-terminal NLS portion, was present in primary
APL cells. In contrast, PML(NLS−) was not detected in M2
and M4 primary leukemia cells. To determine whether there was
sufficient NE activity for this cleavage event, the expression of
this enzyme was examined in primary AML cells. Human primary APL
cells showed abundant NE activity that was much higher than that in
U937 cells. In contrast, neutrophil extracts from normal and NB4
cells did not show PML-RARα cleavage activity or measurable amounts
of NE. Together, these data suggest that PML(NLS−) is
unique for APL and may be a new diagnostic marker of this disease.
PML(NLS−) may also play a role in the pathogenesis of
APL.
NLS-dependent nuclear protein import is an important
regulatory process in the biological information network.
Abnormalities or a deficiency of the NLS may disrupt the
information network and induce disease. To determine whether a
deficiency of the NLS sequence influenced the localization of PML,
we compared the subcellular localization of PML and
PML(NLS−). Immunofluorescence microscopy showed that PML
exhibited a nuclear pattern of expression while
PML(NLS−) aggregates were found in the cytoplasm. In
contrast to the micro-punctate nuclear pattern of PML-RARα in
control NB4 cells, the PML-RARα cleavage activity in NB4/p-NE cells
yielded a cytoplasmic localization. Similarly, PML exhibited a
punctate nuclear pattern in healthy and non-APL cells, while in
cells from APL patients there was mostly a cytoplasmic localization
pattern of PML(NLS−), or a slight micro-punctate nuclear
pattern. These data suggest that the NLS plays a key role in the
nuclear localization of PML and indicate that analysis of the
distribution of PML(NLS−) may provide a potential
diagnostic method for APL.
Exploring how the NLS mediates the transport of PML
into the nucleus, we found that PML and importin α interact in
vivo and co-localize in PML NBs. In contrast,
HA-PML(NLS−) transfected HEK293 and NB4/p-NE cells did
not interact with importin α. While human primary APL cells exhibit
no noticeable co-localization with importin α in the nucleus,
non-APL AML cells and neutrophils from healthy cells containing
wild-type PML protein, exhibit obvious co-localization. These data
suggest that PML is transported into the nucleus through its NLS
sequence and interacts with importin α. The interaction between PML
and importin α could also be regarded as a diagnostic label to
distinguish APL and non-APL leukemia.
APL is a highly aggressive disease that requires
prompt diagnosis and specific early intervention. PML-RARα-positive
APL patients often present with coagulopathy. Without treatment,
~40% of the patients develop pulmonary and cerebral hemorrhages
that can be lethal (26). The
latest research on methods for detecting PML-RARα rearrangements,
including a flow cytometric immunobead assay (27), reverse transcription polymerase
chain reaction (RT-PCR) (28), and
a new high-speed droplet RT-PCR (29), has focused on improved detection of
the PML-RARα fusion gene and protein rather than exploring new
indicators. As the current diagnostic gold standard, RT-PCR for
PML-RARα is time-consuming. There is thus a need for a rapid and
accurate diagnostic test for APL.
Since early promyelocytes produce massive amounts of
NE, cleavage events occur at an early stage of APL. Consequently,
the immunofluorescence assay for assessing the distribution of
PML(NLS−) enables an earlier diagnosis of APL and allows
clinicians to begin treatment sooner. However, this
immunofluorescence assay cannot be used to detect variants of APL
that involve the formation of fusion genes with RARα, or genes
other than PML [e.g. promyelocytic zinc finger; t(11;17) and
nucleophosmin; t(5;17)]. To detect PML proteins in cell lysates,
antibodies were raised against the N-terminal domain of PML,
upstream of the BCR3 break point region. This domain is present in
all known PML-RARα fusion proteins. Thus, these anti-PML antibodies
recognize PML and PML-RARα, as well as PML(NLS−). We are
committed to developing a new anti-PML antibody that is directed
against the NLS domain of PML and is able to distinguish between
PML and PML(NLS−). Such an antibody may be useful
clinically for the diagnosis of APL.
In conclusion, the present study describes a simple,
fast and accurate immunofluorescence assay for the detection of
PML(NLS−) localization that can be used to diagnose APL.
Compared with the traditional morphologic test, sensitivity was
increased by nearly 7.5% and specificity increased from 59.1 to
77.3%. These results indicate that immunofluorescence has
significant potential for detecting PML(NLS−) and may be
useful in diagnosing APL. Two false negatives were found among 27
cases of APL (7.4%). Possible causes of these false negatives
include mutations other than t(15;17), insufficient material on the
slides, or an artifact of prolonged storage (30). All cases were t(15;17)-positive and
stained better immediately rather than following storage. Thus,
false negatives with stored samples may be a limitation of the
test.
Overall, the data reported in the present study
strongly suggest that the NLS plays a critical role in the nuclear
localization of PML by interacting with importin α. NE-mediated
cleavage leading to the formation of PML(NLS−) was
unique in APL patients compared with other non-APL leukemias,
suggesting that PML(NLS−) may represent a novel
biomarker for APL. The detection of cytoplasmic
PML(NLS−) in APL patients by immunofluorescence staining
could be an early diagnostic method for APL.
Acknowledgements
The present study was supported by the National
Natural Science Foundation of China (no. 81171658) and the Natural
Science Foundation Project of CQ CSTC (no. 2011BA5037). We kindly
thank Dr Jianbin Chen of The First Affiliated Hospital of Chongqing
Medical University, for his kind contribution of patient samples to
the present study.
References
|
1
|
Puccetti E and Ruthardt M: Acute
promyelocytic leukemia: PML/RARalpha and the leukemic stem cell.
Leukemia. 18:1169–1175. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wang K, Wang P, Shi J, Zhu X, He M, Jia X,
Yang X, Qiu F, Jin W, Qian M, et al: PML/RARalpha targets promoter
regions containing PU.1 consensus and RARE half sites in acute
promyelocytic leukemia. Cancer Cell. 17:186–197. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Grisolano JL, Wesselschmidt RL, Pelicci PG
and Ley TJ: Altered myeloid development and acute leukemia in
transgenic mice expressing PML-RAR alpha under control of cathepsin
G regulatory sequences. Blood. 89:376–387. 1997.PubMed/NCBI
|
|
4
|
He LZ, Tribioli C, Rivi R, Peruzzi D,
Pelicci PG, Soares V, Cattoretti G and Pandolfi PP: Acute leukemia
with promyelocytic features in PML/RARalpha transgenic mice. Proc
Natl Acad Sci USA. 94:5302–5307. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Westervelt P, Lane AA, Pollock JL,
Oldfather K, Holt MS, Zimonjic DB, Popescu NC, DiPersio JF and Ley
TJ: High-penetrance mouse model of acute promyelocytic leukemia
with very low levels of PML-RARalpha expression. Blood.
102:1857–1865. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Early E, Moore MA, Kakizuka A,
Nason-Burchenal K, Martin P, Evans RM and Dmitrovsky E: Transgenic
expression of PML/RARalpha impairs myelopoiesis. Proc Natl Acad Sci
USA. 93:7900–7904. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lane AA and Ley TJ: Neutrophil elastase
cleaves PML-RARalpha and is important for the development of acute
promyelocytic leukemia in mice. Cell. 115:305–318. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lane AA and Ley TJ: Neutrophil elastase is
important for PML-retinoic acid receptor alpha activities in early
myeloid cells. Mol Cell Biol. 25:23–33. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Gao YM, Zhong L, Zhang X, Hu XX and Liu
BZ: PML(NLS−) inhibits cell apoptosis and promotes
proliferation in HL-60 cells. Int J Med Sci. 10:498–507. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hu XX, Zhong L, Zhang X, Gao YM and Liu
BZ: NLS-RARα promotes proliferation and inhibits differentiation in
HL-60 cells. Int J Med Sci. 11:247–254. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sardiello M, Cairo S, Fontanella B,
Ballabio A and Meroni G: Genomic analysis of the TRIM family
reveals two groups of genes with distinct evolutionary properties.
BMC Evol Biol. 8:2252008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Salomoni P, Ferguson BJ, Wyllie AH and
Rich T: New insights into the role of PML in tumour suppression.
Cell Res. 18:622–640. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Geng Y, Monajembashi S, Shao A, Cui D, He
W, Chen Z, Hemmerich P and Tang J: Contribution of the C-terminal
regions of promyelocytic leukemia protein (PML) isoforms II and V
to PML nuclear body formation. J Biol Chem. 287:30729–30742. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Guan D and Kao HY: The function,
regulation and therapeutic implications of the tumor suppressor
protein, PML. Cell Biosci. 5:602015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ding W, Tong Y, Zhang X, Pan M and Chen S:
Study of arsenic sulfide in solid tumor cells reveals regulation of
nuclear factors of activated T-cells by PML and p53. Sci Rep.
6:197932016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Fu C, Ahmed K, Ding H, Ding X, Lan J, Yang
Z, Miao Y, Zhu Y, Shi Y, Zhu J, et al: Stabilization of PML nuclear
localization by conjugation and oligomerization of SUMO-3.
Oncogene. 24:5401–5413. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Rabellino A, Carter B, Konstantinidou G,
Wu SY, Rimessi A, Byers LA, Heymach JV, Girard L, Chiang CM,
Teruya-Feldstein J, et al: The SUMO E3-ligase PIAS1 regulates the
tumor suppressor PML and its oncogenic counterpart PML-RARA. Cancer
Res. 72:2275–2284. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Weisshaar SR, Keusekotten K, Krause A,
Horst C, Springer HM, Göttsche K, Dohmen RJ and Praefcke GJ:
Arsenic trioxide stimulates SUMO-2/3 modification leading to
RNF4-dependent proteolytic targeting of PML. FEBS Lett.
582:3174–3178. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Jin G, Gao Y and Lin HK: Cytoplasmic PML:
From molecular regulation to biological functions. J Cell Biochem.
115:812–818. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lange A, Mills RE, Lange CJ, Stewart M,
Devine SE and Corbett AH: Classical nuclear localization signals:
Definition, function, and interaction with importin alpha. J Biol
Chem. 282:5101–5105. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Xiao C, Zhong L, Shan Z, Xu T, Gan L, Song
H, Yang R, Li L and Liu B: NLS-RARα inhibits the effects of
all-trans retinoic acid on NB4 cells by interacting with P38α MAPK.
Int J Med Sci. 13:611–619. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Fanelli M, Minucci S, Gelmetti V, Nervi C,
Gambacorti-Passerini C and Pelicci PG: Constitutive degradation of
PML/RARalpha through the proteasome pathway mediates retinoic acid
resistance. Blood. 93:1477–1481. 1999.PubMed/NCBI
|
|
23
|
Jing Y, Xia L, Lu M and Waxman S: The
cleavage product deltaPML-RARalpha contributes to all-trans
retinoic acid-mediated differentiation in acute promyelocytic
leukemia cells. Oncogene. 22:4083–4091. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Le XF, Yang P and Chang KS: Analysis of
the growth and transformation suppressor domains of promyelocytic
leukemia gene, PML. J Biol Chem. 271:130–135. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Gallagher R, Collins S, Trujillo J,
McCredie K, Ahearn M, Tsai S, Metzgar R, Aulakh G, Ting R, Ruscetti
F, et al: Characterization of the continuous, differentiating
myeloid cell line (HL-60) from a patient with acute promyelocytic
leukemia. Blood. 54:713–733. 1979.PubMed/NCBI
|
|
26
|
Taube F, Stölzel F, Thiede C, Ehninger G,
Laniado M and Schaich M: Increased incidence of central nervous
system hemorrhages in patients with secondary acute promyelocytic
leukemia after treatment of multiple sclerosis with mitoxantrone?
Haematologica. 96:e312011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Dekking EH, van der Velden VH, Varro R,
Wai H, Böttcher S, Kneba M, Sonneveld E, Koning A, Boeckx N, Van
Poecke N, et al: EuroFlow Consortium (EU-FP6, LSHB-CT-2006-018708):
Flow cytometric immunobead assay for fast and easy detection of
PML-RARA fusion proteins for the diagnosis of acute promyelocytic
leukemia. Leukemia. 26:1976–1985. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Dolz S, Barragán E, Fuster Ó, Llop M,
Cervera J, Such E, De Juan I, Palanca S, Murria R, Bolufer P, et
al: Novel real-time polymerase chain reaction assay for
simultaneous detection of recurrent fusion genes in acute myeloid
leukemia. J Mol Diagn. 15:678–686. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Sueki A, Matsuda K, Taira C, Yamaguchi A,
Koeda H, Takagi F, Kobayashi Y, Sugano M and Honda T: Rapid
detection of PML-RARA fusion gene by novel high-speed
droplet-reverse transcriptase-polymerase chain reaction:
Possibility for molecular diagnosis without lagging behind the
morphological analyses. Clin Chim Acta. 415:276–278. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Gomis F, Sanz J, Sempere A, Plumé G,
Senent ML, Pérez ML, Cervera J, Moscardó F, Bolufer P, Barragán E,
et al: Immunofluorescent analysis with the anti-PML monoclonal
antibody PG-M3 for rapid and accurate genetic diagnosis of acute
promyelocytic leukemia. Ann Hematol. 83:687–690. 2004. View Article : Google Scholar : PubMed/NCBI
|