Introduction
Ovarian cancer, which is the fifth leading cause of
tumor-associated mortalities in female (1,2),
accounts for 4% of all cancers in women and is still the leading
cause of death in gynecologic malignancies (3). Besides, among all ovarian cancers,
epithelial ovarian cancer (EOC) accounts for 80–90% (4), and has the highest mortality rate of
all gynecological malignancies. Regardless of advances in treatment
techniques, long-term survival rates for patients remain poor
(2), maybe because of the lack of
early symptoms, late diagnosis and invalid chemotherapy (5). Therefore, to ameliorate poor survival
outcomes, obtaining earlier diagnosis and new prognostic indicators
is critical.
Progression of cancer from normal tissue to
carcinomas requires many steps (6),
involving the disharmony of cell division, the inappropriate
activity of a series of different genes and proteins. Cell
invasion, which is a ubiquitous process, is tightly regulated by
multiple signaling proteins, such as plasma membrane receptors,
scaffold and small GTP-binding proteins (7). Through disrupting the program
regulating cell cycle entry and death, activation or upregulation
of oncogenes disrupts these orderly processes (8). Whereby, one of the best characterized
oncogenes is the small GTP-binding protein. It has been reported
that ADP ribosylation factors (ARF), another family of small
GTP-binding proteins, are closely related to cancer progression
(9). Like all other GTPases, ARFs
cycle between their inactive (GDP bound) and active (GTP bound)
form. In most cases, they act as molecular switches to turn on a
couple of signaling cascades associated with vesicle formation,
remodeling of the actin cytoskeleton and transformation of membrane
lipids (10).
ARF1, ADP-ribosylation factor 1, is mainly found in
the Golgi apparatus and acts to promote carrier vesicle biogenesis
by nucleating the assembly of coat protein complexes at sites of
vesicle formation (11). Several
studies have suggested that ARF1 can also localize to the plasma
membrane transmitting signals from transmembrane proteins (12,13).
In addition to their conventional roles of vesicle formation, ARF1
plays important roles in a wide range of physiological and
pathophysiological processes in tumor formation, including inter-
and intracellular signaling, transcription, DNA repair pathways,
cell cycle regulation, and mitosis, as well as necrosis and
apoptosis (14). A role for ARF1 in
the control of integrin-mediated cell adhesion was indicated by
studies on Brefeldin A (BFA)-resistant Arf GEFs and it was shown
that ARF1 mediated paxillin recruitment to the focal adhesions and
potentiated Rho-stimulated stress fiber formation in fibroblasts
(15). Furthermore, inhibition of
ARF1 activation by targeting ARNO impaired ARNO-dependent migration
of MDCK cells (16). In invasive
breast cancer cells, stimulation of the epidermal growth factor
receptor led to the activation of ARF1, the PI3K pathway, and
ultimately cell migration and proliferation (17). Another study also demonstrated that,
phosphorylation of the membrane-associated non-receptor tyrosine
kinase Src was regulated by ARF1 (18). While Src has been shown to induce
the migration, proliferation, adhesion, and growth of cancer cells
and has been reported to be overexpressed or activated in many
cancers including breast cancer (19,20).
In the current study, we found that ARF1 was
significantly overexpressed in EOC specimens and cell lines,
compared with adjacent non-tumorous ones. The expression of ARF1 is
bound up with clinicopathological variables and prognostic
implications. In addition, we detected the effect of ARF1 knockdown
on proliferation and migration of EOC. The results demonstrated
that the small GTPase is critical for both cell proliferation and
migration by directly regulating the activation of the PI3K
pathway, and the PI3K pathway may also be upstream of ARF1. By
defining the signaling pathways regulating proliferation and
migration of EOC cells, we will be able to identify new therapeutic
targets for the treatment of these aggressive types of cancers.
Materials and methods
Patients and tissue samples
The fresh samples were put into liquid nitrogen
immediately after surgical removal and maintained at −80°C until
use for western blot analysis. We investigated 119 cases of EOC
(including 63 serous papillary adenocarcinoma, 19 mucinous
papillary carcinoma, 18 endometrioid adenocarcinoma and 19 clear
cell carcinoma) provided by the Obstetrics and Gynecology
Department of the Affiliated Hospital of Nantong University for
immunohistochemical analysis. All of these patients underwent
surgical resection without preoperative chemotherapy in the surgery
department. All patients were followed up for 1–60 months. The main
clinical and pathologic features of the patients, including age,
tumor grade, the International Federation of Gynecology and
Obstetrics (FIGO) stages, lymph node metastasis, tumor invasion and
survival are shown in Table I.
 | Table I.Expression of ARF1 and related
clinicopathological parameters in the 119 human ovarian cancer
specimens. |
Table I.
Expression of ARF1 and related
clinicopathological parameters in the 119 human ovarian cancer
specimens.
|
|
| ARF1 expression |
|
|
|---|
|
|
|
|
|
|
|---|
| Characteristics | Total | Low | High | P-valuea |
χ2-value |
|---|
| Age (24–80 years,
mean 55 years) |
|
|
|
|
|
|
≤50 | 40 | 17 | 23 | 0.563 | 0.335 |
|
>50 | 79 | 38 | 41 |
|
|
| Histologic
subtype |
|
|
|
|
|
|
Serous | 63 | 26 | 37 | 0.384 | 3.047 |
|
Mucinous | 19 | 12 | 7 |
|
|
|
Endometrioid | 18 | 9 | 9 |
|
|
| Clear
cell | 19 | 8 | 11 |
|
|
| Histological
grade |
|
|
|
|
|
|
Low | 49 | 30 | 19 | 0.006b | 7.546 |
|
High | 70 | 25 | 45 |
|
|
| FIGO stage |
|
|
|
|
|
|
I/II | 60 | 35 | 25 | 0.008b | 7.146 |
|
III/IV | 59 | 20 | 39 |
|
|
| Lymph node
metastasis |
|
|
|
|
|
|
Negative | 86 | 46 | 40 | 0.010b | 6.594 |
|
Positive | 33 | 9 | 24 |
|
|
| Ki-67
expression |
|
|
|
|
|
|
Low | 55 | 33 | 22 | 0.005b | 7.814 |
|
High | 64 | 22 | 42 |
|
|
Antibodies
The antibodies used for western blot analysis
immunohistochemistry and immunofluorescence were as follows: i)
ARF1 antibody (anti-mouse, Santa Cruz Biotechnology, Inc., Santa
Cruz, CA, USA), ii) Ki-67 antibody (anti-mouse, Santa Cruz
Biotechnology, Inc.), iii) PCNA antibody (anti-mouse, Santa Cruz
Biotechnology, Inc.), iv) cyclin D1 antibody (anti-rabbit, Santa
Cruz Biotechnology, Inc.), and v) Akt1 antibody (anti-mouse, Santa
Cruz Biotechnology, Inc.), vi) p-Akt1/2/3(Ser473) antibody
(anti-rabbit, Santa Cruz Biotechnology, Inc.), vii) p-Akt1 (Thr308)
antibody (anti-rabbit, Santa Cruz Biotechnology, Inc.), viii) GAPDH
antibody (anti-rabbit, Santa Cruz Biotechnology, Inc.).
Western blot analysis
Frozen EOC tissues and protein were used for western
blot analysis. They were promptly homogenized in a homogenization
buffer [1 M Tris HCl, pH 7.5, 1% Triton X-100, 1% Nonidet p-40
(NP-40), 10% sodium dodecyl sulfate (SDS), 0.5% sodium
deoxycholate, 0.5 M EDTA, leupeptin 10 µg/ml, aprotinin 10 µg/ml,
and 1 mM PMSF]. The lysates were cleared by centrifugation at
12,000 rpm for 20 min at 4°C. Protein concentrations were
determined with a Bio-Rad protein assay (Bio-Rad Laboratories,
Inc., Hercules, CA, USA). The total cellular protein extracts were
separated by sodium dodecyl sulfate-polyacrylamide gel
electrophoresis (SDS-PAGE) and transferred to
polyvinylidenedifluoride filter (PVDF) membranes (Millipore,
Bedford, MA, USA). After the membranes were blocked in 5% nonfat
milk in PBST for 2 h, they were incubated with appropriate primary
and secondary antibodies. The membrane was detected using an
enhanced chemiluminescence system (ECL; Pierce Biotechnology, Inc.,
Rockford, IL, USA). The experiments were carried out three separate
times.
Immunohistochemistry (IHC)
staining
IHC stain was conducted as previously described
(21). The sections were dewaxed in
xylene for 15 min and rehydrated in graded ethanols. Then,
endogenous peroxidase activity was blocked by soaking in 3%
hydrogen peroxide for 30 min. Thereafter, immunoreactivity was
enhanced by pressure cooking by incubating the tissue sections for
3 min in 0.1 M citrate buffer. After rinsing in phosphate-buffered
saline (PBS, pH 7.2), 10% goat serum was applied to block any
nonspecific reactions for 1.5 h. Then, the sections were incubated
with ARF1 antibody (diluted 1:100) and Ki-67 antibody (diluted
1:800) for 3 h at room temperature (RT). The sections were washed
three times with phosphate-buffered saline (PBS). After rinsing in
water, the sections were counterstained with hematoxylin,
dehydrated, and cover slipped.
Immunohistochemistry analysis
Five high-power views in each specimen were selected
randomly for assessment; and at least 500 cells were counted to
determine the labeling index (LI), representing the percentage of
immunostained cells that is related to the total number of cells.
To evaluate the immunoreaction of ARF1, intensity was estimated in
comparison with the control and scored as follows: 0, negative
staining; 1, weak staining; 2, moderate staining; and 3, strong
staining. Scores which represent the percentage of tumor cells
stained positive were as follows: 0, <1% positive tumor cells;
1, 1–10%; 2, 10–50%; 3, 50–75%; and 4, >75%. Then, we multiplied
the two scores, and a score of 0 was considered negative, 2–3 was
weak, 4–5 was moderate, and 6–7 was considered strong. For
statistical analysis, 0–3 was counted as low expression, and 4–7
were overexpression (15). As for
statistical analysis of Ki-67, a cutoff value was used to
distinguish tumors with a low (<50.7%) or high (≥50.7%) level of
Ki-67 expression.
Immunofluorescence
Cells were plated on glass coverslips and grown for
48 h prior to staining, allowing transfection when needed. Cells
were then rinsed twice in PBS before fixation in 2%
paraformaldehyde for 25 min. They were permeabilized with 0.1%
Triton X-100 in PBS for 25 min. After washing in PBS supplemented
with 0.05% Tween-20 (PBST), cells were incubated for 1 h at room
temperature with the appropriate antibodies in PBST supplemented
with 10% FCS. Coverslips were then rinsed twice with PBST, and
incubated for 1 h at room temperature with the secondary
antibodies.
After that, the coverslips were mounted with an
anti-bleaching glycerol mixture. Samples were viewed using a Zeiss
Axiophot epifluorescence microscope with a 1003 immersion oil
objective. The images, given by the Hamamatsu C4880 cooled CCD
camera connected to the Serie 150/151 from Imaging Technology Inc.,
were acquired by Khoros software package from Khoral Research Inc.
In order to correct an uneven illumination from the mercury lamp, a
shading correction was applied to the images.
Cell culture and cell cycle
analysis
Human ovarian carcinoma cell lines HO-8910 were
purchased from the Shanghai Institute of Cell Biology, and cultured
in RPMI-1640 supplemented with 10% heat-inactivated fetal calf
serum and 100 U/ml penicillin-streptomycin mixture (Gibco-BRL,
Grand Island, NY, USA) at 37°C and 5% CO2. For the
analysis, cells were harvested at the proper time and fixed in 70%
ethanol overnight at 4°C and then incubated with 1 mg/ml RNase for
30 min at 37°C. Subsequently, the cells were stained with propidium
iodide (PI, 50 mg/ml; Becton-Dickinson, San Jose, CA, USA) in PBS,
0.5% Tween-20, then we analyzed the cells using a Becton-Dickinson
BD FACScan flow cytometer and CellQuest acquisition and analysis
software.
Plasmid constructs shRNA and
transfection
Short hairpin RNAs (shRNA) were designed and
provided by GeneChem (Shanghai, China). The target sequences were
as follows: 5′-GAACCAGAAGUGAACGCGA-3′, designated sh-1;
5′-CGGCCGAGATCACAGACAAG-3′, designated sh-2; and
5′-GCCATTAGAAAGCGCAACCAG-3′, designated sh-3. HO-8910 cells were
plated the day before transfection at 50–70% confluency. The cells
were then transfected with siRNAs at final concentrations of 50 nM
with Lipofectamine 2000. The cells were collected after 48 h for
the following assays.
Flow cytometric analysis
Cells were prepared into a single-cell suspension,
and then fixed with 70% ethanol at −20°C for 24 h. Next, cells were
incubated with 1 mg/ml RNase in PBS at 37°C for 30 min and stained
with 0.5 mg/ml propidium iodide. The DNA contents were analyzed by
a FACScan flow cytometer (Becton-Dickinson, Lincoln Park, NJ, USA).
Three independent experiments were conducted to ensure the
results.
Co-immunoprecipitation assay
Co-immunoprecipitation assay was conducted as
previously described (22). HO-8910
cells were harvested and lysed in buffer (50 mmol/l Tris-HCl, pH
7.5, 150 mmol/l NaCl, 5 mmol/l EDTA, 1% NP-40, 0.5% deoxycholate,
0.1% SDS). Supernatants (30 µl) were collected as input. The
remaining liquids were pre-cleared with 30 µl protein A/G agarose
(Santa Cruz Biotechnology, Inc.) on a rocker at 4°C for 2 h. After
that, the pre-cleared supernatants were separated into two and
incubated with the 6 µl ARF1 antibody and 0.3 µl mouse IgG,
respectively, at 4°C overnight with gentle agitation. Then, the
samples were incubated with 30 µl protein A/G at 4°C for 2 h with
gentle agitation, then deposits were collected. The immune
complexes were then analyzed by immunoblotting using antibodies
against ARF1 and PI3K.
Cell viability assay
Cell viablity assay was measured using the Cell
Counting Kit-8 (CCK-8) according to the manufacturer's instructions
(Dojindo, Kumamoto, Japan) to judge the ability of cell
proliferation. Cells transfected with siRNA were seeded into a
96-well plate and cultured in RPMI-1640 medium or control PBS with
100 µl in each well at a concentration of 2×104
cells/well in a volume of 100 µl culture medium and grown
overnight. CCK-8 reagents (Dojindo) were then added to each well
followed by incubation for an additional 2 h at 37°C. The
absorbency was measured at a test wavelength of 490 nm and a
reference wavelength of 630 nm using a microplate reader (Bio-Rad
Laboratories, Inc.). The experiments were repeated at least 3
times.
Wound healing assays
Wound healing was conducted as previously described
(23). Growing in a monolayer in
6-well plates, the cell growth was in nearly complete confluence.
The cells were serum starved for 12 h after 36 h of transfection
with control-shRNA, ARF1-shRNA#2. Afterwards, the monolayer was
scratched with a 10 ml pipette tip, the cells were washed with PBS
and cultured in the 1640 medium with 5% fetal bovine serum (FBS),
and an inverted Leica phase contrast microscope (Leica DFC300 FX)
was used to photograph the cells under ×20 objective lens at 0, 24,
and 36 h.
Statistical analysis
Statistical analysis was performed using SPSS 10.0
software (SPSS Inc., Chicago, IL, USA). The association between
ARF1 and Ki-67 expression and clinicopathological features was
analyzed using the Pearson's χ2 test. ARF1 and Ki-67
expression was studied using the Spearman rank correlation test,
because the data were not normally distributed. For analyzing the
survival data, Kaplan-Meier curves were constructed, and the
log-rank test was performed. Multivariate analysis was performed
using Cox's proportional hazards model, with P<0.05 considered
statistically significant. The results are expressed as the mean ±
SD.
Results
Expression and distribution of ARF1 in
EOC issues and cell lines and its correlation with survival
To verify the tumor entities with frequent aberrant
ARF1 expression in EOC, we first examined ARF1 protein expression
by western blot using several surgical specimens and three kinds of
EOC cell lines (Fig. 1). These
surgical specimens included three normal ovarian tissues and
different grades of ovarian cancer tissues, and the three EOC cell
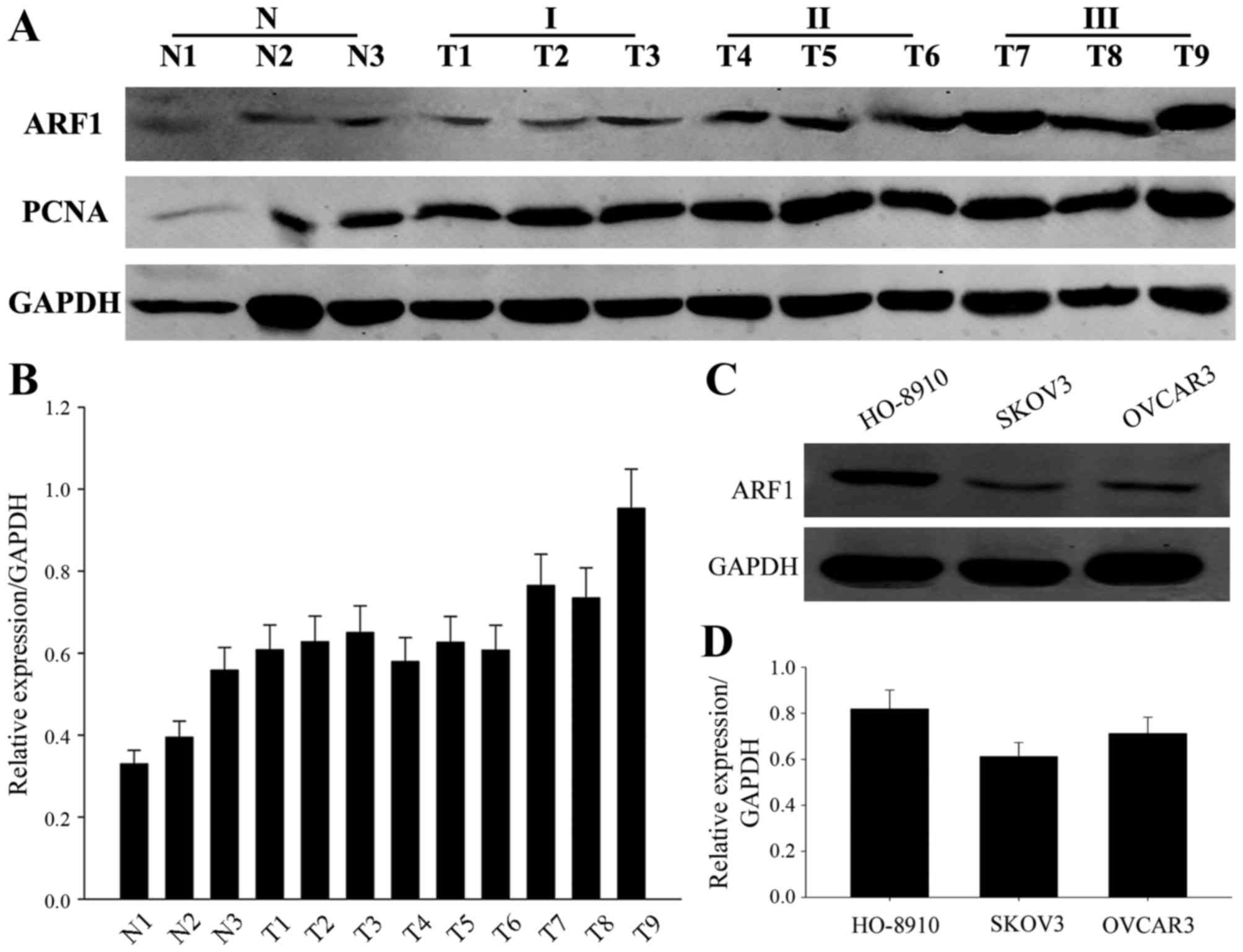
lines were HO-8910, SKOV3 and OVCAR3. As shown in Fig. 1A and B, the expression of ARF1 was
obviously higher in tumors when compared to the normal ovarian
tissues, and its expression was positive correlated with the tissue
grade. Simultaneously, ARF1 was also highly expressed in these
three EOC cell lines (Fig. 1C and
D), especially in the HO-8910 cell lines. Thus, we initially
thought that ARF1 may be associated with EOC progression.
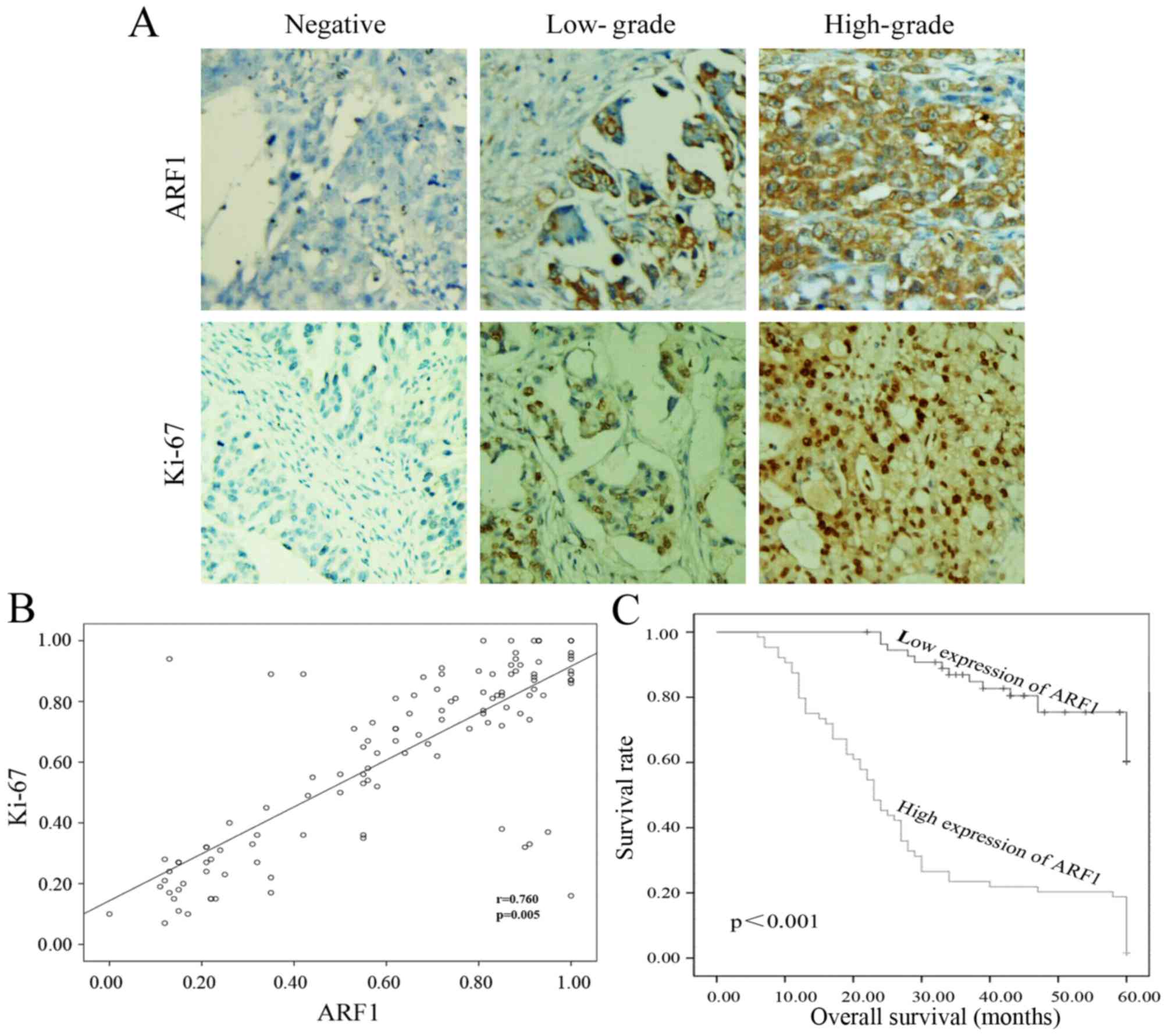
In order to confirm the above, immunohistochemistry
analysis of ARF1 and Ki-67 (a cell proliferation index) was
performed using 119 samples from patient with EOC. As shown in
Fig. 2A, we found that there was no
or little ARF1 or Ki-67 expression among these low-grade EOC
tissues. On the contrary, as the pathological grade increased, the
expression of ARF1 and Ki-67 was upregulated (Fig. 2A). Moreover, we also found that,
ARF1 was expressed mainly in cytomembrane, while the location of
Ki-67 was mainly in the nucleus. Thus, the expression of ARF1 was
higher in EOC tissues and EOC cell lines and it was positively
correlated with grade.
Correlation of ARF1 expression with
clinicopathological features of EOC
To further clarify the clinicopathological
significance of ARF1 expression in EOC, we evaluated the
association of ARF1 expression with clinicopathologic variables of
patients with EOC by Pearson's χ2 test. The
clinicopathological data of patients are summarized in Table I. From the results of statistical
analysis, we could find that the expression of ARF1 was
significantly associated with the histological grade (P=0.006),
FIGO stage (P=0.008), lymph node metastasis (P=0.010), and Ki-67
expression (P=0.005) (Table I).
Besides, we used the scatter plot to further determine the
relationship between the expression of ARF1 and Ki-67. Pearson's
correlation co-efficient revealed that there was a positive
correlation between ARF1 expression and Ki-67-based proliferative
activity (P=0.005, Fig. 2B).
Relationship between ARF1 expression
and clinical prognosis of EOC
119 EOC samples were used for survival analysis.
Pearson's χ2 test showed that the histological grade
(P=0.005), FIGO stage (P=0.012), ARF1 expression (P<0.001) and
Ki-67 (P=0.010) significantly influenced survival, indicating that
these factors were independent prognostic indicators of EOC
(Table II). Moreover, as shown in
Fig. 4, Kaplan Meier analysis was
carried out to calculate the impact of ARF1 expression level on
patient survival. The survival curves revealed that, in all 119
clinical cases, patients with high expression of ARF1 might have a
poorer overall survival (months) than the others (P<0.001,
Fig. 2C). Further, multivariate
analysis using Cox's proportional hazards model showed that the
histological grade (P=0.015), FIGO stage (P=0.043) as well as the
expression level of ARF1 (P=0.001) were 3 independent prognostic
factors of overall survival in EOC patients (Table III).
 | Table II.Survival status and
clinicopathological characteristics in the 119 human ovarian cancer
specimens. |
Table II.
Survival status and
clinicopathological characteristics in the 119 human ovarian cancer
specimens.
|
|
| Survival
status |
|
|
|
|
|
|
|
Characteristics | Total | Died | Alive |
P-valuea |
| Age (24–80 years,
mean 55 years) |
|
|
|
|
|
≤50 | 40 | 23 | 17 | 0.939 |
|
>50 | 79 | 46 | 33 |
|
| FIGO stage |
|
|
|
|
|
I/II | 60 | 28 | 32 | 0.012b |
|
III/IV | 59 | 41 | 18 |
|
| Histological
grade |
|
|
|
|
|
Low | 18 | 21 | 28 | 0.005b |
|
High | 30 | 48 | 22 |
|
| Histologic
subtype |
|
|
|
|
|
Serous | 63 | 41 | 22 | 0.291 |
|
Mucinous | 19 | 8 | 11 |
|
|
Endometrioid | 18 | 9 | 9 |
|
| Clear
cell | 19 | 11 | 8 |
|
| Lymph node
metastasis |
|
|
|
|
|
Negative | 86 | 48 | 38 | 0.439 |
|
Positive | 33 | 21 | 12 |
|
| Ki-67
expression |
|
|
|
|
|
Low | 55 | 25 | 30 | 0.010b |
|
High | 64 | 44 | 20 |
|
| ARF1
expression |
|
|
|
|
|
Low | 55 | 19 | 36 |
<0.001b |
|
High | 64 | 50 | 14 |
|
 | Table III.Contribution of various potential
prognostic factors to survival by Cox's regression analysis in the
119 human ovarian cancer specimens. |
Table III.
Contribution of various potential
prognostic factors to survival by Cox's regression analysis in the
119 human ovarian cancer specimens.
|
Characteristics | Hazard ratio | 95% confidence
interval |
P-valuea |
| Histological
grade | 0.512 | 0.299–0.877 | 0.015b |
| FIGO stage | 0.548 | 0.307–0.981 | 0.043b |
| Lymph node
metastasis | 0.788 | 0.092–6.729 | 0.828 |
| ARF1
expression | 3.260 | 1.814–5.856 | 0.001b |
ARF1 positively participates in the
proliferation progress of EOC cell lines
ARF1 was proven to regulate proliferation of breast
cancer cells by regulating the retinoblastoma protein. We found
ARF1 was positively correlated with the expression of Ki-67 in EOC
specimens. Now, we confirmed that the expression of ARF1 was
obviously increased in EOC cells. We chose HO-8910 cell line, which
had the highest protein level of the EOC cell lines, to construct a
cell serum starvation and releasing model. The result showed that
the HO-8910 cells were clearly arrested in the G1 phase and
re-entered the S phase with flow cytometry analysis following serum
deprivation for 48 h. The cells in G1 phase were up to 67.42%.
After that, the cell population of S phase gradually increased time
dependently, upon continued re-feeding (Fig. 3A and B). Further, we found the
expression of ARF1, PCNA and cyclin D1 was significantly increased
after serum re-addition as assessed with western blot analysis
(Fig. 3C and D). All these results
indicated that ARF1 positively participated in the proliferation
progress of EOC cells.
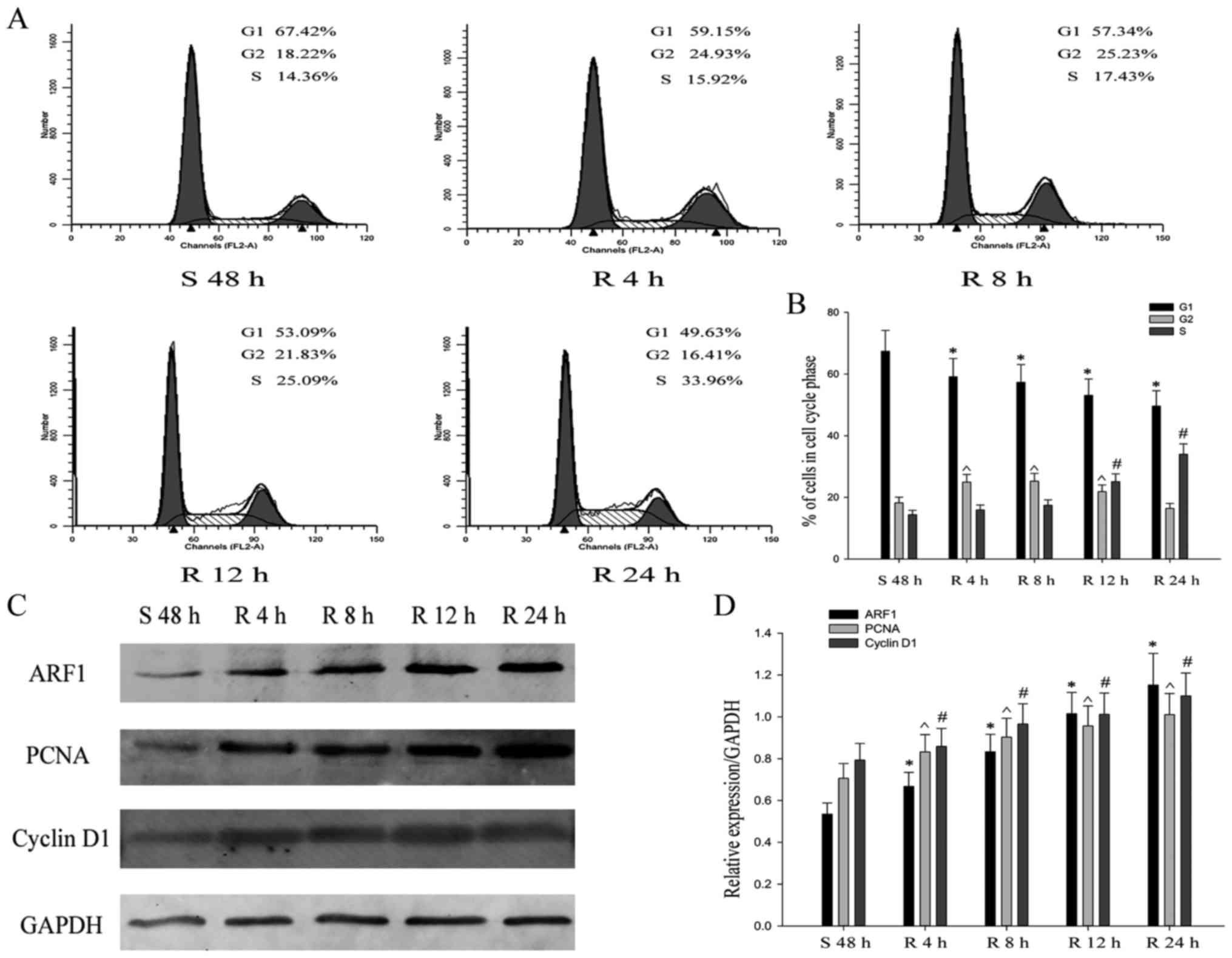 | Figure 3.Expression of ARF1 promotes
proliferation of EOC cells. (A and B) Flow cytometric
quantification of cell cycle progression in HO-8910 cells.
*,#,^P<0.05, compared with G1, G2, and S phase serum starved for
48 h (S48 h). (C and D) The HO-8910 cells were released by
re-feeding with serum, and cell lysates were prepared and analyzed
by western blotting using antibodies against ARF1, PCNA and cyclin
D1 protein to GAPDH for each time point by densitometry. The data
are mean ± SEM of three independent experiments. n=3,
*,#,^P<0.05, compared with serum starved for 48 h (S48 h). SEM,
standard error of the mean; S, serum starvation; R, serum
release. |
Knockdown of ARF1 inhibits
proliferation in EOC cell lines
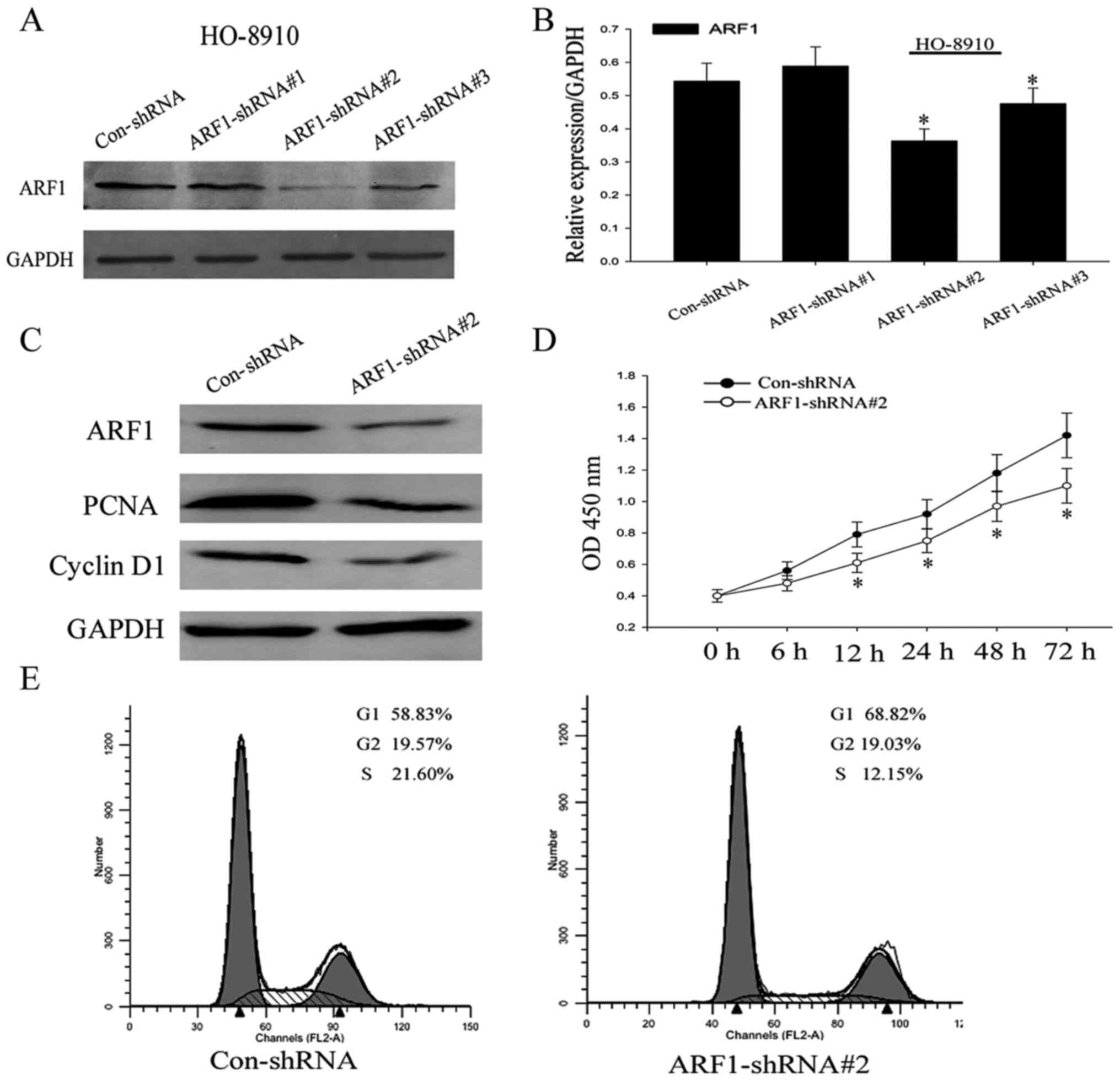
Up to now, we have preliminary cleared the effect of
ARF1 in the process of ovarian cancer, in order to further clarify
its role in cell proliferation, shRNA was then used to knock down
ARF1 expression in the HO8910 cells. HO8910 cells were transiently
transfected with control-shRNA, ARF1-shRNA#1, ARF1-shRNA#2, and
ARF1-shRNA#3. After 48 h, we collected these cell proteins and used
western blot analysis to detect the expression levels of ARF1
(Fig. 4A and B). ARF1 protein
expression levels were substantially decreased in HO-8910 cells
transfected especially with ARF1-shRNA#2 and to a lesser extent
ARF1-shRNA#3 compared to the control-shRNA-transfected cells. Thus,
in the following experiment, we chose ARF1-shRNA#2 as the
experimental model. It was shown that with the significant
downregulation of ARF1 expression levels, the expression of PCNA
and cyclin D1 were also remarkablely decreased (Fig. 4C).
To further investigate whether the activity of ARF1
affected cell viability, CCK-8 assay was used. As shown in Fig. 4D, HO-8910 cells treated with shRNA
suppressed cell proliferation rate compared with the negative
control shRNA. In order to explore relevant causes, we analyzed the
cell cycle of HO8910 cells transfected with control-shRNA and
ARF1-shRNA#2 by flow cytometry. After transfected for the same
period of time, we could see that the proportion of the cells,
which were treated with ARF1-shRNA#2, appeared to be increased in
the G0/G1 phase (from 58.83% to 68.82%), while decreased in the S
phase (from 21.60% to 12.15%) compared to the negative control
(Fig. 4E). This might suggest that
ARF1 is crucial for the G0/G1-S transition and thus the cell
growth. Based on the above results, we came to the conclusion that
ARF1 could certainly promote proliferation in EOC cell lines, while
when knocked down, it was inhibited.
ARF1 significantly affects the ability
of migration as well as proliferation of EOC cells by forming a
PI3K-dependent feedforward signaling pathway
Previously we have made it clear that ARF1 could
obviously promote proliferation of EOC cells. As it was knocked
down, this ability significantly weakened. However, the specific
mechanisms need clarification and whether there was any signal
pathways involved. Considering signaling pathways related to the
ability of proliferation in tumor cells, PI3K signaling pathway
might be the first one to be considerd. In order to further define
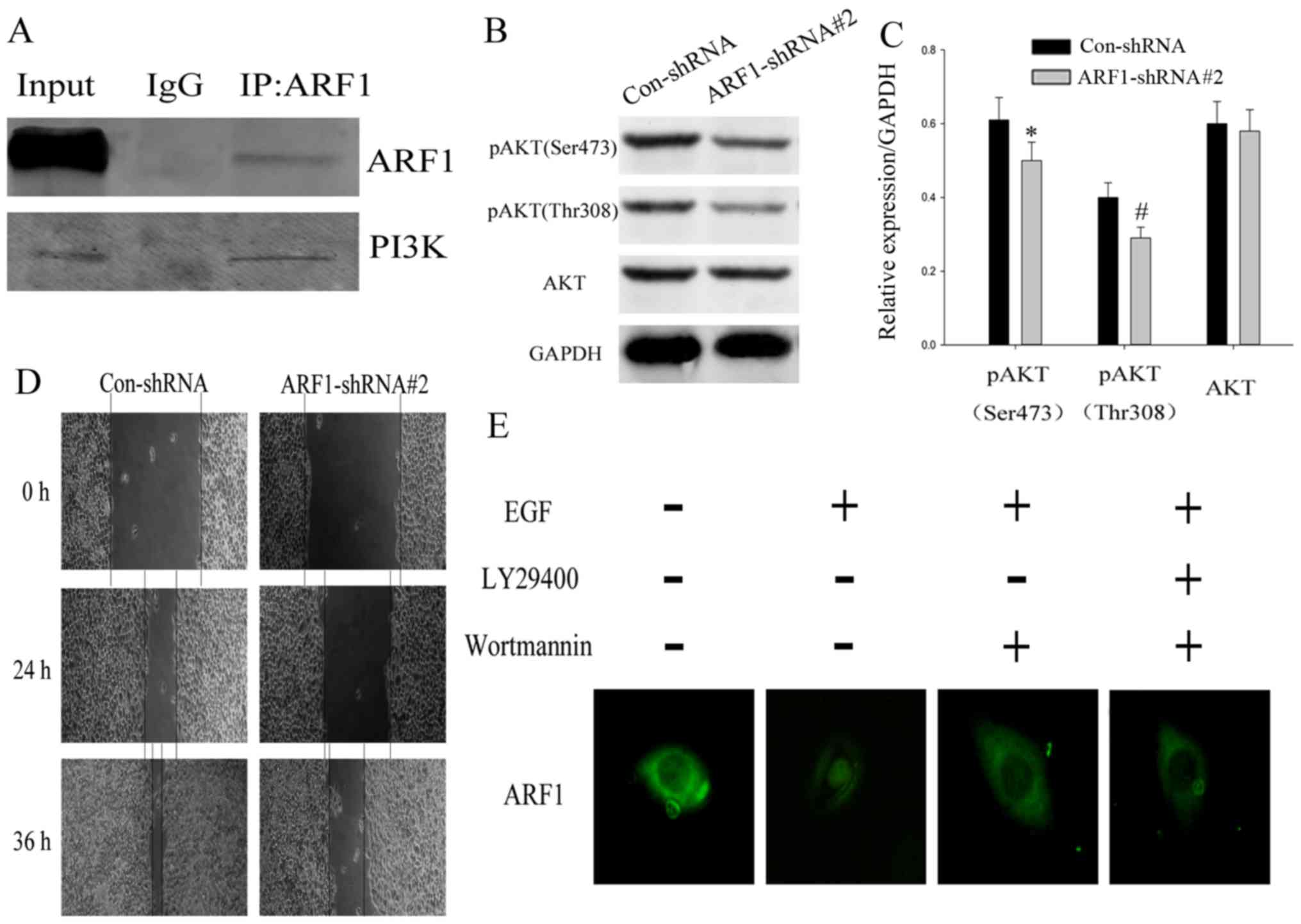
the mechanism of ARF1 in detail, we first attempted to verify
whether it interacted with PI3K, then, co-immunoprecipitation was
used. From the results, we found that ARF1 interacted with PI3K in
EOC cells (Fig. 5A). To further
support the role of ARF1 in regulating the PI3K/Akt pathway, we
investigated if a knockdown of ARF1 would have any effect on the
related molecules in PI3K signaling pathways. After transfection,
we detected the expression levels of phosphorylation of AKT (Ser473
and Thr308) and AKT by western blot analysis.
As shown in Fig. 5B and
C, the expression of pAKT (both Ser473 and Thr308) were notably
decreased in HO-8910 cells treated with ARF1-shRNA#2 compared to
the control, while the AKT in these two kinds of cells seemed to be
consistent. These data support that ARF1 is involved in PI3K
pathway activation. The PI3K pathway is involved in a series of
biological behavior of tumor cells, such as proliferation,
invasion, migration and metastasis. We found, after transfection
with ARF1-shRNA#2 for 24 h, the ability of cell migration also
significantly decreased. After the same treatment for 36 h, this
situation was more obvious (Fig.
5D). These results suggested that ARF1 played a general role in
the proliferation and migration of EOC cell lines through the PI3K
pathway. Then, IHC (Fig. 2A) showed
that ARF1 located in cell membrane, and the epidermal growth factor
receptor (EGFR), also at the plasma membrane is considered a major
oncogenic factor, and its presence in tumors is indicative of poor
prognosis through activating the PI3K signaling pathway. Given this
finding, we hypothesized that the small GTPase ARF1 as a downstream
target molecule of PI3K. We used EGF stimulation and
pharmacological inhibitors of PI3K for confirmation. A
translocation of ARF1 from the cytosol to the membrane compartment
was observed upon EGF stimulation. In addition, cells treated with
wortmannin or LY294002 showed significantly decreased membrane
translocation (Fig. 5E), indicating
that PI3K might regulate ARF1 by adjusting its orientation. Taken
together, all this illustrated that ARF1 significantly affected the
malignant behavior of EOC cells by forming a PI3K-dependent
feed-forward signaling pathway.
Discussion
EOC is the most common and lethal histotype of all
ovarian cancers with a 5-years survival rate of only 30%. Although
traditional therapies such as surgical resection, chemotherapy, and
radiotherapy have been widely applied, there has been little
improvement in overall survival for several decades, emphasizing
the need for new treatments (24).
Thus, a thorough understanding of the molecular events associated
with the process of EOC is necessary. In the present study, we
demonstrate the functional role and clinical significance of ARF1
in EOC.
Previous studies showed that exogenously expressed
ARF1 is mainly localized to the ERGIC and the Golgi (25–27),
and their depletion induces formation of tubules from the ERGIC
(27). Although ARF1 is mostly
known for its role in the regulation of the secretory pathway, the
demonstration that this ARF isoform is present at the plasma
membrane and activated after receptor stimulation provides a new
rationale for its role in different receptor-mediated biological
functions.
Our current study revealed that ARF1 significantly
associated with a poor prognosis in EOC, which was confirmed both
by gene expression and protein expression analyses. Moreover, Cox's
proportional hazards model also proved that the histological grade
as well as the expression level of ARF1 were independent prognostic
factors of overall survival in EOC patients. Furthermore, by serum
starvation and release experiments, CCK-8 assay and wound healing
experiment, it was demonstrated that ARF1 could promote the
proliferation and migration rate of EOC cells. With the knockdown
of its expression by shRNA in an in vitro experiment, the
ability of proliferation and migration and the expression of
related molecules such as PCNA, cyclin D1 and others were
significantly downregulated. These findings showed that ARF1 played
an important role in identifying a poor prognostic result of EOC.
Specifically, we showed that this ARF isoform was present at the
plasma membrane and regulated activation of the PI3K pathway.
It is well known that class IA PI3Ks are
translocated to activate RTKs to regulate cell migration and growth
downstream of RTKs (28).
Activation of the PI3K/Akt pathway by growth factors is associated
with cell survival and growth, as well as proliferation through
multiple downstream targets impinging on cell cycle regulation. In
this study, we demonstrated that depletion of ARF1 markedly
inhibited activation of the PI3K pathway, as assessed by the
phosphorylation of Akt. In either case, our data suggested that
ARF1 might act as a switch to regulate this key event. Taken
together, these findings suggested that ARF1 might contribute to
cancer cell progression and migration by controlling the PI3K
pathway. While, it is reported that, the PI3K binding site of Gab2
regulated the granule-translocation step and that the SHP2 binding
site contributed to plasma-granule membrane fusion. Furthermore,
the small GTPase ARF1 was thought to be a downstream target
molecule of PI3K (29). Combined
with our existing results, we speculate that ARF1 may not only be
upstream of the PI3K pathway to regulate its functions, but also
can be thought as downstream of this pathway, making it regulated,
and this was verified by us. This view also requires further
verification in our following studies.
In summary, our studies confirmed that ARF1 was
highly expressed in both EOC tissues and EOC cell lines, and its
expression was positively correlated with the ability of
proliferation and migration of EOC. Furthermore, the knockdown of
ARF1 could inhibit both the cell proliferation and migration of
EOC. The fact that the depletion of ARF1 had similar effects on EOC
cell lines suggests that the mechanisms that we have uncovered may
be widely used. Taken together, our findings reveal an unsuspected
role for ARF1 and indicates that this small GTPase may be a
potential therapeutic target for the treatment of EOC. It is of
special interest to note the identification of an inhibitor that
specifically prevents ARF1 activation (30). Further studies using such compounds
will provide insights into the precise role of ARF1 in
tumorigenesis.
Glossary
Abbreviations
Abbreviations:
|
ARF1
|
ADP ribosylation factors
|
|
EOC
|
epithelial ovarian cancer
|
References
|
1
|
Lynch HT, Snyder C and Casey MJ:
Hereditary ovarian and breast cancer: What have we learned? Ann
Oncol. 24 Suppl 8:viii83–viii95. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kucukmetin A, Naik R, Galaal K, Bryant A
and Dickinson HO: Palliative surgery versus medical management for
bowel obstruction in ovarian cancer. Cochrane Database Syst Rev.
7:CD0077922010.
|
|
3
|
Keen A, Fitzgerald D, Bryant A and
Dickinson HO: Management of drainage for malignant ascites in
gynaecological cancer. Cochrane Database Syst Rev.
1:CD0077942010.
|
|
4
|
Khosravi-Shahi P and Cabezon-Gutierrez L:
Antiangiogenic drugs in the treatment of advanced epithelial
ovarian cancer. Anticancer Agents Med Chem. 12:982–987. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bhatt P, Vhora I, Patil S, Amrutiya J,
Bhattacharya C, Misra A and Mashru R: Role of antibodies in
diagnosis and treatment of ovarian cancer: Basic approach and
clinical status. J Control Release. 226:148–167. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Shen DF, Liu X, Yang XF, Fang L, Gao Y,
Zhao S, Wu JC, Shi S, Li JJ, Zhao XX, et al: The roles of
parafibromin expression in ovarian epithelial carcinomas: A marker
for differentiation and prognosis and a target for gene therapy.
Tumour Biol. 37:2909–2924. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Li W, Zhou Y, Su Y, Ouyang Y, Xie X, Wu Y,
Mao C and Chen D: IL-23 promotes invasion of esophageal squamous
cell carcinoma cells by activating DLL4/Notch1 signaling pathway.
Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi. 31:812–815, 820. 2015.(In
Chinese). PubMed/NCBI
|
|
8
|
Stuckey A, Fischer A, Miller DH,
Hillenmeyer S, Kim KK, Ritz A, Singh RK, Raphael BJ, Brard L and
Brodsky AS: Integrated genomics of ovarian xenograft tumor
progression and chemotherapy response. BMC Cancer. 11:3082011.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Sabe H, Hashimoto S, Morishige M, Ogawa E,
Hashimoto A, Nam JM, Miura K, Yano H and Onodera Y: The
EGFR-GEP100-Arf6-AMAP1 signaling pathway specific to breast cancer
invasion and metastasis. Traffic. 10:982–993. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Caviston JP, Cohen LA and Donaldson JG:
Arf1 and Arf6 promote ventral actin structures formed by acute
activation of protein kinase C and Src. Cytoskeleton. 71:380–394.
2014. View
Article : Google Scholar : PubMed/NCBI
|
|
11
|
Stearns T, Willingham MC, Botstein D and
Kahn RA: ADP-ribosylation factor is functionally and physically
associated with the Golgi complex. Proc Natl Acad Sci USA.
87:1238–1242. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Robertson DN, Johnson MS, Moggach LO,
Holland PJ, Lutz EM and Mitchell R: Selective interaction of ARF1
with the carboxy-terminal tail domain of the 5-HT2A receptor. Mol
Pharmacol. 64:1239–1250. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Cohen LA, Honda A, Varnai P, Brown FD,
Balla T and Donaldson JG: Active Arf6 recruits ARNO/cytohesin GEFs
to the PM by binding their PH domains. Mol Biol Cell. 18:2244–2253.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hassa PO, Haenni SS, Elser M and Hottiger
MO: Nuclear ADP-ribosylation reactions in mammalian cells: Where
are we today and where are we going? Microbiol Mol Biol Rev.
70:789–829. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Norman JC, Jones D, Barry ST, Holt MR,
Cockcroft S and Critchley DR: ARF1 mediates paxillin recruitment to
focal adhesions and potentiates Rho-stimulated stress fiber
formation in intact and permeabilized Swiss 3T3 fibroblasts. J Cell
Biol. 143:1981–1995. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Viaud J, Zeghouf M, Barelli H, Zeeh JC,
Padilla A, Guibert B, Chardin P, Royer CA, Cherfils J and Chavanieu
A: Structure-based discovery of an inhibitor of Arf activation by
Sec7 domains through targeting of protein-protein complexes. Proc
Natl Acad Sci USA. 104:10370–10375. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Boulay PL, Cotton M, Melançon P and Claing
A: ADP-ribosylation factor 1 controls the activation of the
phosphatidylinositol 3-kinase pathway to regulate epidermal growth
factor-dependent growth and migration of breast cancer cells. J
Biol Chem. 283:36425–36434. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Schlienger S, Ramirez RA and Claing A:
ARF1 regulates adhesion of MDA-MB-231 invasive breast cancer cells
through formation of focal adhesions. Cell Signal. 27:403–415.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Frame MC: Src in cancer: Deregulation and
consequences for cell behaviour. Biochim Biophys Acta.
1602:114–130. 2002.PubMed/NCBI
|
|
20
|
Wheeler DL, Iida M and Dunn EF: The role
of Src in solid tumors. Oncologist. 14:667–678. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ji L, Li H, Gao P, Shang G, Zhang DD,
Zhang N and Jiang T: Nrf2 pathway regulates
multidrug-resistance-associated protein 1 in small cell lung
cancer. PLoS One. 8:e634042013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wang Y, Liu F, Mao F, Hang Q, Huang X, He
S, Wang Y, Cheng C, Wang H, Xu G, et al: Interaction with cyclin
H/cyclin-dependent kinase 7 (CCNH/CDK7) stabilizes C-terminal
binding protein 2 (CtBP2) and promotes cancer cell migration. J
Biol Chem. 288:9028–9034. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wang Y, Yang S, Ni Q, He S, Zhao Y, Yuan
Q, Li C, Chen H, Zhang L, Zou L, et al: Overexpression of forkhead
box J2 can decrease the migration of breast cancer cells. J Cell
Biochem. 113:2729–2737. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kyriakidis I and Papaioannidou P: Estrogen
receptor beta and ovarian cancer: A key to pathogenesis and
response to therapy. Arch Gynecol Obstet. 293:1161–1168. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ara I, Bakir MA and Kudo T: Transfer of
Catellatospora koreensis Lee et al. 2000 as Catelliglobosispora
koreensis gen. nov., comb. nov. and Catellatospora tsunoense Asano
et al. 1989 as Hamadaea tsunoensis gen. nov., comb. nov., and
emended description of the genus Catellatospora Asano and Kawamoto
1986 emend. Lee and Hah 2002. Int J Syst Evol Microbiol.
58:1950–1960. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chun J, Shapovalova Z, Dejgaard SY,
Presley JF and Melançon P: Characterization of class I and II
ADP-ribosylation factors (Arfs) in live cells: GDP-bound class II
Arfs associate with the ER-Golgi intermediate compartment
independently of GBF1. Mol Biol Cell. 19:3488–3500. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ben-Tekaya H, Kahn RA and Hauri HP: ADP
ribosylation factors 1 and 4 and group VIA phospholipase A(2)
regulate morphology and intraorganellar traffic in the endoplasmic
reticulum-Golgi intermediate compartment. Mol Biol Cell.
21:4130–4140. 2010.doi: 10.1091/mbc.E10-01–0022. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Burke JE and Williams RL: Dynamic steps in
receptor tyrosine kinase mediated activation of class IA
phosphoinositide 3-kinases (PI3K) captured by H/D exchange
(HDX-MS). Adv Biol Regul. 53:97–110. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Nishida K, Yamasaki S, Hasegawa A,
Iwamatsu A, Koseki H and Hirano T: Gab2, via PI-3K, regulates ARF1
in FcεRI-mediated granule translocation and mast cell
degranulation. J Immunol. 187:932–941. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Orlichenko L, Stolz DB, Noel P, Behari J,
Liu S and Singh VP: ADP-ribosylation factor 1 protein regulates
trypsinogen activation via organellar trafficking of procathepsin B
protein and autophagic maturation in acute pancreatitis. J Biol
Chem. 287:24284–24293. 2012. View Article : Google Scholar : PubMed/NCBI
|