Introduction
Colorectal cancer (CRC) is a leading cause of
cancer-related deaths worldwide (1). There is a large proportion of CRC
patients who are diagnosed with locally advanced disease with
regional lymph node metastasis. Studying the mechanisms underlying
the metastasis of CRC and identifying new clinical molecular
markers have enabled further subclassification of patients and
thereby a more accurate prediction of biological behavior and
prognosis (2). To the best of our
knowledge, there are a number of molecules that have been recently
reported as CRC biomarker candidates (3). Among these, microRNA-133b (miR-133b)
and TAp63 have attracted much attention due to accumulated evidence
derived from basic and clinical studies supporting their clinical
use (4–6).
miR-133b is one of the most significantly
downregulated miRNAs in CRC (7). It
has been reported to participate in the migration and invasion of
certain types of cancer and is downregulated in tumor tissues, such
as gastric (8), prostate (9) and CRC (10,11).
However, the mechanisms involved in miR-133b downregulation in
cancer remain unclear. TAp63, a protein isoform of the p63
tumor-suppressor gene, is able to activate gene transcription and
is closely related with metastasis (12,13).
In prostate and bladder cancer, TAp63 physically binds to the
caspase-1 promoter and directly regulates caspase-1 expression
(14). However, TAp63 also directly
regulates the expression of miRNAs, for example, miR-205 (15). We previously demonstrated that TAp63
expression leads to the transcriptional regulation of the
expression of miR-133b (16).
Therefore, we speculated that downregulation of TAp63 may be a
mechanism involved in miR-133b dysregulation in CRC. A point
mutation, or single base modification, is a type of mutation that
causes a single nucleotide base substitution, insertion or deletion
of genetic material. Point mutations have been proven to influence
expression of many genes (17–19).
In addition, miRNA gene point mutations have been reported in
almost all types of cancer (20,21).
However, little is known regarding the possible impact of gene
mutations on miR-133b expression.
In the present study, we aimed to understand whether
the expression level of miR-133b is affected by TAp63 and miR-133b
gene point mutations in CRC, and whether TAp63 and miR-133b
expression can independently provide useful information in regards
to the prediction of outcome in CRC.
Materials and methods
Materials
P63 rabbit monoclonal antibodies were purchased from
Abcam (Cambridge, UK). GAPDH rabbit polyclonal antibody was
purchased from ProteinTech Group, Inc. (Chicago, IL, USA).
Tissue specimens
A total of 38 pairs of formalin-fixed,
paraffin-embedded (FFPE) sections of CRC tissues and non-cancerous
tissues (NCTs) were obtained from the Pathology Department of the
Third XiangYa Hospital of Central South University (Hunan, China)
between October 2013 and June 2014. None of these patients had
received radiotherapy or chemotherapy prior to surgery. The
histopathological type and the stage of CRC were determined
according to the criteria of the World Health Organization
classification. All of the patients were staged using the US
National Comprehensive Cancer Network (NCCN) Clinical Guidelines
2014. The present study was approved by the Ethics Committee of the
Third XiangYa Hospital of Central South University. Patient consent
was obtained both from the patients and the families of the
patients.
RNA isolation and RT-qPCR
RT-qPCR was performed as previously described
(16). Briefly, total RNA was
isolated using an E.Z.N.A. Total RNA Kit II (Omega Bio-Tek Inc.,
Norcross, GA, USA). miRNA was isolated using an E.Z.N.A. PF miRNA
Isolation kit (Omega Bio-Tek Inc.). RT-qPCR was performed using
Real-Time Quantitative PCR SYBR-Green detection reagent (CoWin
Biotech Co., Ltd., Beijing, China). miRNA RT-qPCR was performed
using an All-in-One™ miRNA qRT-PCR Detection kit (GeneCopoeia,
Rockville, MD, USA). Primers and the reaction conditions have been
previously described (16). The
relative expression of TAp63 was normalized using the
2−ΔΔCt method relative to GAPDH. The relative expression
of miR-133b was normalized using the 2−ΔΔCt method
relative to U6-snRNA. All PCR reactions were run in triplicate.
Each sample was amplified in triplicate.
Western blotting
Western blot analysis was performed as previously
described using primary antibodies (16). Briefly, cells were lysed in a lysis
buffer and centrifuged at 14,000 × g at 4°C for 10 min. The
supernatants were collected and a BCA protein assay was performed.
Total protein (100 mg) was separated on a 10% polyacrylamide gel
and transferred to polyvinylidene difluoride (PVDF) membranes
(Invitrogen, Carlsbad, CA, USA). The membranes were blocked for 2
h, and were then incubated with a primary antibody at 4°C
overnight. After being washed with PBST, the membranes were
incubated with an HRP-conjugated goat anti-rabbit IgG secondary
antibody for 60 min at room temperature. The images were obtained
using Kodak film (Shanghai, China). All experiments were performed
in triplicate.
Immunohistochemical staining
Immunohistochemistry (IHC) was carried out using
Dako EnVision System (Dako Diagnostics AG, Zug, Switzerland)
following the manufacturers protocol. For TAp63 protein, staining
localized in the nuclei was considered positive. Images were
captured using an Olympus microscope. Image-Pro Plus version 6.0
software (Media Cybernetics, Inc., Rockville, MD, USA) was used to
assess the area and density of the dyed region, and the integrated
optical density (IOD) value of the IHC section. The mean
densitometry of the digital image (magnification, ×400) was
designated as representative TAp63 staining intensity (indicating
the relative TAp63 expression level). The signal density of the
tissue areas from five randomly selected fields were counted in a
blinded manner and subjected to statistical analysis.
In situ hybridization (ISH)
ISH was performed on CRC tissues. A locked nucleic
acid (LNA) detection probe for miR-133b, an LNA U6-positive and an
LNA-negative control probe with a scramble sequence were used in
this analysis. All probes were purchased from Auragene Bioscience
Corporation, Inc. (Changsha, China). All probes were labeled with
digoxigenin. After being washed with phosphate-buffered saline
(PBS) and pre-hybridization, the slides were incubated with
DIG-labeled LNA miR-133b, DIG-labeled LNA U6 probes (positive
control) and DIG-labeled LNA scrambled sequence (negative control)
at 49°C overnight. Washes were carried out at 49°C, and then the
slides were incubated with blocking solution. After PBS washing,
the images were captured using an Olympus microscope. Image-Pro
Plus version 6.0 software was used to assess the area and density
of the dyed region and the IOD value of the ISH section.
Secondary structure prediction
The secondary structure of 446 bp pre-miR-133b
sequences was predicted using RNAfold web server (http://nhjy.hzau.edu.cn/kech/swxxx/jakj/dianzi/Bioinf4/miRNA/miRNA1.htm).
Statistical analysis
We analyzed the data from February 2015. All
statistical calculations were performed using GraphPad Prism
(GraphPad Prism software, version 6.01; GraphPad, San Diego, CA,
USA) and Statistical Package for the Social Sciences (SPSS) PASW
Statistics software, version 20 (SPSS, Inc., Chicago, IL, USA). A
Mann-Whitney U test was used to compare differences between two
groups. The correlation between TAp63 and miR-133b was determined
by the Spearmans rank correlation test. The sensitivity and
specificity in CRC tissues were calculated for each cut-off value.
The area under receiver-operating characteristic curve (AUC) was
also estimated. The level of statistical significance was set at
P<0.05. All experiments were repeated three times.
Results
Clinicopathological characteristics of
the patients
The clinical and histopathological features of the
patients are summarized in Table I.
In total 38 patients were included in the present study, which
comprised 26 men and 12 women, with a combined age range of 55–83
years. Regarding the degree of cell differentiation, out of the 38
cases, 6 (15.79%) were well-differentiated adenocarcinomas, 27
(71.05%) were moderately differentiated and 5 (13.16%) cases were
poorly differentiated adenocarcinomas.
 | Table I.Association of the clinicopathological
characteristics of the CRC cases with TAp63 and miR-133b mRNA
expression. |
Table I.
Association of the clinicopathological
characteristics of the CRC cases with TAp63 and miR-133b mRNA
expression.
|
| TAp63 | miR-133b |
|---|
|
|
|
|
|---|
| Clinicopathological
features | Cases (n=38) | Lowa (n=19) | Higha (n=19) | P-value | Lowa (n=19) | Higha (n=19) | P-value |
|---|
| Mean age (years) |
| 56.63±15.37 | 58.95±10.51 | 0.567 | 59.11±12.40 | 56.47±13.86 | 0.545 |
| Tumor site |
| Left
colon | 10 | 6 | 4 | 0.609 | 5 | 5 | 0.885 |
| Right
colon | 5 | 3 | 2 |
| 3 | 2 |
|
Rectum | 23 | 10 | 13 |
| 11 | 12 |
| Tumor
differentiation |
| Well | 6 | 4 | 2 | 0.549 | 3 | 3 | 0.888 |
|
Moderate | 27 | 12 | 15 |
| 14 | 13 |
| Poor | 5 | 3 | 2 |
| 2 | 3 |
| Staging |
| I | 5 | 2 | 3 | 0.679 | 3 | 2 | 0.731 |
| II | 11 | 6 | 5 |
| 6 | 5 |
|
III | 18 | 8 | 10 |
| 9 | 9 |
| IV | 4 | 3 | 1 |
| 1 | 3 |
| Lymph node
metastasis |
| N0 | 17 | 7 | 11 | 0.016b | 5 | 13 | 0.033b |
| N1 | 12 | 4 | 7 |
| 8 | 3 |
| N2 | 9 | 8 | 1 |
| 6 | 3 |
| Distant
metastasis |
| M0 | 34 | 15 | 19 | 0.034b | 15 | 19 | 0.034b |
| M1 | 4 | 4 | 0 |
| 4 | 0 |
Regarding the evolutionary stage of the studied CRC
cases, the majority of the patients were in the late disease
stages: 5 (11.16%) were diagnosed in stage I, 11 (28.95%) in stage
II, 18 (47.37%) in stage III and 4 (10.53%) in stage IV.
Expression of TAp63 is decreased in
human CRC tissues
To study the expression of TAp63, we measured the
mRNA and protein expression levels of TAp63 in 38 paired human CRC
tissues and NCT samples using qRT-PCR, western blotting and IHC
methods, respectively. Compared with the NCTs, the TAp63 mRNA
expression level was significantly lower in 21 (55.27%) tumor
tissues (Fig. 1A, P<0.05).
Consistent with the RT-qPCR results, a decrease of TAp63 protein
expression was observed in the CRC tissues, compared with the NCTs
(Fig. 1B, P<0.05). IHC also
revealed that TAp63 was downregulated in the CRC tissues (Fig. 1C). Moreover, we evaluated the
density of the staining signal of all 38 cases using the Image-Pro
Plus 6.0 image analysis software. The mean density of cancerous
tissues from all 38 cases was 0.005345±0.0005019, while that of the
adjacent tissue was 0.01483±0.0003317. This result suggests a
significantly lower level of TAp63 expression in CRC tissues
compared to their NCTs (Fig. 1D,
P<0.05).
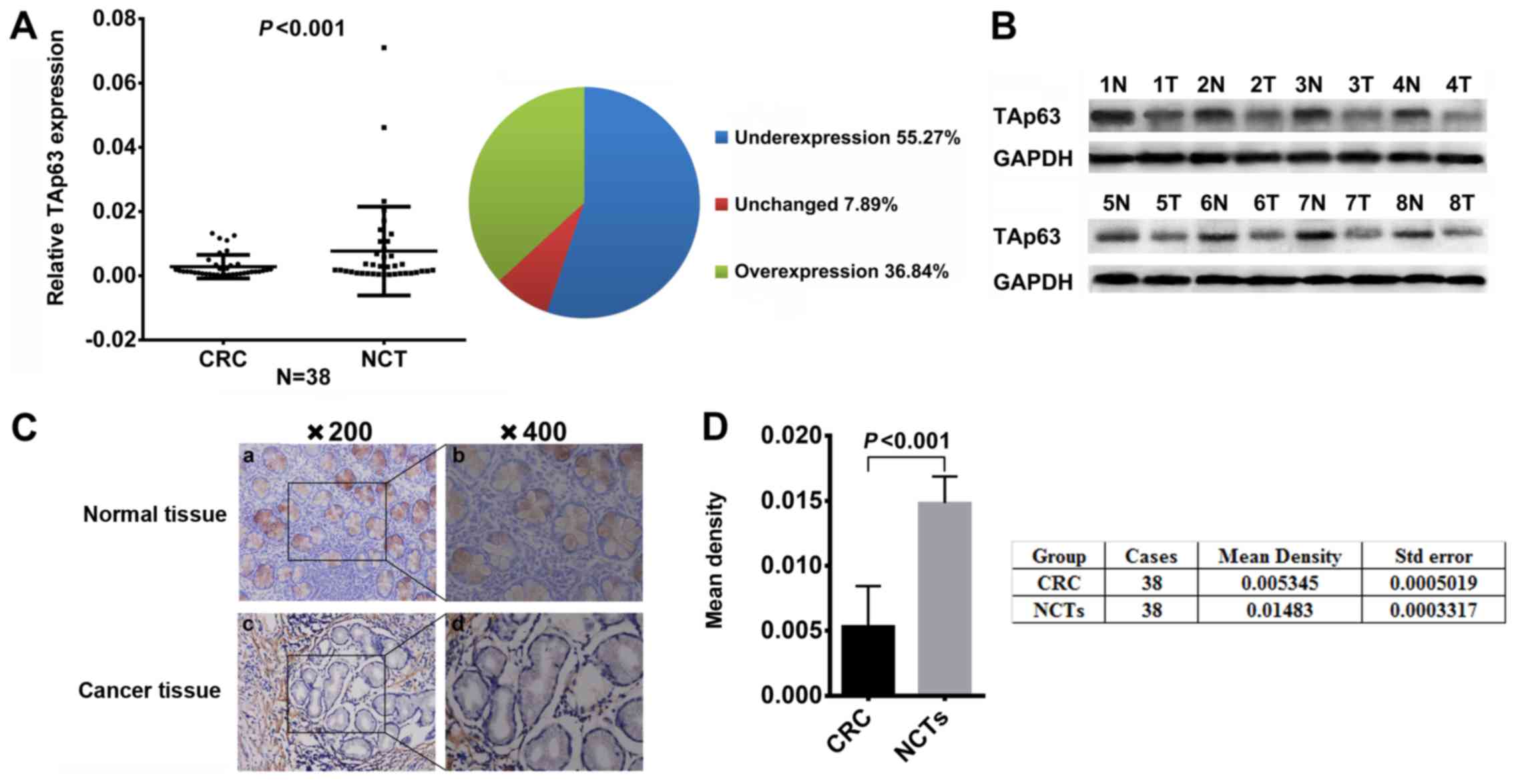 | Figure 1.Expression of TAp63 in CRC patients.
(A) Downregulated mRNA expression of TAp63 in CRC tissues as
assessed by RT-qPCR (n=38; P<0.001). The mean expression of
TAp63 is represented in the left scatter diagram and the expression
distribution is summarized in the right pie chart. (B)
Representative result of TAp63 protein expression in eight paired
CRC and NCTs by IHC (T, tumor tissue; N, normal tissue). (C) IHC
staining of CRC and NCTs from the same patient showed a low TAp63
expression. a, IHC staining of TAp63 in NCT (magnification, ×200);
b, IHC staining of TAp63 in NCT (magnification, ×200); c, IHC
staining of TAp63 in CRC tissue (magnification, ×200); d, IHC
staining of TAp63 in CRC tissue (magnification, ×400). (D) TAp63
expression in CRC and NCTs. The mean density of TAp63 in tumor and
normal tissues from 38 cases is illustrated in the left graph and
is summarized in the table, on the right. A significant difference
was detected between the mean density of TAp63 in CRC
(0.005345±0.0005019) and NCTs (0.01483±0.0003317); P<0.001. |
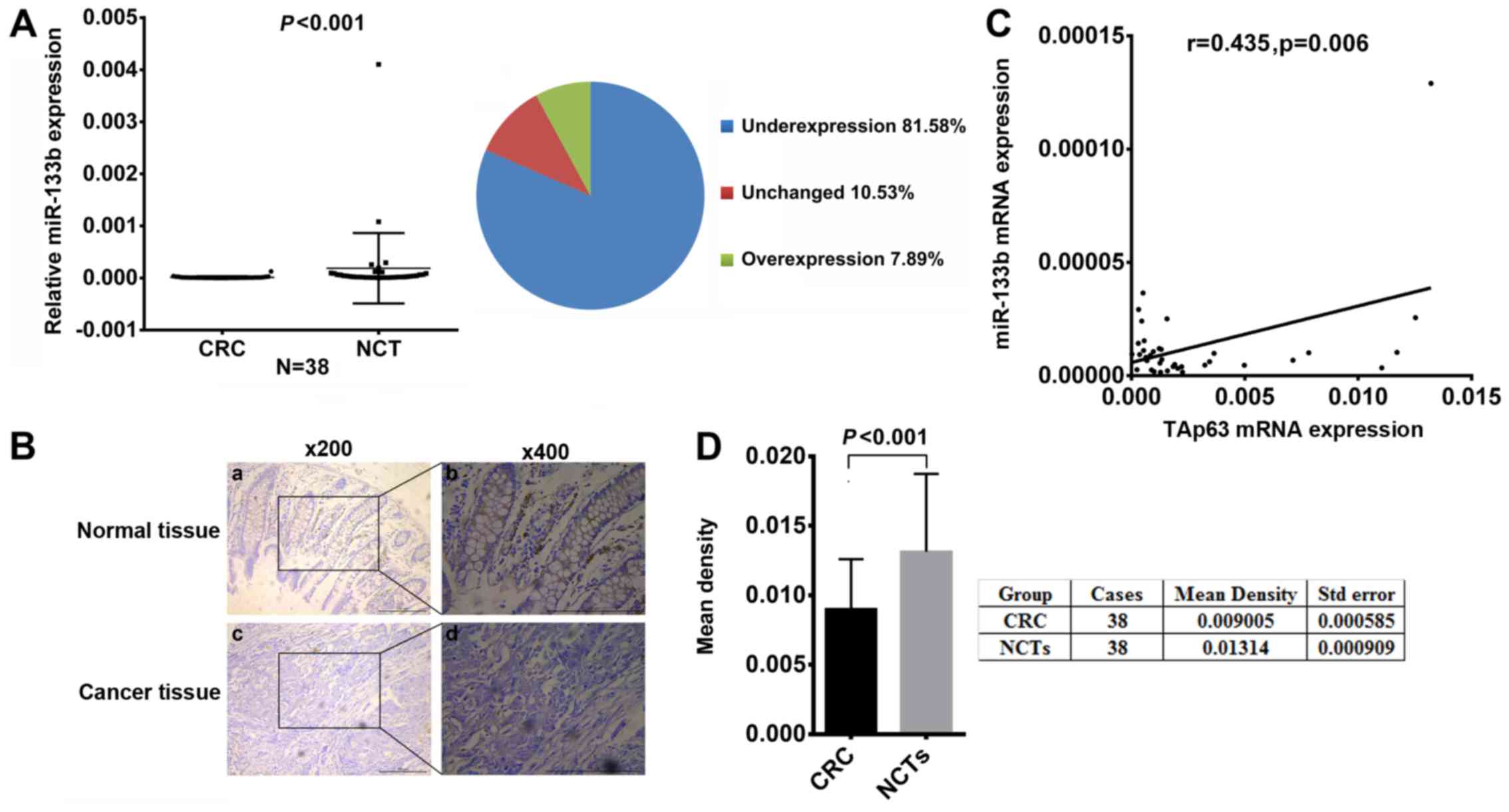
Downregulation of miR-133b is closely
correlated with TAp63 expression
To study the relationship between TAp63 and miR-133b
in human CRC, we assessed the expression levels of miR-133b in the
38 paired CRC and NCT samples using RT-qPCR and ISH. Compared with
the NCTs, the miR-133b expression level was significantly lower in
31 (81.58%) tumor tissues (Fig. 2A,
P<0.05). ISH revealed that miR-133b was downregulated in the CRC
tissues (Fig. 2B). Moreover, we
measured the density of the staining signal in all 38 cases using
Image-Pro Plus 6.0 image analysis software. The mean density of
cancerous tissues from all 38 cases was 0.009005±0.000585, while
that of the adjacent tissue was 0.01314±0.000909. This result
suggests a significantly lower level of miR-133b expression in the
CRC tissues compared to their NCTs (Fig. 2D, P<0.05). In addition, the
expression of miR-133b was found to be positively correlated with
TAp63 (Fig. 2C, P<0.05).
Association of TAp63 and miR-133b with
clinicopathological parameters
The association of TAp63 and miR-133b with various
clinicopathological parameters of the CRC cases are listed in
Table I. An inverse association was
found between TAp63 expression and lymph node and distant
metastases (P<0.05). miR-133b expression was also significantly
correlated with lymph node and distant metastases. No statistically
significant associations were found between the expression of TAp63
or miR-133b and other clinicopathological parameters
(P>0.05).
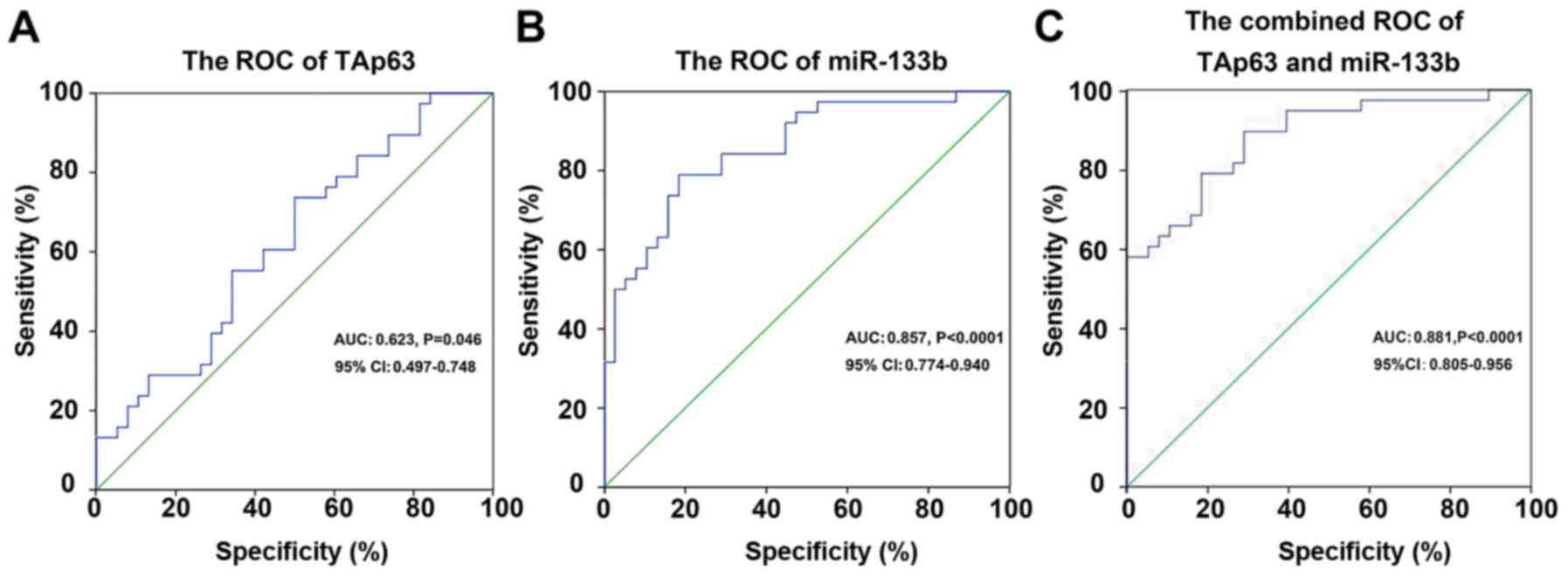
By receiver operating characteristic (ROC) curve
analysis, we proved that TAp63 and miR-133b are suitable predictors
for CRC. The AUC of TAp63 for CRC was 0.623 [95% confidence
interval (CI), 0.497–0.748; P=0.046], with 73.7% sensitivity and
50% specificity, respectively (Fig.
3A). The AUC of miR-133b for CRC was 0.857 (95% CI,
0.774–0.940, P<0.0001), with 78.9% sensitivity and 81.6%
specificity, respectively (Fig.
3B). The combined AUC of TAp63 and miR-133b for CRC was 0.881
(95% CI, 0.805–0.956, P<0.0001), with 89.5% sensitivity and
71.1% specificity, respectively (Fig.
3C).
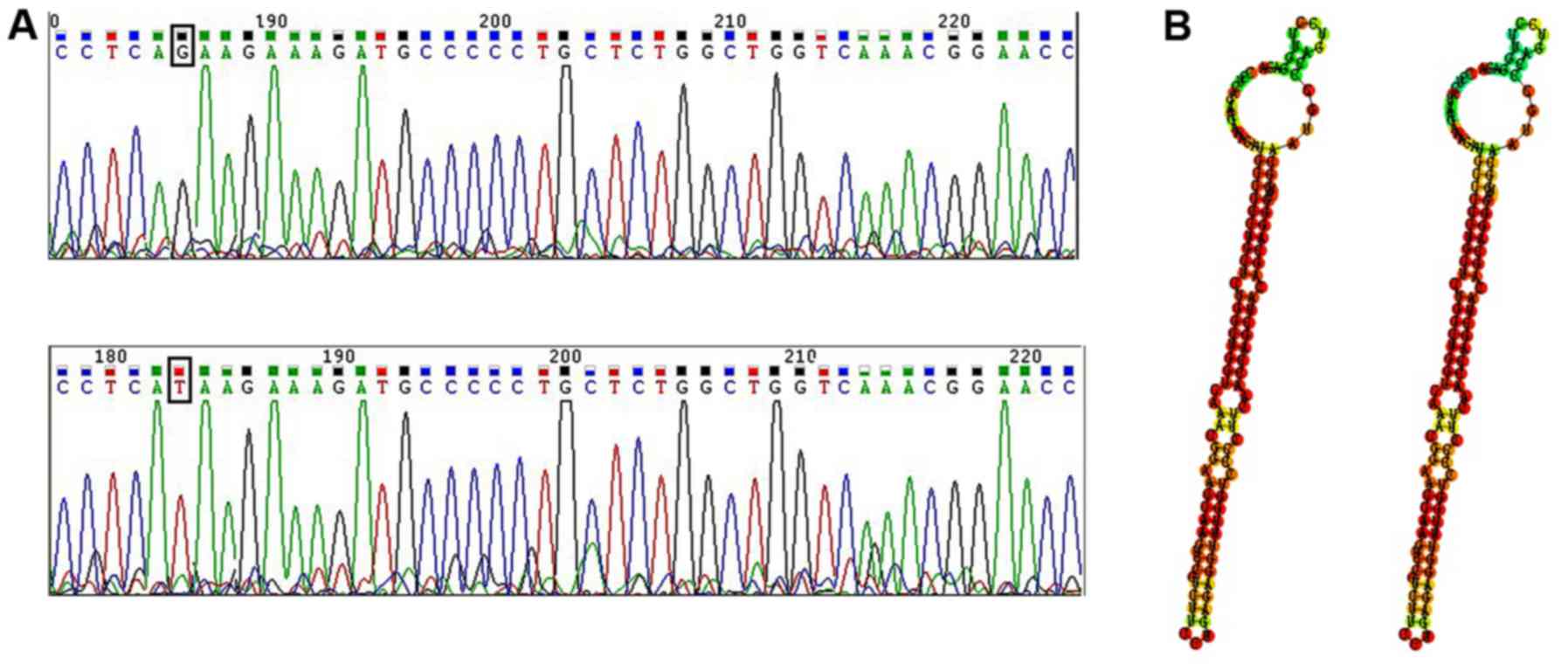
miR-133b point mutation detection by
DNA sequencing
To uncover whether there are point mutations in
miR-133b DNA, we scanned 446 bp of the pre-miR-133b coding region.
The results showed that only one patient had a single nucleotide
change, G to T, located 6 bp from the 3′ end of pre-miR-133b
(Fig. 4A). To further explore the
function of the mutation site, we compared the miR-133b predicted
secondary structure between the gene of the patient and the GenBank
gene. As shown in Fig. 4B, the
nucleotide change did not cause an apparent change in the stem-loop
structure.
Discussion
Several studies have documented a link between the
aberrant expression of TAp63 (22)
and the pathogenesis/prognosis in several types of cancer (23,24).
However, thus far, the expression and clinical significance of
TAp63 in colorectal cancer (CRC) have not been explored. In the
present study, we evaluated the expression of TAp63 in CRC by
RT-qPCR, western blotting and IHC, and analyzed its
clinicopathological and prognostic significance in CRC patients.
Our results showed that the TAp63 mRNA levels were significantly
decreased in 21 (55.27%) tumor tissues. In addition, an inverse
association was found between TAp63 expression and lymph node and
distant metastases. These results support an earlier hypothesis by
Spiesbach et al (25) that
TAp63 may be a cancer suppressor gene and suggest that TAp63 plays
an important role in the tumorigenesis or progression of CRC.
Moreover, compared with the NCTs, the miR-133b expression level was
significantly lower in the 31 (81.58%) tumor tissues and was
positively correlated with TAp63. In addition, low expression of
miR-133b was also significantly correlated with a higher metastasis
of CRC, implying that a decrease in TAp63 and miR-133b expression
may promote tumor metastasis.
Our group has previously shown that miR-133b
promoter hypermethylation is upregulated in CRC tissues, and
miR-133b promoter demethylation increased the expression of
miR-133b (26). Moreover, TAp63
leads to the transcriptional regulation of the expression of
miR-133b (16). However, whether
the downregulation of TAp63 is one of the causes for the low
expression of miR-133b in CRC has not been previously studied and
in particular the role of gene point mutations in miR-133b
expression has not been examined. In the present study we clearly
showed that the expression of miR-133b is positively correlated
with TAp63 and proposed the TAp63-alteration as a potential cause
of miR-133b downregulation.
Notably, we found low expression of miR-133b in 31
(81.58%) of the CRC tissues whereas low expression of TAp63 was
found in 21 (55.27%) of the CRC tissues. Obviously, reduced
expression of the transcription factor is not the only factor that
downregulated the expression of miR-133b in CRC. To the best of our
knowledge, different types of miRNAs have been identified as being
downregulated in breast (27),
gastric (28) and colon cancer
(29). In addition, some of them
were systematically proved to be closely associated with genetic
polymorphisms and mutations (30).
During the present study, we found point mutations within the seed
region of miR-133b in 1 patient. However, the predicting software
showed that the point mutation did not impact the secondary
structure of the pre-miR-133b. This result indicated that point
mutations may not affect the expression level of miR-133b. Thus, in
cancers, miR-133b downregulation tends to occur through miR-133b
promoter hypermethylation and reduced expression of the
transcription factor. Futhermore, the present study has some
limitations. First, the sample size we screened for the sequence
analysis was too small to find a missense mutation. Secondly, we
did not verify the effectiveness of point mutations in CRC
cells.
In conclusion, we demonstrated for the first time
that the expression of TAp63 and miR-133b in CRC tissues is
correlated with metastasis in clinical samples and speculated that
downregulation of TAp63 may be one of the causes for the low
expression of miR-133b in CRC. Our data suggest that TAp63 and
miR-133b may function as potentially valuable diagnostic/prognostic
biomarkers for CRC.
Acknowledgements
The present study was supported by the Science and
Technology Project of Hunan Province (no. 2015SK20206), and the
National Natural Science Foundation of China (no. 81172298).
References
|
1
|
Abu-Amero KK, Helwa I, Al-Muammar A,
Strickland S, Hauser MA, Allingham RR and Liu Y: Screening of the
seed region of MIR184 in keratoconus patients from Saudi Arabia.
Biomed Res Int. 2015:6045082015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Cristobal I, Madoz-Gurpide J,
Martin-Aparicio E, Carames C, Aguilera O, Rojo F and
Garcia-Foncillas J: Comment on ‘TAp63 suppress metastasis via
miR-133b in colon cancer cells’. Br J Cancer. 111:23692014.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kubisch J, Türei D, Földvári-Nagy L, Dunai
ZA, Zsákai L, Varga M, Vellai T, Csermely P and Korcsmáros T:
Complex regulation of autophagy in cancer - integrated approaches
to discover the networks that hold a double-edged sword. Semin
Cancer Biol. 23:252–261. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Shih IM and Kurman RJ: p63 expression is
useful in the distinction of epithelioid trophoblastic and
placental site trophoblastic tumors by profiling trophoblastic
subpopulations. Am J Surg Pathol. 28:1177–1183. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bassampour SA, Abdi R, Bahador R, Shakeri
M, Torkaman A, Yahaghi E and Taheriazam A: Downregulation of
miR-133b/miR-503 acts as efficient prognostic and diagnostic
factors in patients with osteosarcoma and these predictor
biomarkers are correlated with overall survival. Tumour Biol. Aug
16–2015.(Epub ahead of print). PubMed/NCBI
|
|
6
|
Guzel E, Karatas OF, Semercioz A, Ekici S,
Aykan S, Yentur S, Creighton CJ, Ittmann M and Ozen M:
Identification of microRNAs differentially expressed in prostatic
secretions of patients with prostate cancer. Int J Cancer.
136:875–879. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Duan FT, Qian F, Fang K, Lin KY, Wang WT
and Chen YQ: miR-133b, a muscle-specific microRNA, is a novel
prognostic marker that participates in the progression of human
colorectal cancer via regulation of CXCR4 expression. Mol Cancer.
12:1642013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhao Y, Huang J, Zhang L, Qu Y, Li J, Yu
B, Yan M, Yu Y, Liu B and Zhu Z: MiR-133b is frequently decreased
in gastric cancer and its overexpression reduces the metastatic
potential of gastric cancer cells. BMC Cancer. 14:342014.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Li X, Wan X, Chen H, Yang S, Liu Y, Mo W,
Meng D, Du W, Huang Y, Wu H, et al: Identification of miR-133b and
RB1CC1 as independent predictors for biochemical recurrence and
potential therapeutic targets for prostate cancer. Clin Cancer Res.
20:2312–2325. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ellermeier C, Vang S, Cleveland K, Durand
W, Resnick MB and Brodsky AS: Prognostic microRNA expression
signature from examination of colorectal primary and metastatic
tumors. Anticancer Res. 34:3957–3967. 2014.PubMed/NCBI
|
|
11
|
Akçakaya P, Ekelund S, Kolosenko I,
Caramuta S, Ozata DM, Xie H, Lindforss U, Olivecrona H and Lui WO:
miR-185 and miR-133b deregulation is associated with overall
survival and metastasis in colorectal cancer. Int J Oncol.
39:311–318. 2011.PubMed/NCBI
|
|
12
|
Das RK, Anura A, Pal M, Bag S, Majumdar S,
Barui A, Chakraborty C, Ray AK, Sengupta S, Paul RR, et al:
Epithelio-mesenchymal transitional attributes in oral sub-mucous
fibrosis. Exp Mol Pathol. 95:259–269. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang Y, Yan W and Chen X: P63 regulates
tubular formation via epithelial-to-mesenchymal transition.
Oncogene. 33:1548–1557. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Celardo I, Grespi F, Antonov A, Bernassola
F, Garabadgiu AV, Melino G and Amelio I: Caspase-1 is a novel
target of p63 in tumor suppression. Cell Death Dis. 4:e6452013.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Tucci P, Agostini M, Grespi F, Markert EK,
Terrinoni A, Vousden KH, Muller PA, Dötsch V, Kehrloesser S, Sayan
BS, et al: Loss of p63 and its microRNA-205 target results in
enhanced cell migration and metastasis in prostate cancer. Proc
Natl Acad Sci USA. 109:15312–15317. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lin CW, Li XR, Zhang Y, Hu G, Guo YH, Zhou
JY, Du J, Lv L, Gao K, Zhang Y, et al: TAp63 suppress metastasis
via miR-133b in colon cancer cells. Br J Cancer. 110:2310–2320.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Gao QW, Hua LD, Wang J, Fan CX, Deng WY,
Li B, Bian WJ, Shao CX, He N, Zhou P, et al: A point mutation in
SCN1A 5 genomic region decreases the promoter activity and is
associated with mild epilepsy and seizure aggravation induced by
antiepileptic drug. Mol Neurobiol. Mar 11–2016.(Epub ahead of
print). View Article : Google Scholar
|
|
18
|
Brooks MB, Catalfamo JL, MacNguyen R, Tim
D, Fancher S and McCardle JAA: A TMEM16F point mutation causes an
absence of canine platelet TMEM16F and ineffective activation and
death-induced phospholipid scrambling. J Thromb Haemost.
13:2240–2252. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Tan X, Wang H, Luo G, Ren S, Li W, Cui J,
Gill HS, Fu SW and Lu Y: Clinical significance of a point mutation
in DNA polymerase beta (POLB) gene in gastric cancer. Int J Biol
Sci. 11:144–155. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lee HC, Yang CW, Chen CY and Au LC: Single
point mutation of microRNA may cause butterfly effect on alteration
of global gene expression. Biochem Biophys Res Commun.
404:1065–1069. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Palatnik JF, Wollmann H, Schommer C,
Schwab R, Boisbouvier J, Rodriguez R, Warthmann N, Allen E,
Dezulian T, Huson D, et al: Sequence and expression differences
underlie functional specialization of Arabidopsis microRNAs miR159
and miR319. Dev Cell. 13:115–125. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yamaki T, Suenaga Y, Iuchi T, Alagu J,
Takatori A, Itami M, Araki A, Ohira M, Inoue M, Kageyama H, et al:
Temozolomide suppresses MYC via activation of TAp63 to inhibit
progression of human glioblastoma. Sci Rep. 3:11602013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Loljung L, Coates PJ, Nekulova M, Laurell
G, Wahlgren M, Wilms T, Widlöf M, Hansel A and Nylander K: High
expression of p63 is correlated to poor prognosis in squamous cell
carcinoma of the tongue. J Oral Pathol Med. 43:14–19. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Bahnassy AA, Zekri AR, Salem SE, Abou-Bakr
AA, Sakr MA, Abdel-Samiaa AG and Al-Bradei M: Differential
expression of p53 family proteins in colorectal adenomas and
carcinomas: Prognostic and predictive values. Histol Histopathol.
29:207–216. 2014.PubMed/NCBI
|
|
25
|
Spiesbach K, Tannapfel A, Mössner J and
Engeland K: TAp63gamma can substitute for p53 in inducing
expression of the maspin tumor suppressor. Int J Cancer.
114:555–562. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lv LV, Zhou J, Lin C, Hu G, Yi LU, Du J,
Gao K and Li X: DNA methylation is involved in the aberrant
expression of miR-133b in colorectal cancer cells. Oncol Lett.
10:907–912. 2015.PubMed/NCBI
|
|
27
|
Tahiri A, Leivonen SK, Lüders T, Steinfeld
I, Aure M Ragle, Geisler J, Mäkelä R, Nord S, Riis ML, Yakhini Z,
et al: Deregulation of cancer-related miRNAs is a common event in
both benign and malignant human breast tumors. Carcinogenesis.
35:76–85. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ren J, Huang HJ, Gong Y, Yue S, Tang LM
and Cheng SY: MicroRNA-206 suppresses gastric cancer cell growth
and metastasis. Cell Biosci. 4:262014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhang Z, Kim K, Li X, Moreno M, Sharp T,
Goodheart MJ, Safe S, Dupuy AJ and Amendt BA: MicroRNA-26b
represses colon cancer cell proliferation by inhibiting lymphoid
enhancer factor 1 expression. Mol Cancer Ther. 13:1942–1951. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Chen AX, Yu KD, Fan L, Li JY, Yang C,
Huang AJ and Shao ZM: Germline genetic variants disturbing the
Let-7/LIN28 double-negative feedback loop alter breast cancer
susceptibility. PLoS Genet. 7:e10022592011. View Article : Google Scholar : PubMed/NCBI
|