Introduction
Hepatocellular carcinoma (HCC) arises from
hepatocytes and ranks as the fifth most common cancer in men and
the seventh in woman (1). From
etiological factors, chronic HBV infection currently accounts for
more than one-half of HCC cases worldwide, especially in China. The
risk of developing HCC is increased by 10 to 100-fold for HBV
carries (2,3). Among HBV proteins, HBx is a
multifunctional regulator and play a crucial role in heptocellular
carcinogenesis, including in cell cycle regulation, signaling
pathway, and genomic DNA instability. HBx also plays a role in
epigenetic modification, such as DNA methylation and histone
modification (4,5). HBx upregulated DNA methyltransferases
(DNMTs) responsive for DNA methylation leading to inactivation of
tumor suppressor genes (TSGs) (6–8). The
current evidence demonstrates that HBx can affect regulatory
non-coding RNAs, including microRNAs and long ncRNAs (9,10). We
also reported that HBx upregulated miR-21 and promote the
proliferation of hepatoma cells (11).
Our study groups analyzed the different expression
genes between the L02-HBx and L02-Vector control cells using
microarray analysis, and the result showed HBx upregulated 456
genes and downregulated 843 genes. Among them, RIZ1, encoding a
member of PRDM protein family, was found downregulation by more
than 5-fold. RIZ1 (PRDM2) was first isolated by screening for
proteins. RIZ1 (PRDM2) was named retinoblastoma protein-interacting
Zinc finger gene in a screen for proteins binding the
retinoblastoma (Rb) protein (12).
Dysregulation of RIZ1, is associated with cancer, and act as a
tumor suppressor that can induce cell growth arrest and apoptosis,
and are inactivated in various human cancers (13–15).
Inactivation of the RIZ1 (PRDM2) gene by promoter hypermethylation
has been reported in liver and gastric carcinomas (15–19).
To investigate the association of RIZ with HCC, we measured the
expression level of RIZ1 by immunohistochemistry in protein level
and qRT-PCR in transcript level in HCC cases. There was no
significant correlation between RIZ1 protein level and patient age,
gender or tumor size. However, lower protein expression level of
RIZ1 was obvious in tumor tissues compared to peri-carcinoma
tissues. To elucidate the mechanism on downregulation of RIZ1 in
HCC, we reasoned that it should be regulated by some epigenetic
regulation. Earlier studies indicated that HBx attributed to the
downregulation of gene and non-coding RNA transcription through
increasing DNMT activity with the upregulation of DNMT1, or/and
DNMT3A, or/and DNMT3B (5).
Materials and methods
HCC patient samples
All of the HCC tumor tissues, together with paired
non-tumor liver tissues (5 cm away from tumor boarder) from 68 HCC
patients were obtained from the First Affiliated Hospital of
Nanjing Medical University. All the samples were collected with
written consent provided by patients. This study was performed with
ethical consent granted from the medical ethics committee of the
Medical School of Southeast University.
Cell culture
HCC-derived cell lines, SMMC-7721, MHCC-97H, chang
liver, Huh7, QGY-7703, HepG2.2.15), paracancerous cell line
QSG-7701, and immortalized human normal hepatocyte cell line L02
were obtained from TCC Cell Bank (Shanghai, China). All the cell
lines were cultured with RPMI-1640 (Gibco-BRL, Gaithersburg, MD,
USA) supplemented with 10% fetal bovine serum (Gibco-BRL) in 5%
CO2 humidified chamber at 37°C.
Immunohistochemical analysis
Immunohistochemistry (IHC) was used to detect RIZ1
protein expression in HCC 68 tumor and adjacent non-tumor tissues,
and the IHC were performed as previously described (20). Briefly, paraffin embedded tissue
specimens from all 68 patients were incubated with specific primary
antibodies against RIZ1 (Abcam, ab3790) overnight at 4°C after
antigen retrieval, followed by incubation with biotinylated
secondary antibodies anti-goat IgG. The sections were then treated
with Vectastain Elite ABC reagent (Vector Laboratories, Mountain
View, CA, USA). All sections were counterstained with haematoxylin.
Images were captured using Nikon Eclipse TE2000-S microscope system
(Nikon, Melville, NY, USA).
Reverse transcription polymerase chain
reaction (RT-PCR) and real-time quantitative RT-PCR (qRT-PCR)
Total RNA was isolated using TRIzol reagent
(Invitrogen, Carlsbad, CA, USA). cDNA was synthesized from 1 µg of
total RNA using Oligo(dT) as primers and reverse-transcribed
according to the kit's instructions (Takara, Dalian, China). The
qPCR was carried out using the SYBR Premix Ex Taq (Takara)
according to the manufacturer's protocol by StepOne Plus system
(Applied Biosystems, Foster City, CA, USA). The real-time PCR
reactions were performed in triplicate and β-actin was used as the
internal control. The relative expression was evaluated by the
comparative CT method (2−∆∆Ct method). The primer
sequences of each gene are shown in Table I.
 | Table I.Primers and annealing temperature of
genes and the sequence of siRNA. |
Table I.
Primers and annealing temperature of
genes and the sequence of siRNA.
| Gene | Primers (5′ -
3′) | Temperature
(°C) | Amplification size
(bp) |
|---|
| β-actin | F:
AAAGACCTGTACGCCAACAC | 60 | 220 |
|
| R:
GTCATACTCCTGCTTGCTGAT |
| RIZ1 | F:
GAACACTACTGAGCCTGTGG | 62 | 124 |
|
| R:
ACACCAATCCGGGTCTTGTC |
| HBx | F:
GACGTCCTTTGTCTACGTCC | 64 | 97 |
|
| R:
GAACGGCAGATGAAGAAGGG |
| DNMT1 | F:
CCGAGTTGGTGATGGTGTGTAC | 61 | 324 |
|
| R:
AGGTTGATGTCTGCGTGGTAGC |
| DNMT3A | F:
TATTGATGAGCGCACAAGAGAGC | 65 | 110 |
|
| R:
GGGTGTTCCAGGGTAACATTGAG |
| DNMT3B | F:
GACTTGGTGATTGGCGGAA | 64 | 270 |
|
|
R:GGCCCTGTGAGCAGCAGA |
| RIZ1-Promoter | F:
GCGTTGGTCACTGCTCCTGG | 60 | 249 |
|
| R:
CAAGTCCGACCGCACCCTA |
| miR-152 | Stem-loop |
|
|
GTCGTATCCAGTGCAGGGTCCGAGGTATT
CGCACTGGATACGACCCAAGT |
|
| F:
GTCGTCAGTGCATGACAGAACTT | 58 | 60 |
|
| R:
GTGCAGGGTCCGAGGT |
| U6 | F:
CGCTTCGGCAGCACATATACTA | 64 | 90 |
|
| R:
CGCTTCGGCAGCACATATACTA |
| RIZ1 | MF:
GTGGTGGTTATTGGGCGACGGC | 57 | 225 |
|
| MR:
GCTATTTCGCCGACCCCGACG |
| RIZ1 | UF:
TGGTGGTTATTGGGTGATGGT | 58 | 221 |
|
| UR:
ACTATTTCACCAACCCCAAGA |
| miR-152 mimics |
UCAGUGCAUGACAGAACUUGGAAGUUCUGUCAUGCACUGAUU |
| Negative
control |
UUCUCCGACGUGUCACGUTTACGUGACACGUUCGGAGAATT |
Western blot analysis
Cells (5×106) were harvested and lysed in
0.2 ml of ice-cold lysis buffer [10 mmol/l Tris-HCl (pH 8.0),
containing 150 mmol/l NaCl, 1 mmol/l phenylmethylsulfonyl fluoride,
and 1% Triton X-100]. An equal amount of cellular proteins were
separated by sodium dodecyl sulfate-polyacrylamide gel
electrophoresis (SDS-PAGE) on 10 or 12% resolving gels. Proteins
were electrophoretically transferred onto nitrocellulose membranes
and incubated overnight at 4 in block buffer [(5% non-fat milk in
Tris-Base Tween-20 (TBST)]. Membranes were incubated for 1 h at
room temperature with specific monoclonal antibodies for RIZ1
(1:1000, Sigma), for His tag (Fusion expression with HBx) (1:1000,
Cell Signaling Technology) for DNMT1 (1:1000, Santa) and β-actin
(1:8000, Sigma) antibodies followed by six 5-min washes with TBST.
After washing, the membrane was incubated with horseradish
peroxidase-conjugated secondary antibodies (1:80000, Sigma), and
the immune complexes were detected with enhanced chemiluminescence
(Pierce). β-actin protein levels were used as a control to ensure
equal protein loading. Each experiment was repeated at least 3
times. The intensity ratios of bands were determined and calculated
by Gel-Pro Analyzer 3.0.
Transfection
The miR-152 mimics and specific siRNAs targeting
DNMT1 and the control scrambled sequence siRNA were synthesized by
GenePharma (Shanghai, China), the sequences are listed in Table I. The cells were transfected with
mimics and siRNA using the Lipofectamine 2000 kit (Invitrogen).
After 48 or 72 h, cells were harvested for analysis. SMMC-7721
cells were transiently transfected with the pcDNA4/TO-HBx1,
pcDNA4/TO-HBx2 construct, or the control pcDNA4/TO using the FuGene
HD transfection reagent (Roche, Mannheim, Germany). The
transfection process followed the manufacturer's protocol. L02-HBx
no. 1 (L02-HBx #1) and L02-HBx no. 8 (L02-HBx #8) were the two
isolated HBx transfectants (gifts from Dr Xin-Yuan Guan, The
University of Hong Kong).
Methylation-specific PCR (MSP) and
treatment of 5′-Aza-dC
Genomic DNA was extracted from the cells using the
phenol-chloroform method followed by methylation-specific PCR were
performed as described previously (1). Modified DNA from L02-Vector, L02-HBx
#1, L02-HBx #8, 7721-Control and 7721-si-DNMT1 cells was amplified
to determine the methylation status of the promoter region of gene
by MSP. The primers used for amplifying methylated and unmethylated
RIZ1 are listed in Table I. Cell
lines were treated with 5′-Aza (Sigma) to determine whether
demethylation could restore RIZ1 expression. Cells were plated and
treated with 0, 5 and 25 µM of 5′-Aza for 3 days, changing the
medium containing 5′-Aza every day before extracting the genomic
DNA and total RNA.
Chromatin immunoprecipitation
(ChIP)
The ChIP experiments were performed using an EZ
ChIP™ Chromatin Immunoprecipitation kit for cell line samples
(Millipore, Billerica, MA, USA) according to the manufacturer's
instructions. Briefly, the crosslinked chromatin DNA were sonicated
into 200–1000 bp fragments and fixed with 1% formaldehyde. Then
immunoprecipitation followed using an anti-DNMT1 antibody and
normal mouse IgG as the negative control. The primers used for the
amplification of RIZ1 promoter DNA fragments are listed in Table I.
Statistical analysis
The independent Student's t-test was used to compare
the results expressed as mean ± SD and Chi-square (χ2)
test for categorical variables between any two pre-selected groups.
A P-value <0.05 was considered to indicate a statistically
significant difference.
Results
HBx represses RIZ1 expression in HCC
cells
We analyzed the differential expression profiles in
L02-HBx and control L02-Vector cells by cDNA microarray analysis in
a previous study (11). Of these
altered genes, RIZ1, a typical downregulated gene in L02-HBx
cells, was selected for further research because of its location on
1p36 region, which commonly undergoes loss of heterozygosity (LOH)
in HCC (15). However, it is
unclear whether RIZ1 expression level is related to HCC.
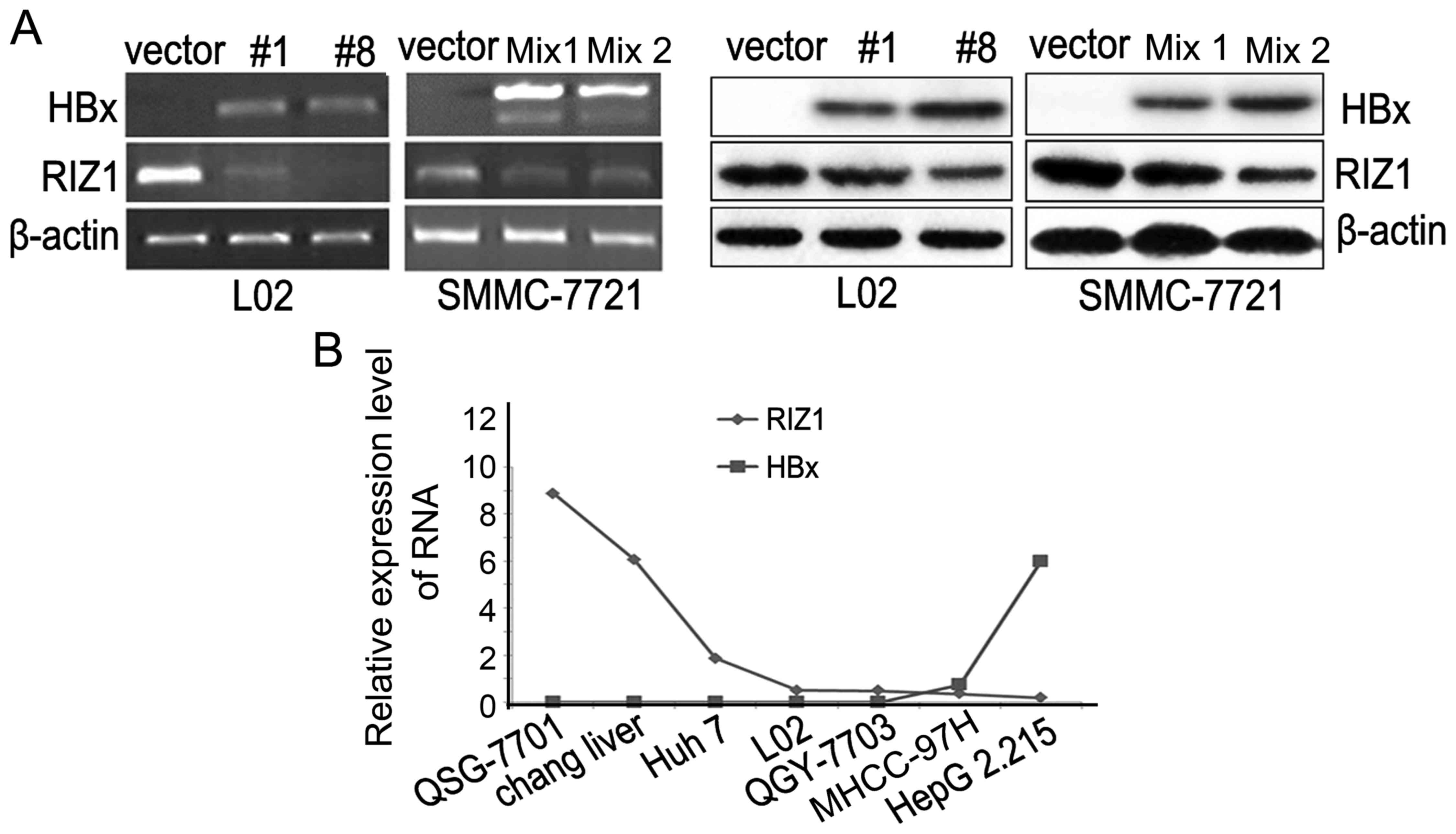
The L02 and SMMC-7721 cells were transfected with
the pcDNA4-HBx and pcDNA4 vectors. Results showed that RIZ1 was
downregulated in HBx transfected cells compared with the control
cells (vector) both at RNA level and protein level (Fig. 1A). To further verify whether this
downregulation of RIZ1 is correlated with HBx expression, we
examined the expression level of RIZ1 and HBx in HCC cell lines.
The expression of RIZ1 was relatively low in HBx positive cells
MHCC-97H and HepG2.215 compared to HBx negative ones (Fig. 1B). Hence, to some extent, this
negative correlation between the expression of RIZ1 and HBx in HCC
cell lines confirmed that the RIZ1 expression is related to
HBx.
Hypermethylation contributes to the
downregulation of RIZ1 induced by HBx
Hypermethylation of the RIZ1 promoter is one of the
most frequent epigenetic inactivation events, which lead to
silencing of RIZ1 expression in various tumors (21–23).
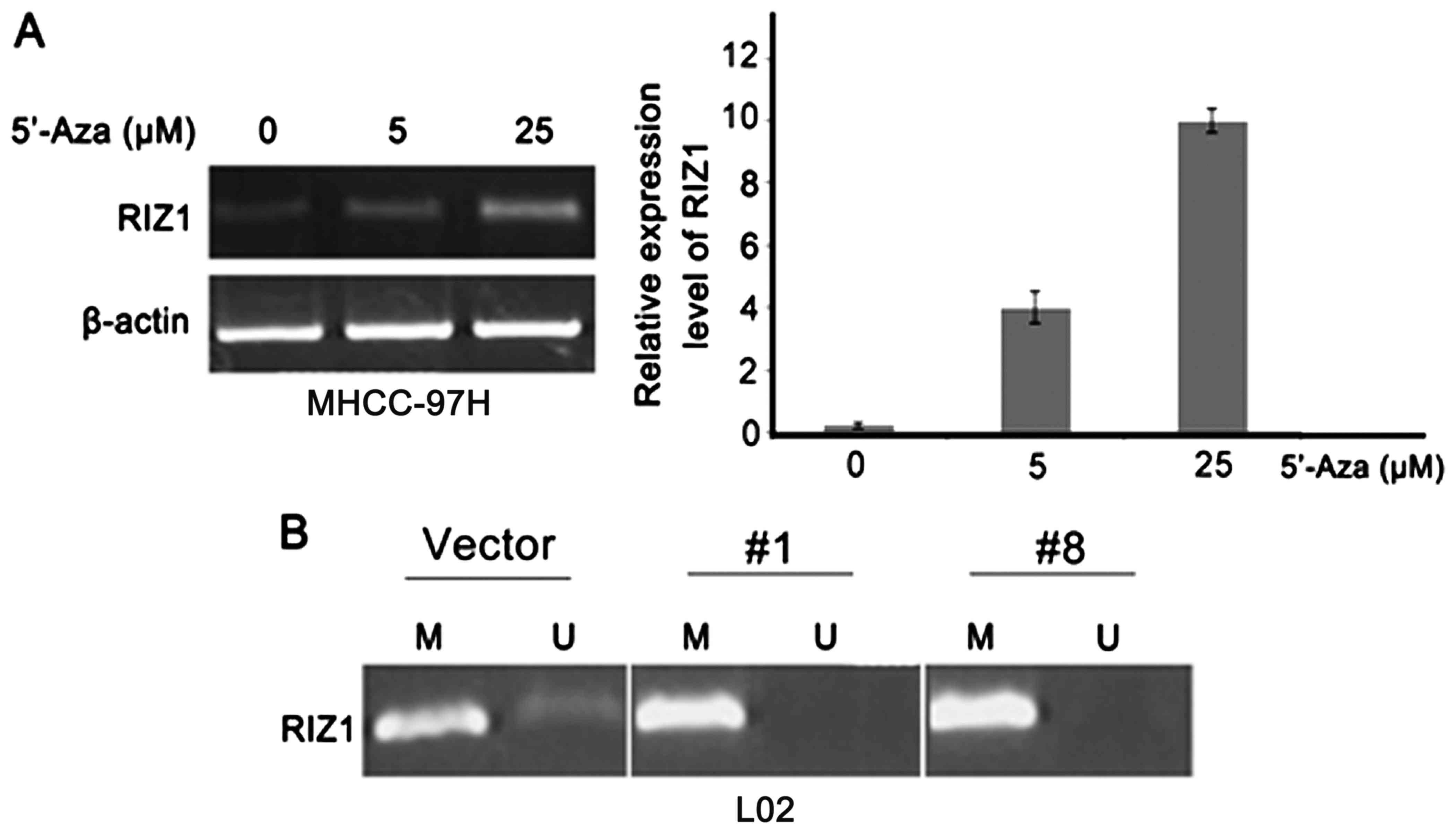
5′-Aza, an inhibitor of DNMTs, was used to treat the HCC cell
lines. After treatment of 5′-Aza, RIZ1 expression level was
restored depending on the dose of the drug (Fig. 2A). Next, we investigated whether HBx
induces the hypermethylation of RIZ1 promoter by
methylation-specific PCR (MSP). The methylation status of RIZ1
promoter region were examined between L02-Vec and L02-HBx 1# and
L02-HBx 8# cells (Fig. 2B). The
results showed that HBx was able to, at least partly, increase the
methylation level of RIZ1 promoter in HBx transfected L02
cells.
HBx represses RIZ1 expression through
increasing DNMT1 level
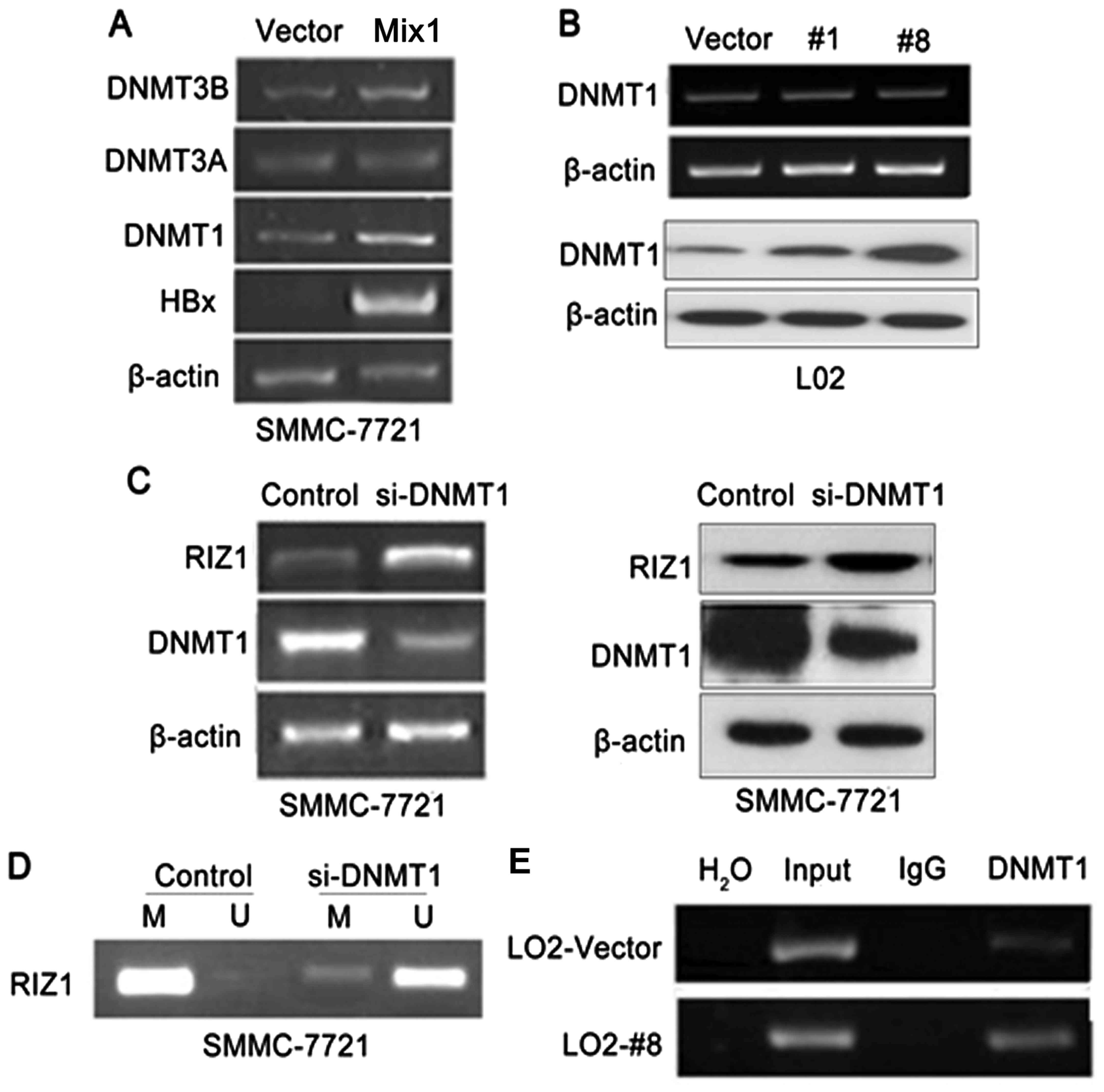
As DNA methylation is catalyzed by DNA
methyltransferase including DNMT1, DNMT3A and DNMT3B, we aimed to
investigate whether the HBx-mediated RIZ1 inactivation induced by
hypermethylation is related to DNMTs. In HBx transfected SMMC-7721
cells and L02 cells, DNMT1 were upregulated rather than DNMT3A and
DNMT3B. DNMT1 expression were significantly upregulated in
SMMC-7721 cells transfected with HBx in both RNA and protein levels
(Fig. 3A). DNMT1 expression was
upregulated in L02-HBx 1# and L02-HBx 8# cells in protein level,
however, in RNA level, DNMT1 was no significant upregulated in
these cells (Fig. 3B). In order to
explore whether DNMT1 is responsible for the expression of RIZ1, we
depleted the DNMT1 expression using siRNA in HCC cell lines and
observed that the RIZ1 is upregulated after knocking down of DNMT1
(Fig. 3C). Moreover, knockdown of
DNMT1 reduced partly the methylation level of RIZ1 promoter
(Fig. 3D). ChIP analysis using an
antibody against DNMT1 revealed that the binding of DNMT1 to RIZ1
promoter regions is enriched in L02-HBx cells (Fig. 3E), which suggested that DNMT1
mediates the repression of RIZ1 by HBx. Collectively, these data
suggested that DNMT1 modulated RIZ1 by directly binding to and
silencing RIZ1 gene via promoter hypermethylation in HBx expression
cells.
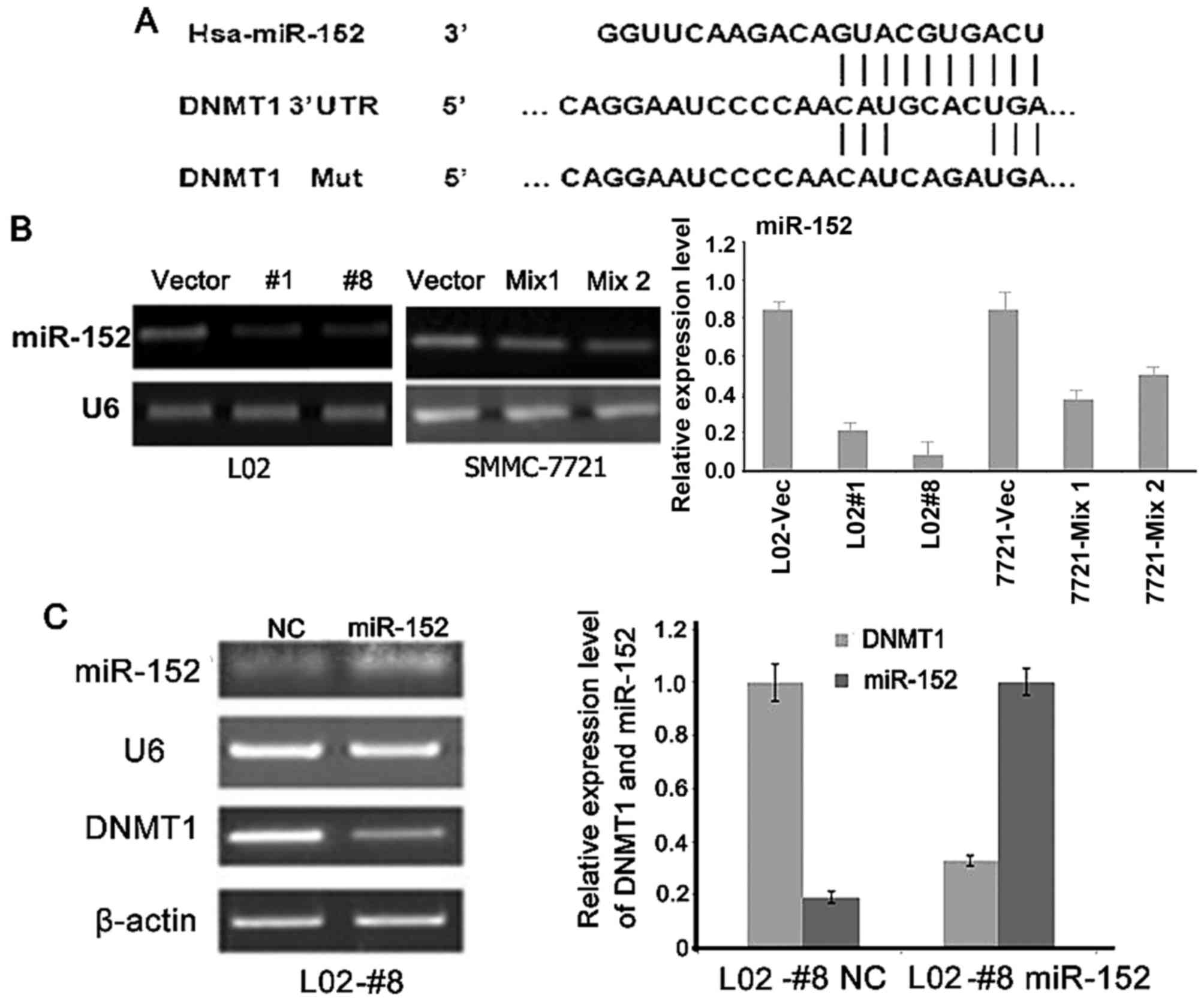
HBx increases DNMT1 level via
suppressing miR-152
Next, we investigated how HBx induced DNMT1
expression. It is well known that HBx can deregulate the expression
of cellular miRNAs (8,24,25).
Therefore, we examined whether HBx could deregulate a specific
candidate miRNA and regulate DNMT1 expression. Several studies
previously demonstrated that DNMT1 3′-UTR is partially
complementary to miR-152 (26–28)
and is a direct target gene of miR-152 in several cancer types
(Fig. 4A). In the present study, we
found that overexpression of HBx decreases miR-152 expression
(Fig. 4B), while overexpression of
miR-152 reduces DNMT1 expression (Fig.
4C).
Downregulation of RIZ1 in tumor
tissues correlates to HBV-related HCC
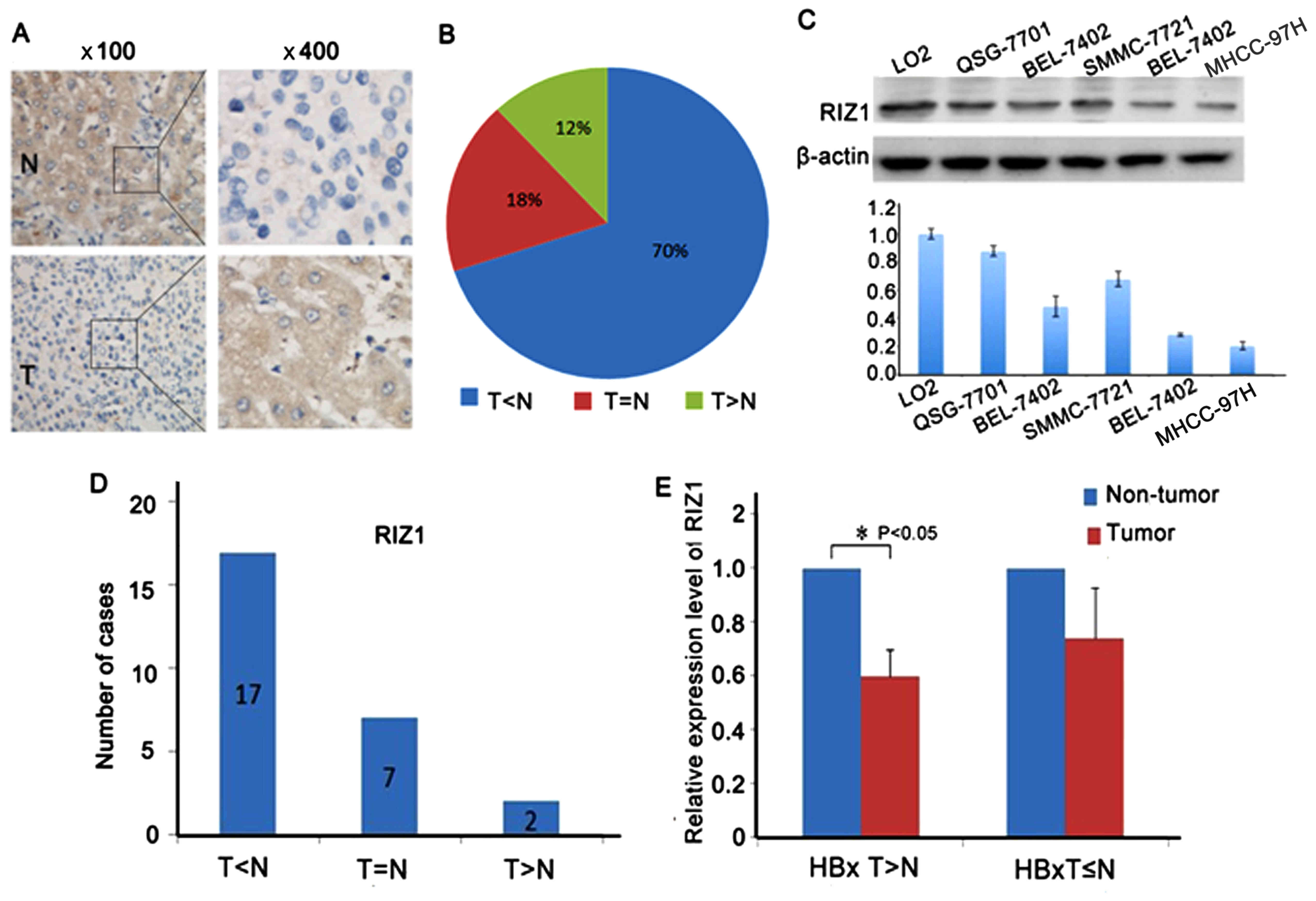
RIZ1 expression was analyzed by RT-PCR in 26 pairs
of HCC cases and immunohistochemistry (IHC) in 68 pairs of cases.
Seven representative results are shown in Fig. 5A. Compared to the paired
corresponding non-tumor tissues, 70% of HCC cases showed
downregulation of RIZ1 at the protein level (Fig. 5B). Downregulated expression of RIZ1
protein was detected by western blotting in HCC cell lines compared
with L02 cells which are immortal hepatic cells (Fig. 5C). HCC cases (65%) showed a
decreased expression of RIZ1 in RNA level (Fig. 5D). Then, HCC patients were divided
into sub-groups based on their relative HBx mRNA status.
Interestingly, in the clinical cases with high HBx group, the
expression of RIZ1 was significantly decreased in tumor tissues
compared to non-tumor tissues (Fig.
5E). Collectively, the relationship between HBx and RIZ1 mRNA
levels in HCC samples revealed a negative correlation between RIZ1
and HBx expression in HCC patients.
Discussion
HBV is a major risk factor worldwide for developing
HCC and contributes to HCC by multiple mechanisms. Among HBV
proteins, HBx is highly conserved among the different subtypes of
the virus and play a crucial role in hepatocellular carcinogenesis
as a multifunctional regulator. Recent studies have shown that HBx
was able to influence gene expression involve in various signaling
pathways, as well as epigenetic and genetic processes. RIZ (PRDM2),
a candidate HCC suppressor gene, is a member of the PR
(PRDI-BF1-RIZ homology)-domain-containing zinc finger gene family.
The PR-domain-containing RIZ1 product of the RIZ locus, in contrast
to the PR-domain-minus product RIZ2, is commonly lost or
underexpressed in HCC (15).
RIZ1 has tumor suppressor activity and is frequently
loss of its function in many human cancers. Some frameshift
mutation, LOH or promoter methylation is the main cause responsive
for inactivation RIZ1 in human gastric cancer (16,17).
DNA methylation is a common mechanism for the inactivating of the
RIZ1 tumor suppressor gene in human liver and breast cancers
(29). YY1 protein binds and
cooperates to positive regulation of the RIZ1 promoter in human
osteosarcoma cells and tissues (30). Reduced expression of RIZ1 may play
an important role in the pathogenesis and/or development of
cervical cancer, and is considered to be caused in part by aberrant
DNA methylation (31). Epigenetic
silencing of RIZ1 gene may play an important role in the early
stage human ESCC (32). The
expression of RIZ1 genes are associated with meningiomas
progression by inhibiting cellular proliferation and arrested the
cells in the G2/M phase of the cell cycle (13). Induced expression of RIZ1 in glioma
cells increased apoptosis and arrested cells in G2-M phase by
increased p53 and caspase-3 expression and decreased p-IKBα and
p-AKT protein levels, suggesting that RIZ1 may stimulate
p53-mediated apoptosis and inhibit p-IKBα and p-AKT signaling
pathways (33). In HCC, RIZ1 can
induce cell cycle arrest, apoptosis, or both and suppress HCC
tumorigenicity in nude mice. The promoter methylation and H3K9
modifications together to silence the RIZ1 gene in HCC (18). These studies have focused on the
methylation status of RIZ1 and its expression. However, the
underlying mechanisms involved in RIZ1 silencing, especially in HBX
related HCC, still remain unknown.
In the present study, we found RIZ1 mRNA levels were
lower in HBX transfected L02-HBX cells, than in the control. DNA
methylation is a common mechanism in inactivating the RIZ1 tumor
suppressor gene in human liver and breast cancers (29). The objective of the present study
was to investigate the mechanism of decreased expression of RIZ1
induced by HBX in HCC cells. HBx recombinant transfection decreased
RIZ1 expression, however, 5′-Aza restored RIZ1 expression in
MHCC-97H cell lines. As is known, DNA methylation is catalyzed by
DNMTs, it is necessary to investigate the expression level of
DNMT1, DNMT3A and DNMT3B in HBx-induced HCC cells. The result
showed that the expression of DNMT1 rather than DNMT3A and/or
DNMT3B is significant upregulated. Subsequently, DNMT1 siRNA
restored RIZ1 expression in SMMC-7721 cells with demethylation of
CpG sites in RIZ1 promoter region. ChIP results showed DNMT1
protein can binds to the RIZ1 promoter in HCC cells transfected
with HBX recombinant. This is the first report that DNMT1 is
involved in epigenetic inactivation of RIZ1 in tumor cells.
Accumulating evidence implied that HBx can
deregulate a set of miRNAs (8).
Large-scale miRNA expression profiling of human cancers have
revealed that miRNA deregulation is frequently associated with a
variety of tumors including human HCC (9,11,34,35).
These findings highlight the role of miRNA in HBV relative HCC
carcinogenesis. Several studies have previously demonstrated that
DNMT1 is a direct target gene of miR-152 (27,28).
Thus, we present a hypothesis where miR-152 may contribute to the
HBx-mediated upregulation of DNMT1. We first found that miR-152
levels were downregulated in HBx stably transfected L02 cells.
Furthermore, miR-152 minic inhibited DNMT1 in L02-HBx #8 cells.
Enhanced miR-152 was able to abolish the HBx-mediated upregulation
of DNMT1 in L02-HBx cells. This finding suggests that HBx
downregulates RIZ1 expression in hepatoma cells by enhancing partly
the expression of DNMT1, via decreased miR-152. Upregulation of
DNMT1 mediated by HBx might be one of mechanisms suppressing RIZ1
expression in HCC. The present study also showed that the
expression of RIZ1 was downregulated in HCC tissues and cell lines.
In HCC cases, the decreased expression rates of RIZ1 were 70%. A
significant negative relationship between the expression of HBx and
the expression of RIZ1 was observed in HCC patients and HCC cell
lines.
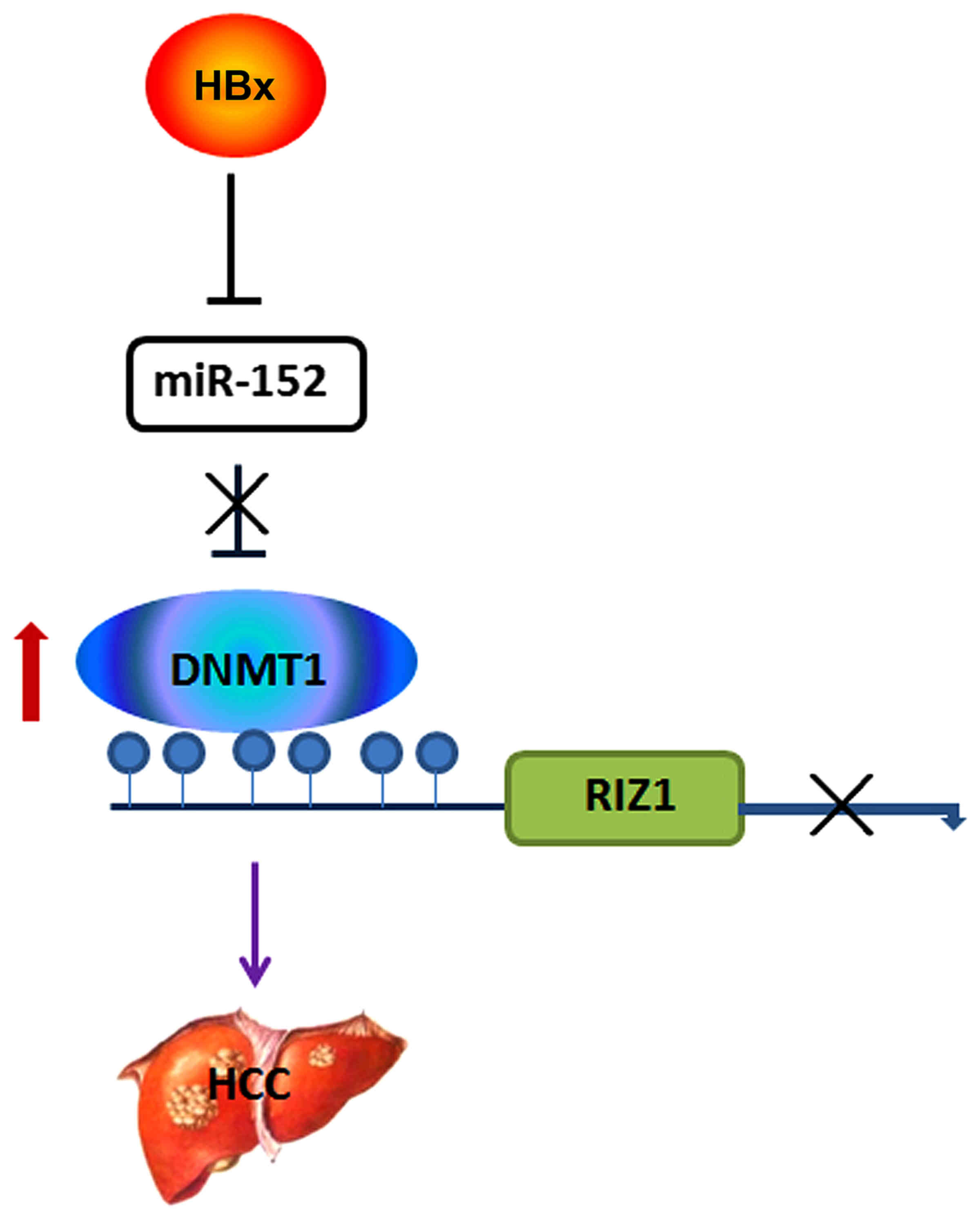
In summary (Fig. 6),
our results reveled that the transfection of HBX can reduce miR-152
and upregulate the expression of DNMT1. DNMT1 regulates the
expression of RIZ1, partly, by modulating the methylation status of
RIZ1 promoter. We are the first to report that HBx downregulates
RIZ1 expression, at least partly, associated with miR-152.
Acknowledgements
This work was supported by the National Natural
Science Foundation of China (no. 91229107, no. 81472548 and no.
81672414), Innovative Scientific Research Projects for College
Graduates of Jiangsu Province (no. 3231005701). We thank Professor
Xinyuan Guan, University of Hong Kong, for providing the HBx
construct.
Glossary
Abbreviations
Abbreviations:
|
HCC
|
hepatocellular carcinoma
|
|
HBV
|
hepatitis B virus
|
|
HBx
|
hepatitis B virus×protein
|
|
miRNA
|
microRNA
|
|
DNMTs
|
DNA methyltransferases
|
|
MSP
|
methylation-specific PCR
|
|
qPCR
|
quantitative real time PCR
|
|
siRNA
|
small interfering RNA
|
|
TSG
|
tumor suppressor gene
|
|
RIZ1
|
retinoblastoma protein-interacting
zinc finger gene 1
|
References
|
1
|
Llovet JM, Burroughs A and Bruix J:
Hepatocellular carcinoma. Lancet. 362:1907–1917. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Beasley RP, Hwang LY, Lin CC and Chien CS:
Hepatocellular carcinoma and hepatitis B virus. A prospective study
of 22 707 men in Taiwan. Lancet. 2:1129–1133. 1981. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wang Y, Cui F, Lv Y, Li C, Xu X, Deng C,
Wang D, Sun Y, Hu G, Lang Z, et al: HBsAg and HBx knocked into the
p21 locus causes hepatocellular carcinoma in mice. Hepatology.
39:318–324. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Soejima K, Fang W and Rollins BJ: DNA
methyltransferase 3b contributes to oncogenic transformation
induced by SV40T antigen and activated Ras. Oncogene. 22:4723–4733.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Park IY, Sohn BH, Yu E, Suh DJ, Chung YH,
Lee JH, Surzycki SJ and Lee YI: Aberrant epigenetic modifications
in hepatocarcinogenesis induced by hepatitis B virus X protein.
Gastroenterology. 132:1476–1494. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Tsai CN, Tsai CL, Tse KP, Chang HY and
Chang YS: The Epstein-Barr virus oncogene product, latent membrane
protein 1, induces the downregulation of E-cadherin gene expression
via activation of DNA methyltransferases. Proc Natl Acad Sci USA.
99:10084–10089. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Qiu X, Zhang L, Lu S, Song Y, Lao Y, Hu J
and Fan H: Upregulation of DNMT1 mediated by HBx suppresses RASSF1A
expression independent of DNA methylation. Oncol Rep. 31:202–208.
2014.PubMed/NCBI
|
|
8
|
Wang Y, Lu Y, Toh ST, Sung WK, Tan P, Chow
P, Chung AY, Jooi LL and Lee CG: Lethal-7 is down-regulated by the
hepatitis B virus×protein and targets signal transducer and
activator of transcription 3. J Hepatol. 53:57–66. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Murakami Y, Yasuda T, Saigo K, Urashima T,
Toyoda H, Okanoue T and Shimotohno K: Comprehensive analysis of
microRNA expression patterns in hepatocellular carcinoma and
non-tumorous tissues. Oncogene. 25:2537–2545. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lin L, Yin X, Hu X, Wang Q and Zheng L:
The impact of hepatitis B virus×protein and microRNAs in
hepatocellular carcinoma: A comprehensive analysis. Tumour Biol.
35:11695–11700. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Qiu X, Dong S, Qiao F, Lu S, Song Y, Lao
Y, Li Y, Zeng T, Hu J, Zhang L, et al: HBx-mediated miR-21
upregulation represses tumor-suppressor function of PDCD4 in
hepatocellular carcinoma. Oncogene. 32:3296–3305. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Buyse IM, Shao G and Huang S: The
retinoblastoma protein binds to RIZ, a zinc-finger protein that
shares an epitope with the adenovirus E1A protein. Proc Natl Acad
Sci USA. 92:4467–4471. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Liu ZY, Wang JY, Liu HH, Ma XM, Wang CL,
Zhang XP, Tao YQ, Lu YC, Liao JC and Hu GH: Retinoblastoma
protein-interacting zinc-finger gene 1 (RIZ1) dysregulation in
human malignant meningiomas. Oncogene. 32:1216–1222. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Suzuki M, Shigematsu H, Shivapurkar N,
Reddy J, Miyajima K, Takahashi T, Gazdar AF and Frenkel EP:
Methylation of apoptosis related genes in the pathogenesis and
prognosis of prostate cancer. Cancer Lett. 242:222–230. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Fang W, Piao Z, Simon D, Sheu JC and Huang
S: Mapping of a minimal deleted region in human hepatocellular
carcinoma to 1p36.13-p36.23 and mutational analysis of the RIZ
(PRDM2) gene localized to the region. Genes Chromosomes Cancer.
28:269–275. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Tokumaru Y, Nomoto S, Jerónimo C, Henrique
R, Harden S, Trink B and Sidransky D: Biallelic inactivation of the
RIZ1 gene in human gastric cancer. Oncogene. 22:6954–6958. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Oshimo Y, Oue N, Mitani Y, Nakayama H,
Kitadai Y, Yoshida K, Chayama K and Yasui W: Frequent epigenetic
inactivation of RIZ1 by promoter hypermethylation in human gastric
carcinoma. Int J Cancer. 110:212–218. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhang C, Li H, Wang Y, Liu W, Zhang Q,
Zhang T, Zhang X, Han B and Zhou G: Epigenetic inactivation of the
tumor suppressor gene RIZ1 in hepatocellular carcinoma involves
both DNA methylation and histone modifications. J Hepatol.
53:889–895. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Nishida N, Nagasaka T, Nishimura T, Ikai
I, Boland CR and Goel A: Aberrant methylation of multiple tumor
suppressor genes in aging liver, chronic hepatitis, and
hepatocellular carcinoma. Hepatology. 47:908–918. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Fan H, Liu D, Qiu X, Qiao F, Wu Q, Su X,
Zhang F, Song Y, Zhao Z and Xie W: A functional polymorphism in the
DNA methyltransferase-3A promoter modifies the susceptibility in
gastric cancer but not in esophageal carcinoma. BMC Med. 8:122010.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chappell G, Kutanzi K, Uehara T, Tryndyak
V, Hong HH, Hoenerhoff M, Beland FA, Rusyn I and Pogribny IP:
Genetic and epigenetic changes in fibrosis-associated
hepatocarcinogenesis in mice. Int J Cancer. 134:2778–2788. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Hutajulu SH, Indrasari SR, Indrawati LP,
Harijadi A, Duin S, Haryana SM, Steenbergen RD, Greijer AE and
Middeldorp JM: Epigenetic markers for early detection of
nasopharyngeal carcinoma in a high risk population. Mol Cancer.
10:482011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Mori N, Yoshinaga K, Tomita K, Ohwashi M,
Kondoh T, Shimura H, Wang YH, Shiseki M, Okada M and Motoji T:
Aberrant methylation of the RIZ1 gene in myelodysplastic syndrome
and acute myeloid leukemia. Leuk Res. 35:516–521. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhang X, Liu S, Hu T, Liu S, He Y and Sun
S: Up-regulated microRNA-143 transcribed by nuclear factor kappa B
enhances hepatocarcinoma metastasis by repressing fibronectin
expression. Hepatology. 50:490–499. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kong G, Zhang J, Zhang S, Shan C, Ye L and
Zhang X: Upregulated microRNA-29a by hepatitis B virus X protein
enhances hepatoma cell migration by targeting PTEN in cell culture
model. PLoS One. 6:e195182011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Xiang Y, Ma N, Wang D, Zhang Y, Zhou J, Wu
G, Zhao R, Huang H, Wang X, Qiao Y, et al: MiR-152 and miR-185
co-contribute to ovarian cancer cells cisplatin sensitivity by
targeting DNMT1 directly: A novel epigenetic therapy independent of
decitabine. Oncogene. 33:378–386. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ji W, Yang L, Yuan J, Yang L, Zhang M, Qi
D, Duan X, Xuan A, Zhang W, Lu J, et al: MicroRNA-152 targets DNA
methyltransferase 1 in NiS-transformed cells via a feedback
mechanism. Carcinogenesis. 34:446–453. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Huang J, Wang Y, Guo Y and Sun S:
Down-regulated microRNA-152 induces aberrant DNA methylation in
hepatitis B virus-related hepatocellular carcinoma by targeting DNA
methyltransferase 1. Hepatology. 52:60–70. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Du Y, Carling T, Fang W, Piao Z, Sheu JC
and Huang S: Hypermethylation in human cancers of the RIZ1 tumor
suppressor gene, a member of a histone/protein methyltransferase
superfamily. Cancer Res. 61:8094–8099. 2001.PubMed/NCBI
|
|
30
|
Abbondanza C, de Nigris F, De Rosa C,
Rossiello R, Puca GA and Napoli C: Silencing of YY1 downregulates
RIZ1 promoter in human osteosarcoma. Oncol Res. 17:33–41.
2008.PubMed/NCBI
|
|
31
|
Cheng HY, Gao Y and Lou G: DNA methylation
of the RIZ1 tumor suppressor gene plays an important role in the
tumorigenesis of cervical cancer. Eur J Med Res. 15:20–24. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Dong SW, Zhang P, Liu YM, Cui YT, Wang S,
Liang SJ, He Z, Sun P and Wang YG: Study on RIZ1 gene promoter
methylation status in human esophageal squamous cell carcinoma.
World J Gastroenterol. 18:576–582. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhang C, Zhu Q, He H, Jiang L, Qiang Q, Hu
L, Hu G, Jiang Y, Ding X and Lu Y: RIZ1: A potential tumor
suppressor in glioma. BMC Cancer. 15:9902015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Varnholt H, Drebber U, Schulze F,
Wedemeyer I, Schirmacher P, Dienes HP and Odenthal M: MicroRNA gene
expression profile of hepatitis C virus-associated hepatocellular
carcinoma. Hepatology. 47:1223–1232. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Ladeiro Y, Couchy G, Balabaud C,
Bioulac-Sage P, Pelletier L, Rebouissou S and Zucman-Rossi J:
MicroRNA profiling in hepatocellular tumors is associated with
clinical features and oncogene/tumor suppressor gene mutations.
Hepatology. 47:1955–1963. 2008. View Article : Google Scholar : PubMed/NCBI
|