Introduction
Esophageal cancer is a main cancer in China and the
sixth leading cause of cancer-related death worldwide, with
considerable geographical variations in incidence (1). Shantou is a high-risk area of
esophageal squamous cell carcinoma. The clinicopathological
features of ESCC are different from the esophageal adenocarcinoma
in Western countries (2). The
molecular mechanisms of esophageal cancer remain unclear. According
to encode project, ~80% of the human genome is transcribed to RNA,
of which ~90% genome transcribes into non-coding RNA. These
non-coding RNAs are increasingly recognized and defined as
endogenous regulatory RNA involved in maintenance of cellular
homeostasis during growth, development and pathogenesis disease
(3,4). Maternally expressed gene 3 (MEG3),
which is an imprinted gene, located on chromosome 14q32. MEG3 was
first identified as the ortholog of gene trap locus 2 (Gtl2) in
mice (5). It is an imprinted gene
expressed from the maternal allele with a length of ~1.6-kb
nucleotides. Although MEG3 is expressed in many human normal
tissues, the loss of MEG3 expression has been found in various
types of human tumors, including non-small cell lung cancer,
hepatoma, gastric, prostate and other cancers (6–9).
Furthermore, MEG3 can inhibit cancer cell proliferation or induce
apoptosis in vitro and in vivo by stimulating
p53-dependent transcription (10–12).
Endoplasmic reticulum (ER) is the site of protein
synthesis, protein folding, maintainance of calcium homeostasis,
synthesis of lipids and sterols (13). Various stimuli can disturb ER
homeostasis and result in the accumulation of unfolded or misfolded
proteins leading to pathological consequences, generally known as
‘ER stress’. Homeostatic regulation in the ER is under the control
of three evolutionary conserved pathways: IRE1a-XBP-1, PERK-eIF2a
and ATF6. These pathways, also referred to as the unfolded protein
response (UPR), are activated in response to ER stress (14). UPR promotes cell adapt stress.
However, when the response is not sufficient and ER stress
persists, UPR changes its function from acting to promote cellular
survival to commit the cell to apoptosis through upregulation of
pro-apoptotic factors, such as CHOP and caspase-4/12 (15).
Although the expression of MEG3 has been shown to be
downregulated in some tumor tissues, the biological role of MEG3
and its mechanism in ESCC remain largely unknown. Especially, very
little is known about the regulation relationship between MEG3 and
ER stress in ESCC. In the present study, we characterized the
expression of MEG3 in ESCC tissues and adjacent normal esophageal
tissues and explored its effects on EC109 cells.
Materials and methods
Clinical samples and tissue
processing
Twenty-eight paired esophageal cancer tissues and
adjacent normal tissues were obtained from patients who underwent
surgery at the Second Affiliated Hospital of Shantou University
Medical College between 2011 and 2015. Among them, 20 males and 8
females, aged from 42 to 75 years. No local or systemic treatment
was conducted in these patients before surgery. All of the
specimens were collected within 30 min after tumor resection and
stored in liquid nitrogen immediately until RNA extraction. Written
consents were obtained from all patients prior to surgery and the
study protocol was approved by the Ethics Committee of the Second
Affiliated Hospital of Shantou Medical College.
Cell culture and experimental
groups
EC109 cells were purchased from the Shanghai
Institute of Biochemistry and Cell Biology (Shanghai, China). Cells
were cultured in Dulbecco's modified Eagle's medium (DMEM)
containing 10% (v/v) fetal bovine serum (FBS; Invitrogen Life
Technologies, Carlsbad, CA, USA), penicillin (final concentration,
100 U/ml), and streptomycin (final concentration, 100 mg/ml), under
a humidified atmosphere with 5% CO2 at 37°C. The cell
culture medium was changed every one or two days, and cells were
passaged at 80–90% confluence. EC109 cells were transfected with
pcDNA3.1 empty plasmids or pcDNA3.1-MEG3 plasmids and divided into
two groups: pcDNA3.1 (control group, Ctrl) and pcDNA3.1-MEG3
(overexpressing MEG3 group, MEG3).
Plasmid identification and cell
transfection and bacterial transformation
The MEG3 gene was designed and synthesized by means
of PCR according to the GenBank of Human MEG3 (NR_002766) gene
sequence as previously described (16). The MEG3 gene fragment was identified
by restriction endonuclease analysis and inserted into a plasmid
(pcDNA3.1). Plasmids containing MEG3 sequence were then transfected
into competent cell DH5α. To obtain plasmids stably expressing
MEG3, pcDNA3.1-MEG3 plasmids were extracted from E. coli
(DH5α).
For ectopic MEG3 expression, pcDNA3.1-MEG3 plasmids
were transiently transfected into EC109 cells using Lipofectamine
2000 transfection reagent (Invitrogen Life Technologies, Shanghai,
China) according to the manufacturer's instructions. pcDNA3.1 empty
plasmids were transfected to cells in parallel as a control. RT-PCR
was performed to examine mRNA expression level of MEG3.
RNA extraction and qRT-PCR assay
Total RNA was extracted from tissues or cultured
cells with TRIZol reagent (Invitrogen Life Technologies) according
to the manufacturers protocol. The isolated RNA was reverse
transcribed into cDNA using a reverse transcription kit (Takara
Biotechnology, Dalian, China). The expression was quantified by
qRT-PCR using a standard protocol from the SYBR-Green PCR kit
(Toyobo, Co., Ltd., Osaka, Japan) on the StepOnePlus qRT-PCR System
(Applied Biosystems, Foster City, CA, USA). The following gene
specific primers: forward, 5′-CTGCCCATCTACACCTCACG-3 and reverse,
5-CTCTCCGCCGTCTGCGCTAGGGGCT-3 for MEG3; forward, 5′
GTCAACGGATTTGGTCTGTATT3′ and reverse, 5′-AGTCTTCTGGGTGGCAGTGAT-3′
for GAPDH. Forward, 5′-AAACGGCTACCACATCCAAG-3′ and reverse
5′-CAATTACAGGGCCTCGAAAG-3′ for 18S. The PCR reaction was conducted
at 95°C for 30 sec followed by 40 cycles of 95°C for 5 sec and 60°C
for 30 sec. Each sample was analyzed in triplicate and the relative
expression was calculated using the 2−ΔΔCt method
relative to GAPDH or 18S.
Cell viability assay by CCK-8
The viability of cells transfected with
pcDNA3.1-MEG3 or pcDNA3.1 was determined by using Cell Counting
kit-8 (CCK-8). EC109 cells were inoculated into 96-well plates at
5×103 cells/well for 24 h. Cells were transfected with
pcDNA3.1-MEG3 or pcDNA3.1 for 48 h. The culture medium was then
discarded and the cells were washed with phosphate-buffered saline
(PBS), and then bred with 0.5 mg/ml of
2-(2-methoxy-4-nitrophenyl)-3-(4-nitrophenyl)-5-(2,4-disulfoanilino)-2H-tetrazolium
monosodium salt (WST-8) at 37°C for 4 h. After incubation, the
concentration of product was determined by measuring the absorbance
at 450 nm using an enzyme-linked immunosorbent assay reader
(FilterMax F5; Molecular Devices, Sunnyvale, CA, USA).
Fluorescence microscope and flow
cytometric analysis of apoptosis
Apoptotic nuclei staining. DAPI is a fluorescent dye
which specifically conjugates to ds-DNA and is thus used to
visualize nuclear morphological features; the nuclei of apoptotic
cells demonstrate condensation and fragmentation.EC109 cells
transfected with pcDNA3.1-MEG3 or pcDNA3.1 were harvested at 48 h.
Cells were fixed in 4% paraformaldehyde for 30 min at room
temperature and then permeabilized with 0.2% Triton X-100 in PBS
for 5 min at room temperature. DAPI (500 ng/ml) staining was
performed at room temperature for 10 min. The morphology of the
nuclei was viewed and captured with a fluorescence microscope
(Olympus BX51; Olympus, Tokyo, Japan).
EC109 cells transfected with pcDNA3.1-MEG3 or
pcDNA3.1 were harvested at 48 h. Cells were washed twice with
ice-cold PBS and resuspended in 1X binding buffer to a
concentration of 1×106/ml. The cells were then bled with
5 µl Annexin V/FITC and 10 µl propidium iodide (PI) (20 µg/ml) for
15 min at room temperature keeping out of light. Analyses were
performed with a BD Accuri™ C6 flow cytometer (BD Biosciences, San
Diego, CA, USA) with the FL1 and FL3 detector.
Western blot analysis
EC109 cells transfected with pcDNA3.1-MEG3 or
pcDNA3.1 were harvested at 48 h and washed with ice-cold PBS three
times. The cellular protein lysates were prepared with RIPA buffer
containing PMSF. Protein fractions were prepared using a nuclear
and cytoplasmic protein extraction kit (Beyotime Institute of
Biotechnology, Haimen, China). Protein concentration was measured
by using a BCA protein assay kit (Thermo Fisher Scientific,
Waltham, MA, USA) according to the manufacturer's instructions. For
western blot assay, protein (50 µg) was subjected to
electrophoresis on SDS-PAGE and then transfered onto nitrocellulose
membranes. The membranes were blocked with 5% non-fat dry milk in
Tris-buffered saline (pH 7.6, TBS) containing 0.5% Tween-20 at room
temperature for 60 min. Then membranes were incubated with primary
antibodies against: β-actin (1:5,000), GRP78 (1:500), ATF6 (1:500),
IRE1 (1:500), PERK (1:500), CHOP (1:1,000), caspase-9 (1:1,000) and
cleaved caspase-3 (1:1,000) in TBST at 4°C overnight. The next day,
membranes then were washed with TBST and incubated with fluorescent
secondary antibodies (LI-COR Biosciences, Lincoln, NE, USA) coupled
to the first antibody at room temperature out of light for 1 h,
followed by TBST washing three times. Drying with neutral absorbent
paper and scanned by Odyssey Detection System (LI-COR Biosciences).
Expression of proteins was analyzed using the Quantity One software
(Bio-Rad Laboratories, Hercules, CA, USA) and was normalized to
that of β-actin (for total cell fraction).
Statistical analysis
All experiments were independently performed at
least three times. The values are expressed as mean ± SD. The
statistical significance of the data was calculated using Student's
t-test, Chi-square test or one-way analysis of variance (ANOVA)
with a post-hoc test of multiple comparisons. A P<0.05 was
considered statistically significantly.
Results
MEG3 expression is downregulated in
human esophageal cancer tissues
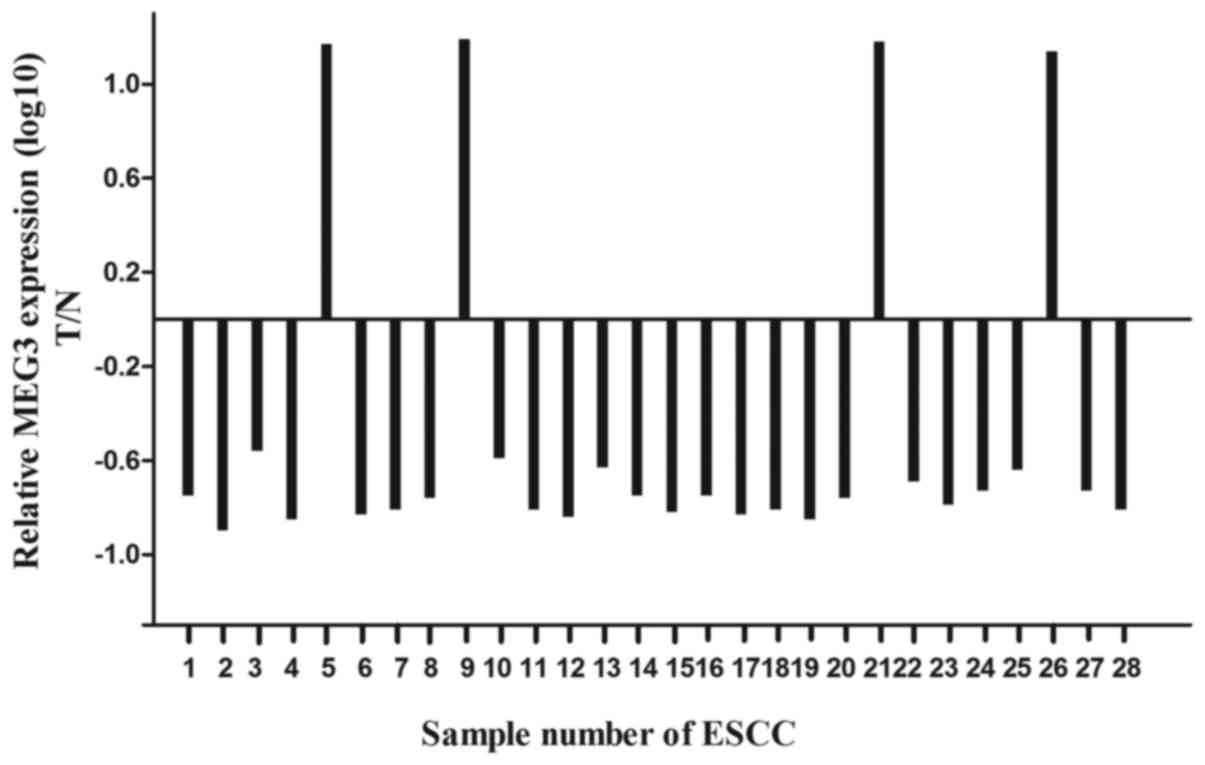
To find whether lncRNA MEG3 was differentially
expressed in the ESCC tissues, a total of 28 paired esophageal
cancer tissues and adjacent normal tissues were analyzed for MEG3
expression using qRT-PCR and normalized to GAPDH. Our results
showed that the expression level of MEG3 was significantly lower in
ESCC tissues compared with the normal tissues (Fig. 1; P<0.05), and the expression
level of MEG3 was negatively correlated with prognosis (data not
shown).
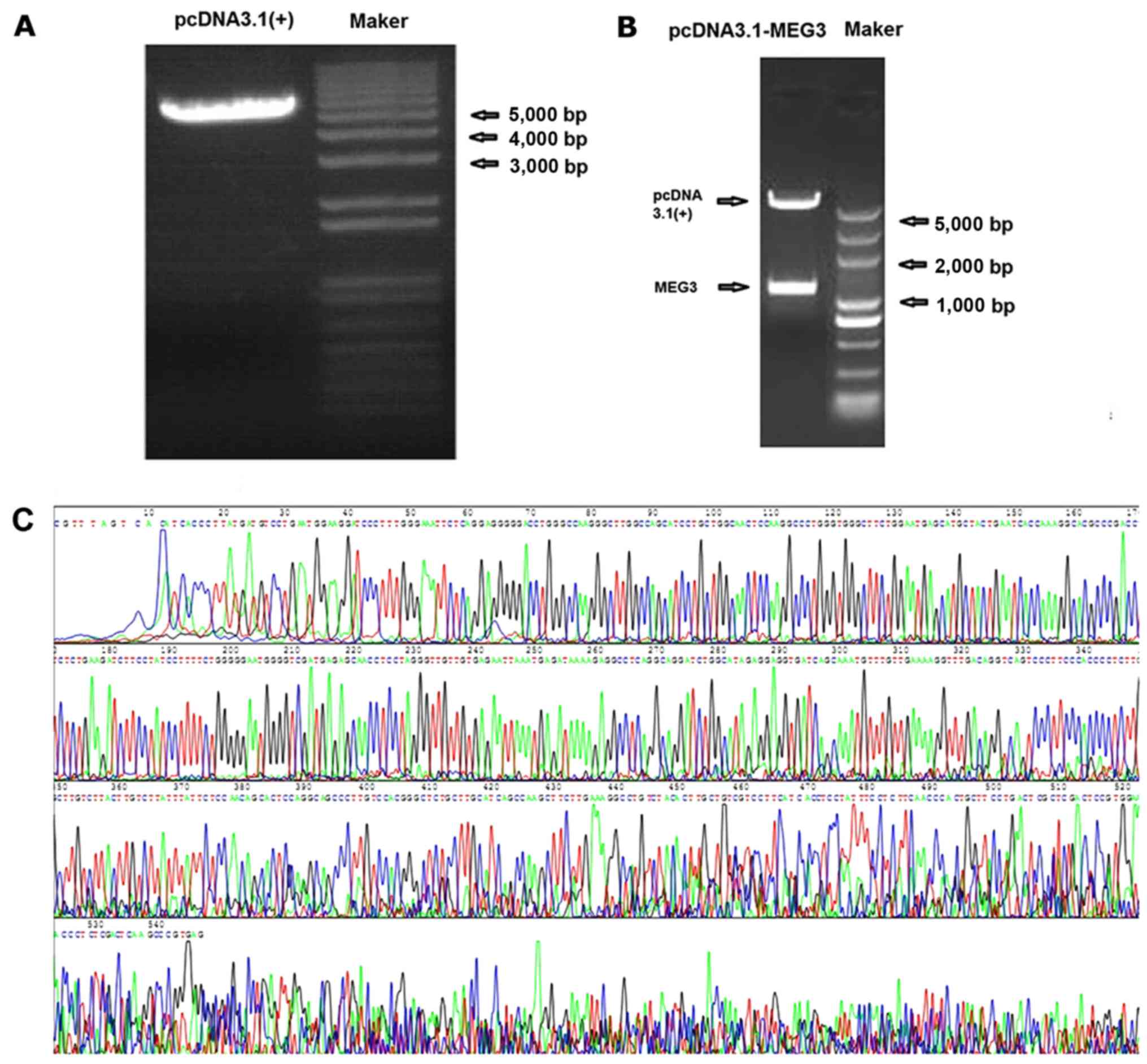
pcDNA3.1-MEG3 plasmid construction and
identification
The MEG3 sequence was synthesized and subcloned into
pcDNA3.1 (Invitrogen Life Technologies) to generate pcDNA3.1-MEG3.
LncRNA MEG3 gene was confirmed by XhoI and BamHI
restriction enzyme digestion, then inserted into the pcDNA3.1
vector. The particular sequence of MEG3 inserted into vectors was
the same as designed. The fragment did not appear in the control
vector, showing that pcDNA3.1-MEG3 plasmid was successfully
constructed (Fig. 2).
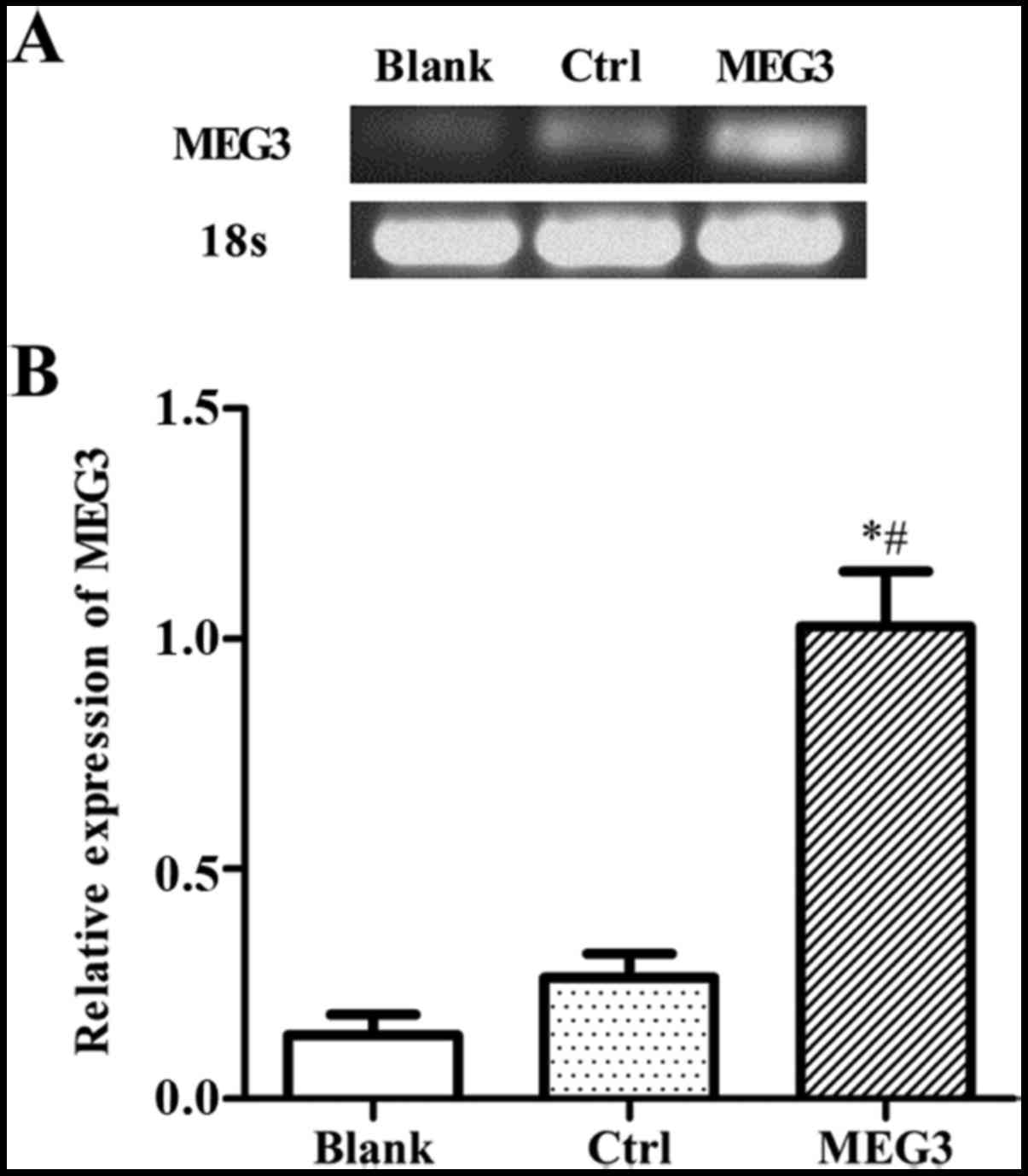
MEG3 expression increases after
pcDNA3.1-MEG3 transfection in EC109 cells
To explore the effect of MEG3 in EC in vitro,
we first measured its expression in EC109 cell lines. EC109 cells
were transiently transfected with either pcDNA3.1 empty plasmids
(control) or pcDNA3.1-MEG3 (overexpressing MEG3). The expression
levels of MEG3 mRNA were measured by RT-PCR. As shown in Fig. 3A, the expression level of MEG3 is
low in normal EC109 cells. Compared to the blank group (cells were
not treated by any plasmids) or control group (pcDNA3.1), the mRNA
expression level of MEG3 was significantly increased after
transfection with pcDNA3.1-MEG3. It demonstrated that pcDNA3.1-MEG3
was successfully transfected and high level mRNA expression of MEG3
was obtained in EC109 cells (Fig.
3).
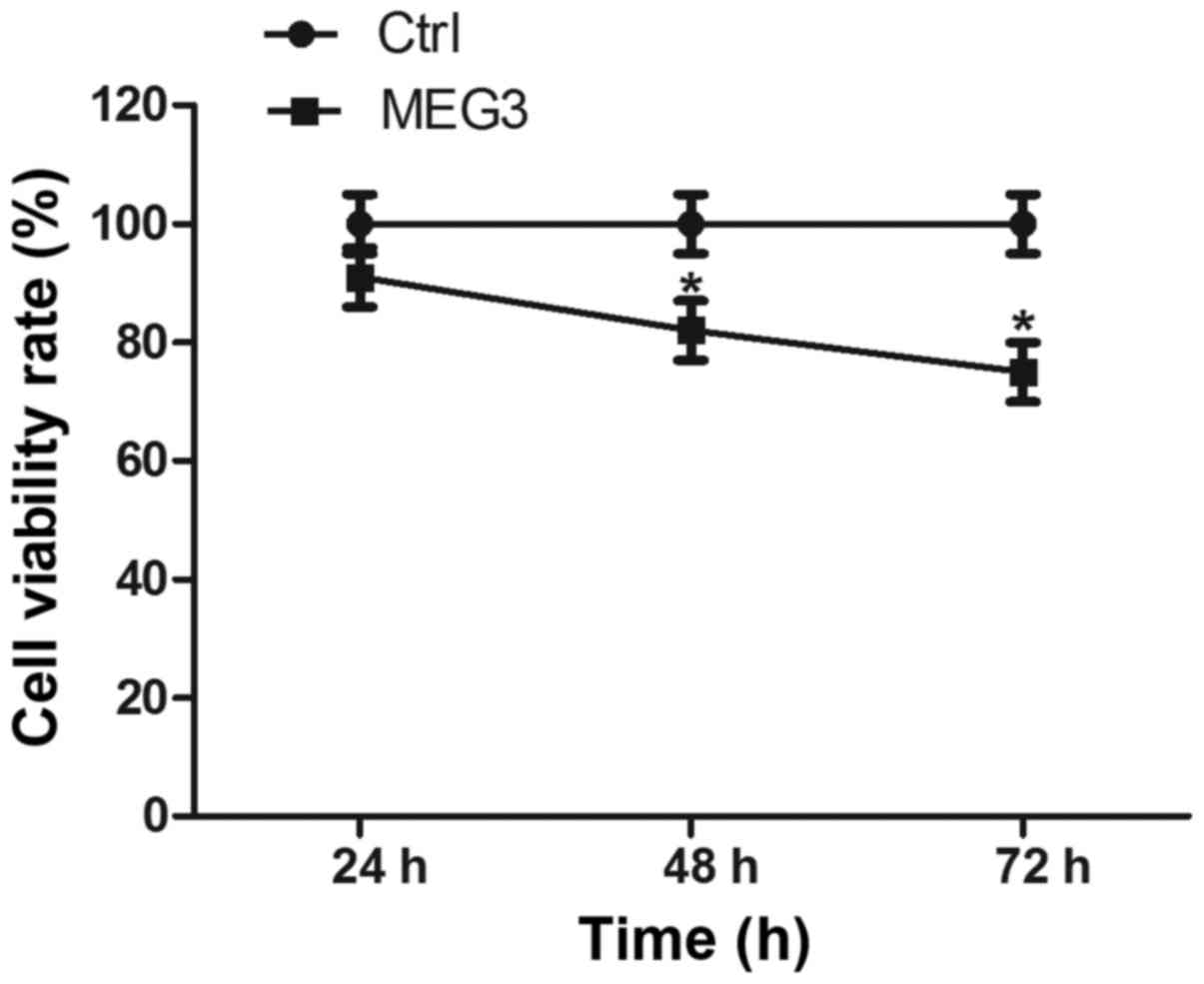
Ectopic expression of MEG3 inhibits
cell proliferation
Then, we attempted to determine whether MEG3 affects
EC109 cell proliferation in vitro. Cell proliferation was
measured by the CCK-8 assay. Ectopic expression of MEG3
significantly decreased cell vitality rate by 16.9% and 22.6% after
48 and 72 h, respectively, compared with the control group
(Fig. 4; both P<0.05). These
results suggest that overexpressing MEG3 can inhibit EC109 cell
growth.
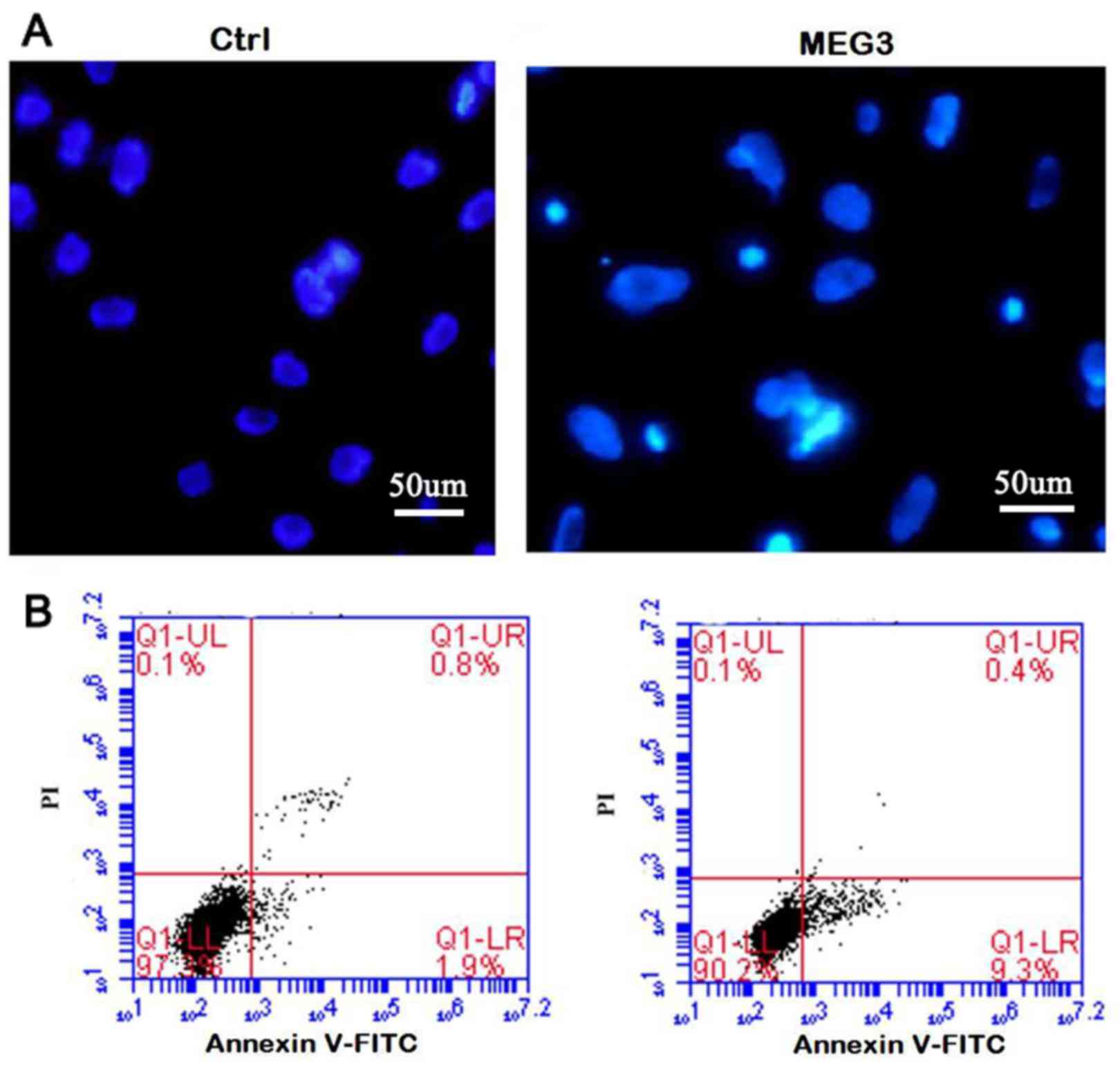
Ectopic expression of MEG3 induces
cell apoptosis
In order to clarify whether the inhibition of cell
proliferation is related to cell death or apoptosis, DAPI staining
and the flow cytometric (Annexin V/PI) assay were performed. The
morphologic hallmarks of apoptosis include chromatin margination,
nuclear condensation and fragmentation. As shown in Fig. 5A-MEG3, in overexpressing MEG3 cells,
some cell nuclei became condensed and shrunken and typical
apoptotic bodies appeared. However, normal cell nuclei were uniform
and without condensation or fragmentation in the control group
(Fig. 5A-Ctrl). Flow cytometric
assay further confirmed the DAPI staining results. The apoptosis
rate in MEG3 overexpression group is significantly higher than that
of the control (Fig. 5B, 9.56±2.6
vs. 1.41±0.37%; P<0.05), showing that ectopic expression of MEG3
induces cell apoptosis.
Ectopic expression of MEG3 activates
the ER stress pathway
To explore the mechanism by which MEG3 induced
growth arrest and apoptosis, western blot assay was carried out to
examine the expression of UPR and apoptosis-associated proteins
CHOP, caspase-9 and cleaved caspase-3. Ectopic expression of MEG3
significantly increased the expression of molecular chaperone GRP78
and three transmembrane proteins of UPR (IRE1, PERK and ATF6), as
well as CHOP, caspase-9 and cleaved caspase-3 (Fig. 6), showing that two key apoptotic
pathways of ER stress (CHOP and caspase-9 pathway) were
activated.
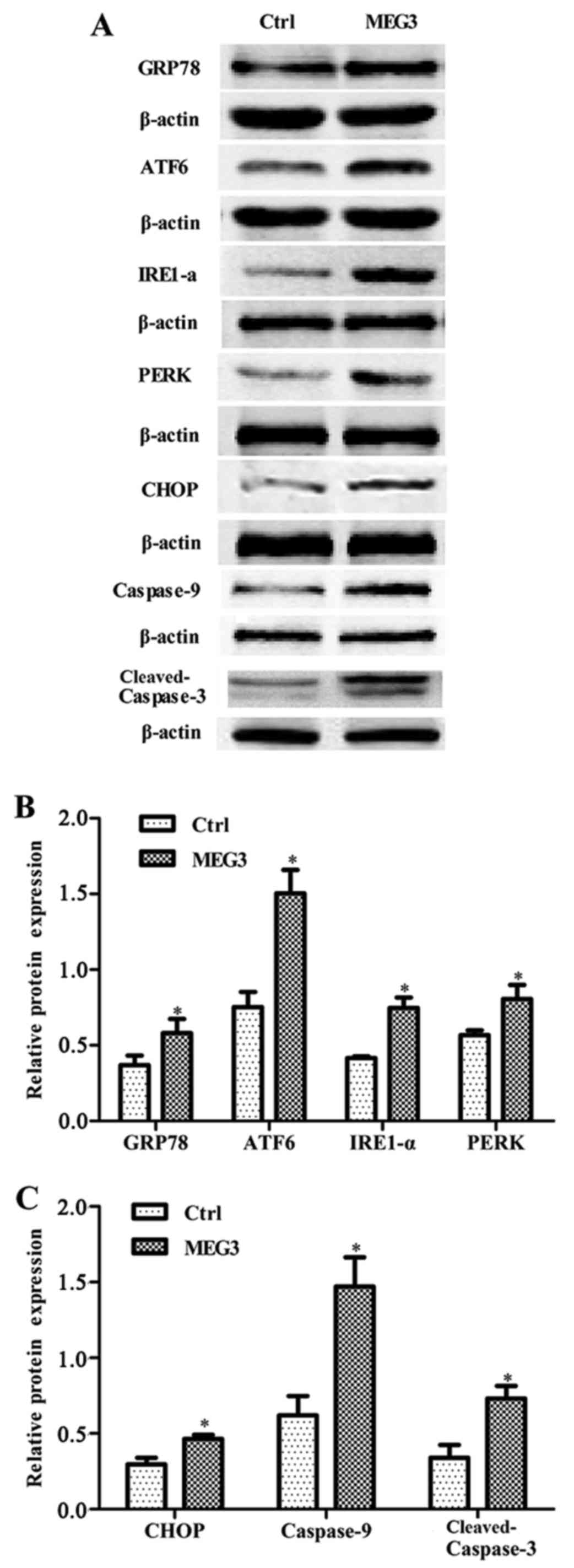 | Figure 6.Effects of ectopic MEG3 on expression
of ER stress-related proteins (GRP78, ATF6, IRE1-α, PERK, CHOP,
caspase-9 and cleaved-caspase-3). (A) Representative images of
western blot analysis. The total cellular proteins derived from
pcDNA3.1-MEG3-transfected or pcDNA3.1-transfected EC109 cells were
immunoblotted with a panel of antibody specific for GRP78, ATF6,
IRE1-α, PERK, CHOP, caspase-9, cleaved caspase-3, CHOP and β-actin.
(B and C) The expression of each index was normalized to expression
level of β-actin, and the relative change was expressed as a ratio
or fold. Data represent means ± SD of three independent
experiments. *P<0.05 vs. control. |
Discussion
lncRNAs were once considered to be nucleic acids
without any functions and regarded as transcriptional noise. But
recent studies have indicated that lncRNAs play important roles in
various cellular development and human diseases (17,18).
To date, many lncRNAs have been identified, and their involvement
in all kinds of tumors has been reported. However, the molecular
basis is still not well established. Therefore, the relationship
between proteins and specific lncRNAs is a hot topic in the field
of tumor biology. Although the expression of MEG3 has been shown to
be downregulated in some tumor tissues (6–9), it
still needs to be further confirmed in ESCC patients. The paired
normal adjacent esophageal tissues were also collected and were
used as the controls. The results showed that MEG3 expression was
significantly decreased compared to the levels of matching normal
specimens (Fig. 1). This finding
was consistent with recent studies in ESCC and breast cancer
(19,20). Therefore, the present study once
again confirmed the abnormal expression of MEG3 in ESCC.
In order to further identify the biological function
of MEG3, MEG3 gene was designed according to the GenBank and
synthesized by Invitrogen Life Technologies. pcDNA3.1-MEG3 plasmid
vectors were constructed. The MEG3 gene was confirmed by
XhoI and BamHI restriction enzyme digestion, then
inserted into the pcDNA3.1 vector and transfected into EC109 cells
(Fig. 2). The results showed that
the MEG3 mRNA expression level is low in EC109 cells (Fig. 3) and ectopic expression of MEG3
inhibited cell proliferation (Fig.
4). Moreover, fluorescence microscopy and Annexin V/PI staining
analysis confirmed that overexpressing MEG3 induced cell apoptosis
(Fig. 5), which demonstrates that
MEG3 functions as a tumor suppressor in EC.
Our previous study showed that adenosine, a
cytotoxic drug, can cause ER stress in EC109 cells in which GRP78
displays an anti-apoptotic effect (14). In this study, we found that ectopic
expression of MEG3 also increased the molecular chaperone GRP78
expression, which was consistent with our previous results in
hepatoma HepG2 cells (11), showing
that MEG3-induced ER stress might not be a rare phemenon in tumor
cells.
As previously described, the activation of UPR
starts from dissociation of GRP78 from ATF6, IRE1 and PERK. The
activation of ATF6, IRE1 and PERK can enhance the degradation of
unfolded and misfolded proteins through proteasomes (14,21),
if the stress is temporary (22).
However, if the cells suffer from prolonged or severe stress,
additional responses are initiated, involving IRE1/Ask1/JNK,
caspase-12/caspase-9/caspase-3, ERK/ATF-4/CHOP pathways and these
pathways can promote cell apoptosis (23,24).
All three branches of the UPR regulate the activation of CHOP
(25). In the present study, the
expression of the three key proteins of UPR (IRE1, ATF6 and PERK)
was increased in MEG3 overexpression cells. Furthermore, the
expression of CHOP, caspase-9 and cleaved caspase-3 was also
upregulated in MEG3 overexpression group (Fig. 6), showing that MEG3 activates UPR
downstream pathways. Overexpressing MEG3 inhibited cell growth and
induced cell apoptosis (Figs. 4 and
5). Taken together, these results
demonstrate that MEG3 triggers apoptosis via the ER stress pathway
in EC109 cells. In our study, caspase-9 was significantly activated
and the change of mitochondrial membrane potential was observed in
MEG3 overexpression group (data not shown). Since caspase-9 is also
an important factor of the mitochondrial apoptotic pathway, we
postulate that other mechanisms may also be involved in
MEG3-induced apoptosis and it need further investigation in
different esophageal cancer cell lines.
In conclusion, the present study presents that MEG3
is significantly downregulated in esophageal squamous cell
carcinoma tissues and exerts tumor-suppressive functions. The
mechanism of MEG3-induced apoptosis may be related to the
activation of the ER stress pathway.
Acknowledgements
The present study was supported by the Guangdong
Natural Science Foundation in China (no. 2014A030313470) and the
Collaborative and Creative Center, Molecular Diagnosis and
Personalized Medicine, Shantou University, Guangdong, China. This
study was also supported by the Department of Education, Guangdong
Government under the Top-tier University Development Scheme for
Research and Control of Infectious Diseases.
References
|
1
|
Vihinen M: Establishment of an
international database for genetic variants in esophageal cancer.
Ann N Y Acad Sci. 1381:45–49. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Gandon A, Gronnier C, Renaud F, Borde P,
Vanderbeken M, Hec F, Piessen G, Adenis A, Mirabel X and Mariette
C: Esophageal adenocarcinoma: Impact of a large hiatal hernia on
outcomes after surgery. Ann Surg. 264:862–870. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lin CY and Xu HM: Novel perspectives of
long non-coding RNAs in esophageal carcinoma. Carcinogenesis.
36:1255–1262. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
He Y, Meng XM, Huang C, Wu BM, Zhang L, Lv
XW and Li J: Long noncoding RNAs: Novel insights into hepatocelluar
carcinoma. Cancer Lett. 344:20–27. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Miyoshi N, Wagatsuma H, Wakana S,
Shiroishi T, Nomura M, Aisaka K, et al: Identification of an
imprinted gene, Meg3/Gtl2 and its human homologue MEG3, first
mapped on mouse distal chromosome 12 and human chromosome 14q.
Genes Cells. 5:211–220. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Su L, Han D, Wu J and Huo X: Skp2
regulates non-small cell lung cancer cell growth by Meg3 and
miR-3163. Tumour Biol. 37:3925–3931. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhu J, Liu S, Ye F, Shen Y, Tie Y, Zhu J,
Wei L, Jin Y, Fu H, Wu Y, et al: Long noncoding RNA MEG3 interacts
with p53 protein and regulates partial p53 target genes in hepatoma
cells. PLoS One. 10:e01397902015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Sun M, Xia R, Jin F, Xu T, Liu Z, De W and
Liu X: Downregulated long noncoding RNA MEG3 is associated with
poor prognosis and promotes cell proliferation in gastric cancer.
Tumour Biol. 35:1065–1073. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ribarska T, Goering W, Droop J, Bastian
KM, Ingenwerth M and Schulz WA: Deregulation of an imprinted gene
network in prostate cancer. Epigenetics. 9:704–717. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hu D, Su C, Jiang M, Shen Y, Shi A, Zhao
F, Chen R, Shen Z, Bao J and Tang W: Fenofibrate inhibited
pancreatic cancer cells proliferation via activation of p53
mediated by upregulation of LncRNA MEG3. Biochem Biophys Res
Commun. 471:290–295. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chen RP, Huang ZL, Liu LX, Xiang MQ, Li
GP, Feng JL, Liu B and Wu LF: Involvement of endoplasmic reticulum
stress and p53 in lncRNA MEG3-induced human hepatoma HepG2 cell
apoptosis. Oncol Rep. 36:1649–1657. 2016.PubMed/NCBI
|
|
12
|
Zhou Y, Zhong Y, Wang Y, Zhang X, Batista
DL, Gejman R, Ansell PJ, Zhao J, Weng C and Klibanski A: Activation
of p53 by MEG3 non-coding RNA. J Biol Chem. 282:24731–24742. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Piperi C, Adamopoulos C, Dalagiorgou G,
Diamanti-Kandarakis E and Papavassiliou AG: Crosstalk between
advanced glycation and endoplasmic reticulum stress: Emerging
therapeutic targeting for metabolic diseases. J Clin Endocrinol
Metab. 97:2231–2242. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wu LF, Guo YT, Zhang QH, Xiang MQ, Deng W,
Ye YQ, Pu ZJ, Feng JL and Huang GY: Enhanced antitumor effects of
adenoviral-mediated siRNA against GRP78 gene on adenosine-induced
apoptosis in human hepatoma HepG2 cells. Int J Mol Sci. 15:525–544.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zinszner H, Kuroda M, Wang X, Batchvarova
N, Lightfoot RT, Remotti H, Stevens JL and Ron D: CHOP is
implicated in programmed cell death in response to impaired
function of the endoplasmic reticulum. Genes Dev. 12:982–995. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Liu LX, Deng W, Zhou XT, Chen RP, Xiang
MQ, Guo YT, Pu ZJ, Li R, Wang GF and Wu LF: The mechanism of
adenosine-mediated activation of lncRNA MEG3 and its antitumor
effects in human hepatoma cells. Int J Oncol. 48:421–429.
2016.PubMed/NCBI
|
|
17
|
Li C, Liang G, Yao W, Sui J, Shen X, Zhang
Y, Ma S, Ye Y, Zhang Z, Zhang W, et al: Differential expression
profiles of long non-coding RNAs reveal potential biomarkers for
identification of human gastric cancer. Oncol Rep. 35:1529–1540.
2016.PubMed/NCBI
|
|
18
|
Ernst C and Morton CC: Identification and
function of long non-coding RNA. Front Cell Neurosci. 7:1682013.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lv D, Sun R, Yu Q and Zhang X: The long
non-coding RNA maternally expressed gene 3 activates p53 and is
downregulated in esophageal squamous cell cancer. Tumour Biol.
37:16259–16267. 2016. View Article : Google Scholar
|
|
20
|
Sun L, Li Y and Yang B: Downregulated long
non-coding RNA MEG3 in breast cancer regulates proliferation,
migration and invasion by depending on p53s transcriptional
activity. Biochem Biophys Res Commun. 478:323–329. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Travers KJ, Patil CK, Wodicka L, Lockhart
DJ, Weissman JS and Walter P: Functional and genomic analyses
reveal an essential coordination between the unfolded protein
response and ER-associated degradation. Cell. 101:249–258. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lee AS: The ER chaperone and signaling
regulator GRP78/BiP as a monitor of endoplasmic reticulum stress.
Methods. 35:373–381. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Liu G, Sun Y, Li Z, Song T, Wang H, Zhang
Y and Ge Z: Apoptosis induced by endoplasmic reticulum stress
involved in diabetic kidney disease. Biochem Biophys Res Commun.
370:651–656. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hitomi J, Katayama T, Taniguchi M, Honda
A, Imaizumi K and Tohyama M: Apoptosis induced by endoplasmic
reticulum stress depends on activation of caspase-3 via caspase-12.
Neurosci Lett. 357:127–130. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Bromati CR, Lellis-Santos C, Yamanaka TS,
Nogueira TC, Leonelli M, Caperuto LC, Gorjão R, Leite AR, Anhê GF
and Bordin S: UPR induces transient burst of apoptosis in islets of
early lactating rats through reduced AKT phosphorylation via
ATF4/CHOP stimulation of TRB3 expression. Am J Physiol Regul Integr
Comp Physiol. 300:R92–R100. 2011. View Article : Google Scholar : PubMed/NCBI
|