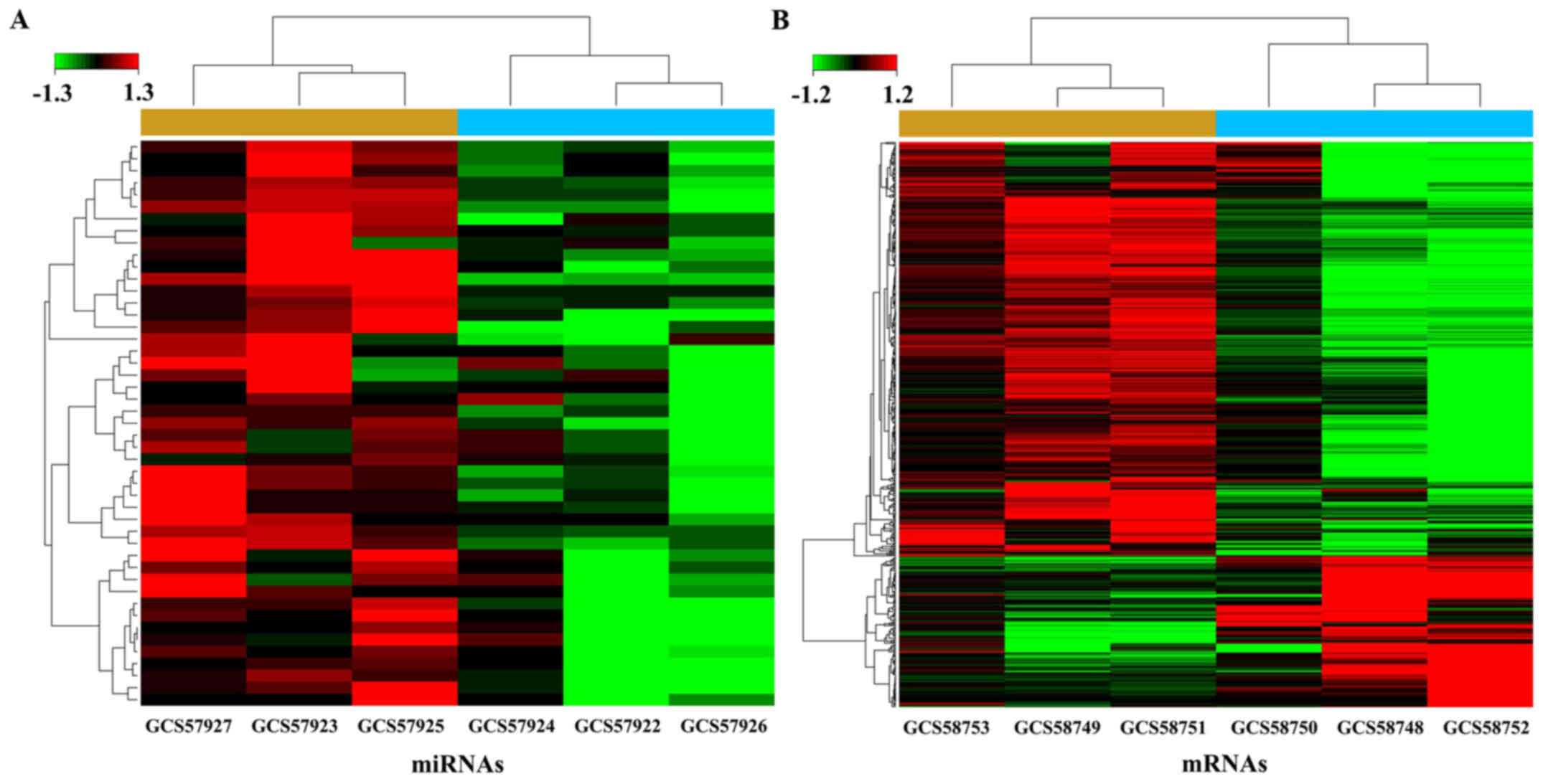
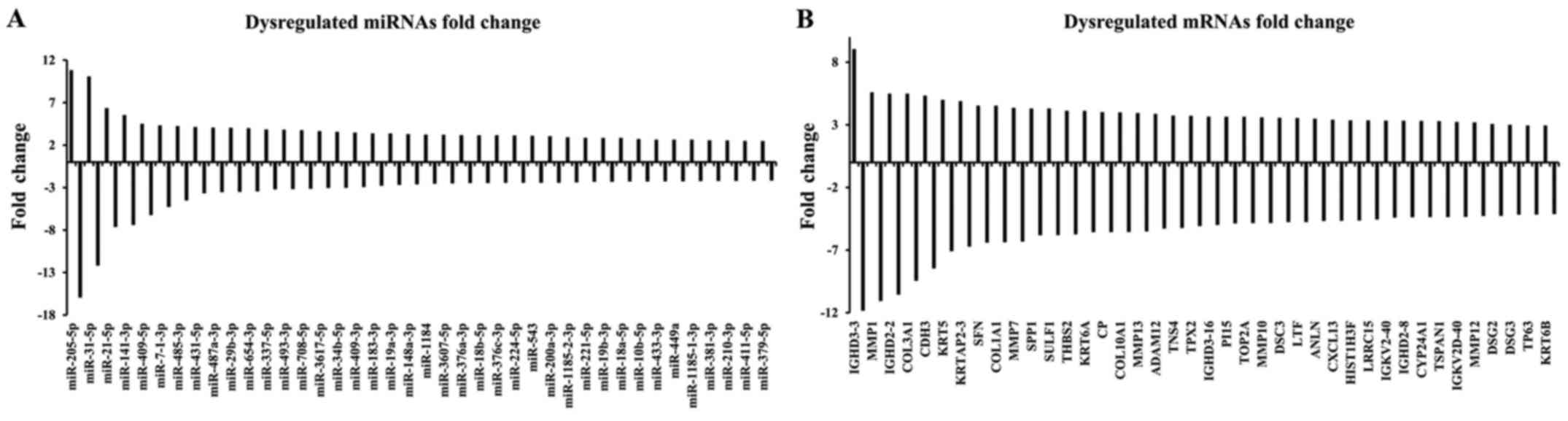
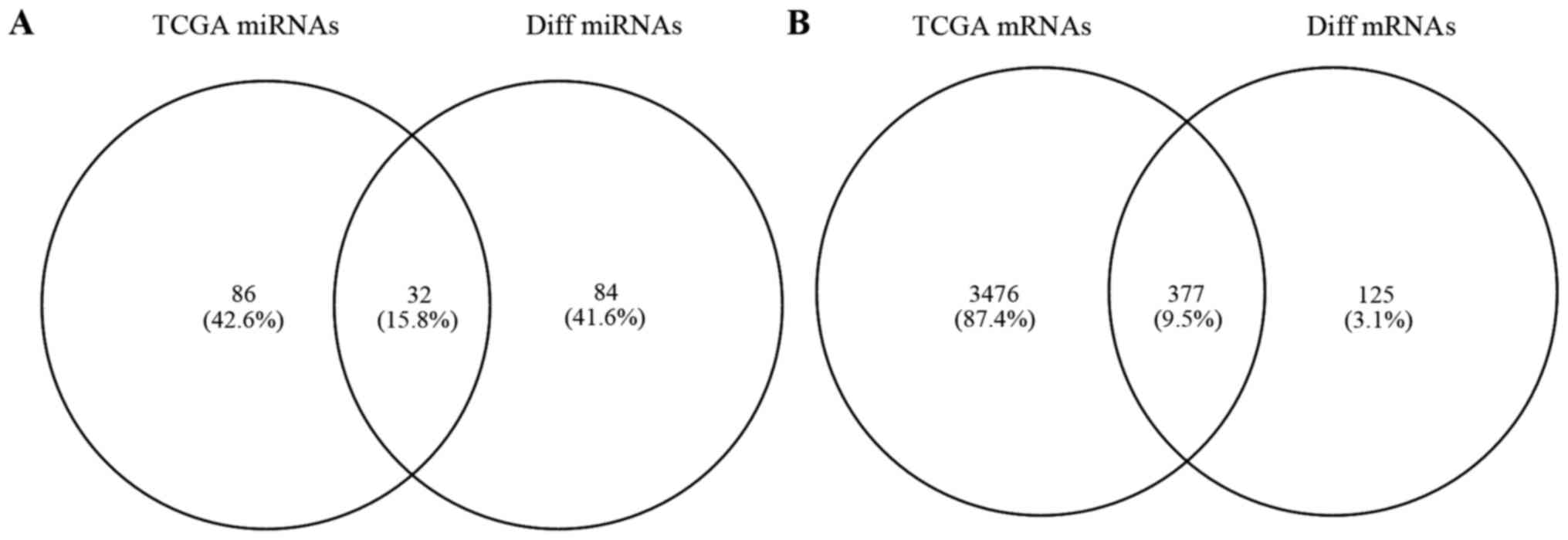
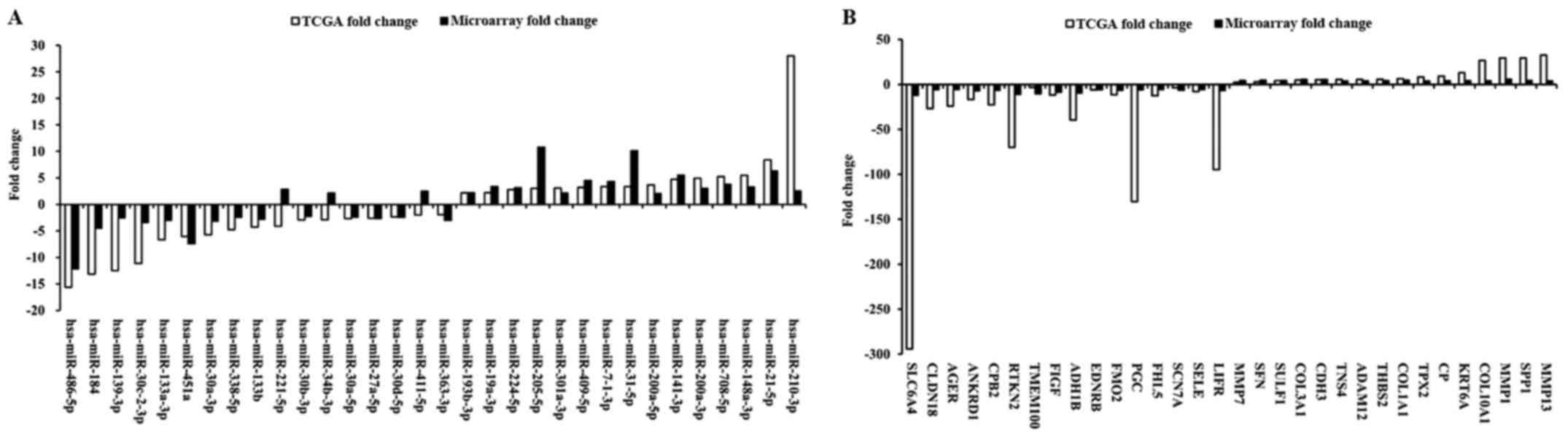
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Vrijens K, Bollati V and Nawrot TS:
MicroRNAs as potential signatures of environmental exposure or
effect: A systematic review. Environ Health Perspect. 123:399–411.
2015.PubMed/NCBI
|
|
4
|
Zhang H, Li W, Nan F, Ren F, Wang H, Xu Y
and Zhang F: MicroRNA expression profile of colon cancer stem-like
cells in HT29 adenocarcinoma cell line. Biochem Biophys Res Commun.
404:273–278. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Xia S, Huang CC, Le M, Dittmar R, Du M,
Yuan T, Guo Y, Wang Y, Wang X, Tsai S, et al: Genomic variations in
plasma cell free DNA differentiate early stage lung cancers from
normal controls. Lung Cancer. 90:78–84. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Pérez-Ramírez C, Cañadas-Garre M,
Jiménez-Varo E, Faus-Dáder MJ and Calleja-Hernández MA: MET: A new
promising biomarker in non-small-cell lung carcinoma.
Pharmacogenomics. 16:631–647. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Fan L, Qi H, Teng J, Su B, Chen H, Wang C
and Xia Q: Identification of serum miRNAs by nano-quantum dots
microarray as diagnostic biomarkers for early detection of
non-small cell lung cancer. Tumour Biol. 37:7777–7784. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Geng Q, Fan T, Zhang B, Wang W, Xu Y and
Hu H: Five microRNAs in plasma as novel biomarkers for screening of
early-stage non-small cell lung cancer. Respir Res. 15:1492014.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Inamura K and Ishikawa Y: MicroRNA in lung
cancer: Novel biomarkers and potential tools for treatment. J Clin
Med. 5:52016. View Article : Google Scholar :
|
|
10
|
Kim JO, Gazala S, Razzak R, Guo L, Ghosh
S, Roa WH and Bédard EL: Non-small cell lung cancer detection using
microRNA expression profiling of bronchoalveolar lavage fluid and
sputum. Anticancer Res. 35:1873–1880. 2015.PubMed/NCBI
|
|
11
|
Wang Y, Zhang X, Liu L, Li H, Yu J, Wang C
and Ren X: Clinical implication of microRNA for lung cancer. Cancer
Biother Radiopharm. 28:261–267. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Solomides CC, Evans BJ, Navenot JM,
Vadigepalli R, Peiper SC and Wang ZX: MicroRNA profiling in lung
cancer reveals new molecular markers for diagnosis. Acta Cytol.
56:645–654. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Singh DK, Bose S and Kumar S: Role of
microRNA in regulating cell signaling pathways, cell cycle, and
apoptosis in non-small cell lung cancer. Curr Mol Med. 16:474–486.
2016. View Article : Google Scholar
|
|
14
|
Razzak R, Bédard EL, Kim JO, Gazala S, Guo
L, Ghosh S, Joy A, Nijjar T, Wong E and Roa WH: MicroRNA expression
profiling of sputum for the detection of early and locally advanced
non-small-cell lung cancer: A prospective case-control study. Curr
Oncol. 23:e86–e94. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Pu Q, Huang Y, Lu Y, Peng Y, Zhang J, Feng
G, Wang C, Liu L and Dai Y: Tissue-specific and plasma microRNA
profiles could be promising biomarkers of histological
classification and TNM stage in non-small cell lung cancer. Thorac
Cancer. 7:348–354. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Krutakova M, Sarlinova M, Matakova T,
Dzian A, Hamzik J, Pec M, Javorkova S and Halasova E: The role of
dysregulated microRNA expression in lung cancer. Adv Exp Med Biol.
911:1–8. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Li JH, Liu S, Zhou H, Qu LH and Yang JH:
starBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA
interaction networks from large-scale CLIP-Seq data. Nucleic Acids
Res. 42:D92–D97. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hsu SD, Tseng YT, Shrestha S, Lin YL,
Khaleel A, Chou CH, Chu CF, Huang HY, Lin CM, Ho SY, et al:
miRTarBase update 2014: An information resource for experimentally
validated miRNA-target interactions. Nucleic Acids Res. 42:D78–D85.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li CY, Liang GY, Yao WZ, Sui J, Shen X,
Zhang YQ, Peng H, Hong WW, Ye YC, Zhang ZY, et al: Integrated
analysis of long non-coding RNA competing interactions reveals the
potential role in progression of human gastric cancer. Int J Oncol.
48:1965–1976. 2016.PubMed/NCBI
|
|
20
|
Yang YL, Xu LP, Zhuo FL and Wang TY:
Prognostic value of microRNA-10b overexpression in peripheral blood
mononuclear cells of nonsmall-cell lung cancer patients. Tumour
Biol. 36:7069–7075. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Xu C, Zheng Y, Lian D, Ye S, Yang J and
Zeng Z: Analysis of microRNA expression profile identifies novel
biomarkers for non-small cell lung cancer. Tumori. 101:104–110.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wiwanitkit V: MicroRNA assays for
diagnosis lung cancer biopsy. J Thorac Oncol. 10:e52–53. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wang P, Yang D, Zhang H, Wei X, Ma T,
Cheng Z, Hong Q, Hu J, Zhuo H, Song Y, et al: Early detection of
lung cancer in serum by a panel of microRNA biomarkers. Clin Lung
Cancer. 16:313–319 e1. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Chen X, Hu Z, Wang W, Ba Y, Ma L, Zhang C,
Wang C, Ren Z, Zhao Y, Wu S, et al: Identification of ten serum
microRNAs from a genome-wide serum microRNA expression profile as
novel noninvasive biomarkers for nonsmall cell lung cancer
diagnosis. Int J Cancer. 130:1620–1628. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yin QW, Sun XF, Yang GT, Li XB, Wu MS and
Zhao J: Increased expression of microRNA-150 is associated with
poor prognosis in non-small cell lung cancer. Int J Clin Exp
Pathol. 8:842–846. 2015.PubMed/NCBI
|
|
26
|
Wu C, Cao Y, He Z, He J, Hu C, Duan H and
Jiang J: Serum levels of miR-19b and miR-146a as prognostic
biomarkers for non-small cell lung cancer. Tohoku J Exp Med.
232:85–95. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Molina-Pinelo S, Pastor MD, Suarez R,
Romero-Romero B, González De la Peña M, Salinas A, García-Carbonero
R, De Miguel MJ, Rodríguez-Panadero F, Carnero A, et al: MicroRNA
clusters: Dysregulation in lung adenocarcinoma and COPD. Eur Respir
J. 43:1740–1749. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Yuan Y, Zheng S, Li Q, Xiang X, Gao T, Ran
P, Sun L, Huang Q, Xie F, Du J, et al: Overexpression of miR-30a in
lung adenocarcinoma A549 cell line inhibits migration and invasion
via targeting EYA2. Acta Biochim Biophys Sin (Shanghai).
48:220–228. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Chen D, Guo W, Qiu Z, Wang Q, Li Y, Liang
L, Liu L, Huang S, Zhao Y and He X: MicroRNA-30d-5p inhibits tumour
cell proliferation and motility by directly targeting CCNE2 in
non-small cell lung cancer. Cancer Lett. 362:208–217. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Hu J, Ni S, Cao Y, Zhang T, Wu T, Yin X,
Lang Y and Lu H: The angiogenic effect of microRNA-21 targeting
TIMP3 through the regulation of MMP2 and MMP9. PLoS One.
11:e01495372016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Jiang M, Zhang P, Hu G, Xiao Z, Xu F,
Zhong T, Huang F, Kuang H and Zhang W: Relative expressions of
miR-205-5p, miR-205-3p, and miR-21 in tissues and serum of
non-small cell lung cancer patients. Mol Cell Biochem. 383:67–75.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wang F, Lu J, Peng X, Wang J, Liu X, Chen
X, Jiang Y, Li X and Zhang B: Integrated analysis of microRNA
regulatory network in nasopharyngeal carcinoma with deep
sequencing. J Exp Clin Cancer Res. 35:172016.doi:
10.1186/s13046-016-0292-4. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Stoecklin-Wasmer C, Guarnieri P, Celenti
R, Demmer RT, Kebschull M and Papapanou PN: MicroRNAs and their
target genes in gingival tissues. J Dent Res. 91:934–940. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wang W, Lin H, Zhou L, Zhu Q, Gao S, Xie
H, Liu Z, Xu Z, Wei J, Huang X, et al: MicroRNA-30a-3p inhibits
tumor proliferation, invasiveness and metastasis and is
downregulated in hepatocellular carcinoma. Eur J Surg Oncol.
40:1586–1594. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Moch H and Lukamowicz-Rajska M:
miR-30c-2-3p and miR-30a-3p: New pieces of the jigsaw puzzle in
HIF2α regulation. Cancer Discov. 4:22–24. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Li W, Liu C, Zhao C, Zhai L and Lv S:
Downregulation of β3 integrin by miR-30a-5p modulates cell adhesion
and invasion by interrupting Erk/Ets-1 network in triple-negative
breast cancer. Int J Oncol. 48:1155–1164. 2016.PubMed/NCBI
|
|
37
|
Li WF, Dai H, Ou Q, Zuo GQ and Liu CA:
Overexpression of microRNA-30a-5p inhibits liver cancer cell
proliferation and induces apoptosis by targeting MTDH/PTEN/AKT
pathway. Tumour Biol. 37:5885–5895. 2016. View Article : Google Scholar : PubMed/NCBI
|