Introduction
Gastric cancer (GC) is the fourth most common
malignancy and third leading cause of cancer-related death
worldwide, thereby representing a global cancer burden (1,2).
Although its incidence is decreased, the outcomes for GC patients
remain unsatisfactory due to lack of effective biomarkers to detect
early GC and predict both chemosensitivity and relapse. During the
early disease, GC patients experience non-specific symptoms, and
most patients are diagnosed with advanced GC because of lacking
early-stage symptoms; late-stage diagnoses are generally
considerably late for effective treatment and result in 5-year
survival rate of <20% (3).
Therefore, new therapies toward molecular targets should be
developed to improve the clinical outcome in GC.
Protein phosphatase 2A (PP2A), one of the main
serine-threonine phosphatases, negatively regulates numerous signal
transduction pathways and functions as a tumor suppressor in
several cancers (4). Consistent
with its role as a tumor suppressor, PP2A is crucial in the
regulation of survival, cell cycle progression, and differentiation
by negatively regulating the PI3K/Akt pathway (5), and dephosphorylating and inactivating
ERK and MEK1 family kinases. Aberrant expression, mutations, and
somatic alterations of PP2A scaffold and regulatory subunits are
frequently found in human breast, lung, colon, and skin cancers
(6). Thus, reactivation of PP2A
activity based on its tumor suppressor properties is considered an
attractive therapeutic strategy for human cancer treatment
(7,8).
SE translocation (SET) oncoprotein, an endogenous
inhibitor of PP2A (9), was
initially identified as a component of SET-CAN fusion gene produced
by somatic translocation in acute, undifferentiated leukemia
(10). It is a multifunctional
protein, which belongs to the NAP1 family of histone chaperones.
SET binds to nucleosomal histones and inhibits histone acetylation
by masking histone tails as component of the INHAT complex
(11). Recent reports revealed
aberrant upregulation of SET in multiple cancer types; this protein
is also a potential therapeutic target for cancer (12–18).
SET is also involved in cell proliferation, apoptosis, and invasive
behavior because it controls histone acetylation, β-adrenergic
receptor phosphorylation, and granzyme activity (11–18).
However, the role of SET in GC tumorigenesis and metastasis remains
elusive.
In this study, we analyzed the expression of SET in
GC specimens and established GC cell lines by western blot analysis
and immunohistochemistry. We manipulated SET levels in different
types of GC cells and measured their effect on tumorigenesis and
metastasis in vitro and in vivo. To address possible
mechanisms, the effect of SET on metastasis morphology and function
was investigated. On the basis of our results, we propose that SET
is essential in tumorigenesis and metastasis of GC by inhibiting
PP2A.
Materials and methods
Patients
Two independent GC cohorts in tissue microarray
(TMA) were utilized in this study. The training cohort TMA was
purchased from Wuhan Iwill Biological Technology Co., Ltd. (Wuhan,
China). It included 102 tissues of patients, and 25 paired
non-cancerous normal tissues from these patients were also
obtained. The array dot diameter was 1.5 mm, and each dot
represented a tissue spot from one individual specimen that was
selected and pathologically confirmed. Six pairs of tumor and
adjacent normal gastric tissues were collected immediately after
surgical resection and stored in liquid nitrogen until further use.
For western blot assay, tissue specimens were ground in liquid
nitrogen-cooled mortar, tissue powder was suspended in lysis buffer
[50 mM Tris-HCl (pH 7.4), 150 mM NaCl, 1% Triton X-100, 1% sodium
deoxycholate, 0.1% SDS, 1 mM PMSF, complete protease inhibitor
cocktail] and cleared by centrifugation. The samples were used by
the approval of the Institutional Review Board of Hubei University
of Medicine and Dongfeng General Hospital Affiliated to Hubei
University of Medicine. All tissue samples were obtained with
written informed consent from patients at the Dongfeng General
Hospital.
Cell culture
The GC cell lines MKN74, SGC7901, BGC823, and MGC803
and normal human gastric epithelial cell line GES-1 were purchased
from the American Tissue Culture Collection (ATCC, Manassas, VA,
USA). SGC7901, BGC823, and GES-1 were cultured in Dulbecco's
modified Eagle's medium (DMEM) containing 10% fetal bovine serum
(FBS; Hyclone Laboratories, Inc., Logan, UT, USA), 100 U/ml
penicillin, 100 mg/ml streptomycin. MKN74 and MGC803 cells were
cultured in RPMI-1640 (Hyclone Laboratories, Inc.) supplemented
with 10% FBS, 100 U/ml penicillin (Amresco, Cleveland, OH, USA),
streptomycin (100 mg/ml) (Amresco). All cells were incubated in a
humidified atmosphere with CO2 at 37°C.
Real-time quantitative PCR (qPCR)
Expression of the SET gene was examined by
real-time polymerase chain reaction (PCR) normalized to expression
of β-actin. Total RNA was extracted from cells using TRIzol
reagent (Invitrogen; Thermo Fisher Scientifc, Inc., Waltham, MA,
USA) according to the manufacturer's protocol. qPCR analysis of
SET was performed with 2 µg of total RNA and ReverTra Ace
qPCR RT kit (Toyobo Co., Ltd. Life Science Department, Osaka
Japan). Mixed 2 µg RNA, 4 µl 5X RT buffer, 1 µl RT enzyme mix, 1 µl
primer mix, and nuclease-free water ≤20 µl volume. The reverse
transcription step as follows: 37°C for 15 min; 98°C for 5 min,
then stored at −20°C. For qPCR, we used SET gene forward
primer, 5′-AAATATAACAAACTCCGCCAACC-3′; reverse primer,
5′-CAGTGCCTCTTCATCTTCCTC-3′. β-actin forward primer,
5′-GGCCAGGTCATCACCATTG-3′; reverse primer,
5′-GGATGTCCACGTCACACTTCA-3′. RT-qPCR was performed in an ABI
StepOnePlus™ Real-Time PCR system (ABI; Thermo Fisher Scientifc,
Inc.) using SYBR® Green Real-time PCR Master Mix (Toyobo
Co., Ltd. Life Science Department). Mixed SYBR Green PCR Master Mix
10 µl, forward and reverse primers 200 nM, cDNA template 100 ng,
and ddH2O ≤20 µl volume. PCR conditions consisted of the
following: 95°C for 3 min for denaturation; 95°C for 15 sec for
annealing; and 60°C for 1 min for extension, for 40 cycles. The
threshold cycle for each sample was selected from the linear range
and converted to a starting quantity by interpolation from a
standard curve generated on the same plate for each set of primers.
The SET mRNA levels were normalized for each well to the
β-actin mRNA levels using the 2−∆∆Cq method (19). Each experiment was repeated three
times.
Western blot analysis
Cell pellets were lysed in RIPA buffer containing 50
mM Tris, pH 8.0, 150 mM NaCl, 0.1% SDS, 0.5% deoxycholate, 1%
NP-40, 1 mM DTT, 1 mM NaF, 1 mM sodium vanadate, 1 mM PMSF
(Sigma-Aldrich; Merck Millipore, Darmstadt, Germany), and 1%
protease inhibitors cocktail (Merck, Millipore). Lysates were
normalized for total protein (25 µg) and loaded on 8–12% sodium
dodecyl sulfate polyacrylamide gel, electrophoresed, and
transferred to a PVDF membrane (Millipore, Kenilworth, NJ, USA),
followed by blocking with 5% skimmed milk at room temperature for 1
h. The membrane was incubated with primary antibodies overnight at
4°C and rinsed with Tris-buffered saline with Tween-20. The primary
antibodies used were anti-β-actin (1:50,000 dilution; cat. no.
A3854) (Sigma-Aldrich); anti-SET (1:2,000 dilution; cat. no.
55201–1-AP), anti-c-Myc (1:2,000 dilution; cat. no. 10828-1-AP)
(Proteintech); anti-MMP2 (1:2,000 dilution; cat. no. 2763-1)
(Epitomics); anti-cyclin D1 (1:500 dilution; cat. no. sc-246),
anti-phospho-Akt (S473) (1:500 dilution; cat. no. sc-7985),
anti-Akt (1:500 dilution; cat. no. sc-8312) (Santa Cruz
Biotechnology, Santa Cruz, CA, USA).
Immunohistochemistry
Immunohistochemical analysis as well as the scoring
of immunoreactivity was performed using the rabbit polyclonal
anti-SET antibody. Evaluation of immunostaining positive SET is
found mainly in the cytoplasm. It was graded according to both the
intensity and percentage of cells with positive staining. SET
immunopositivity was graded in one to three tumor scores for each
patient based on the intensity of the immunoreactivity in the
cancer cells, that is, 3 (+++) was strong, 2 (++) was moderate, 1
was weak (+), and 0 was negative. An optimal cutoff value was
identified: a staining index of 2 was used to define tumors of high
expression, and 1 or lower for low expression.
RNA interference
Using Lipofectamine® 3000 Transfection
reagent (Invitrogen, Carlsbad, CA, USA) according to the
manufacturer's instructions, GC cells were transfected with shRNA
expression vector pGPU6/GFP/Neo-SET (GenePharma, Shanghai, China).
The sequences were as follows: NC short hairpin RNA,
5′-GTTCTCCGAACGTGTCACGT-3′; shSET 1#,
5′-GCCCTTCCTGTCTGAACAAAAAT-3′; shSET 2#,
5′-GTGTACACAAAGGATTTGATGT-3′; and shSET 3#,
5′-GGTGATCCATCTTCGAAGTCCT-3′. Also, 48 h after transfection the
cells were harvested for western blot analysis, cell viability, and
MTT assay. Cells were transfected with the lentivirus system
(GenePharma). Transfection efficiency was assessed by western blot
analysis and cell sorting, which was also used to select stably
transfected cells.
Cytotoxic assay and cell
viability
Cells were seeded into a 96-well plate and
pre-cultured for 24 h, and then transfected with plasmid or siRNA
for 24 or 48 h. Cell cytotoxicity was determined by MTT assay. The
absorbance was measured at 570 nm by automated microplated reader
(Bio-Tek, VT, USA), and the cell death rate was calculated as
followed: inhibition rate (%) = (average A570 of the
control group - average A570 of the experimental
group)/(average A570 of the control group - average
A570 of the blank group) × 100%. Cell viability was
estimated by trypan blue dye exclusion (20).
Soft-agar colony formation assay
Cells were suspended in 1 ml of RPMI-1640 or DMEM
containing 0.3% low-melting-point agarose (Amresco) and 10% FBS,
and plated on a bottom layer containing 0.6% agarose and 10% FBS in
6-well plate in triplicate. After 2 weeks, plates were stained with
0.2% gentian violet and the colonies were counted under light
microscope (IX70; Olympus Corp., Tokyo, Japan) (20).
Murine models
Equal number of female and male nude immunodeficient
mice (nu/nu), 6–8 weeks old, were purchased from Hunan SJA
Laboratory Animal Co., Ltd., and maintained and monitored in a
specific pathogen-free environment. All animal studies were
conducted according to protocols approved by the Hubei University
of Medicine Animal Care and Use Committee, complying with the rules
of Regulations for the Administration of Affairs Concerning
Experimental Animals (Approved by the State Council of China). The
mice were injected subcutaneously with SGC7901 (5×106
cells/mice) stably transfected with shSET or NC vectors in 100 µl
PBS were injected subcutaneously into the right flank of each
mouse. Tumors were measured with a caliper and the volume
calculated as follows: 4π/3 × (width/2)2 × (length/2),
representing the 3-dimensional volume of an ellipse. After 30 days,
the mice were euthanized by cervical dislocation and tumor tissues
were excised, imaged, and weighed.
Invasion assay
An invasion assay was carried out using 24-well
plate (Corning, Inc., Corning, NY, USA). A
polyvinyl-pyrrolidone-free polycarbonate filter (8-µm pore size)
(Corning) was coated with Matrigel (BD Biosciences, Franklin Lakes,
NJ, USA). The lower chamber was filled with medium containing 20%
FBS as chemoattractant. The coated filter and upper chamber were
laid over the lower chamber. Cells (1×104 cells/well)
were seeded onto the upper chamber wells. After incubation for 20 h
at 37°C, the filter was fixed and stained with 2% ethanol
containing 0.2% crystal violet (15 min). After being dried, the
stained cells were enumerated under light microscope at 10x
objective. For quantification, the invaded stained cells on the
other side of the membrane were extracted with 33% acetic acid. The
absorbance of the eluted stain was determined at 570 nm.
Wound healing assay
Cells (4×105 cells/2 ml) were seeded in a
6-well plate and incubated at 37°C until 90–100% confluence. After
the confluent cells were scratched with a 200-µl pipette tip,
followed by washing with PBS, and then treated with serum-free
medium. After 24 h of incubation, the cells were fixed and stained
with 2% ethanol containing 0.2% crystal violet powder (15 min), and
randomly chosen fields were photographed under a light microscope
at 4x objective. The number of cells migrated into the scratched
area was calculated.
PP2A activity assay
PP2A immunoprecipitation phosphatase assay kit
(Upstate, Temecula, CA, USA) was used to measure phosphate release
as an index of phosphatase activity according to the manufacturer's
instructions. Briefly, 100 µg protein isolated from cells was
incubated with 4 µg anti-PP2A monoclonal antibody overnight.
Protein A agarose beads (40 µl) were added and the mixture was
incubated at 4°C for 2 h. Subsequently, the beads were collected
and washed three times with 700 µl of ice-cold TBS and one time
with 500 µl Ser/Thr assay buffer. The beads were further incubated
with 750 mM phosphopeptide in assay buffer for 10 min at 30°C with
constant agitation. of Malachite Green Phosphate Detection solution
(100 µl) was added and the absorbance at 650 nm was measured on a
microplate reader (21).
Statistical analysis
All experiments were repeated at least three times
and the data are presented as the mean ± SD unless noted otherwise.
Differences between data groups were evaluated for significance
using Student's t-test of unpaired data or one way analysis of
variance and Bonferroni post hoc test. P-values <0.05 indicate
statistical significance.
Results
SET is overexpressed and associated
with poor prognosis in GC
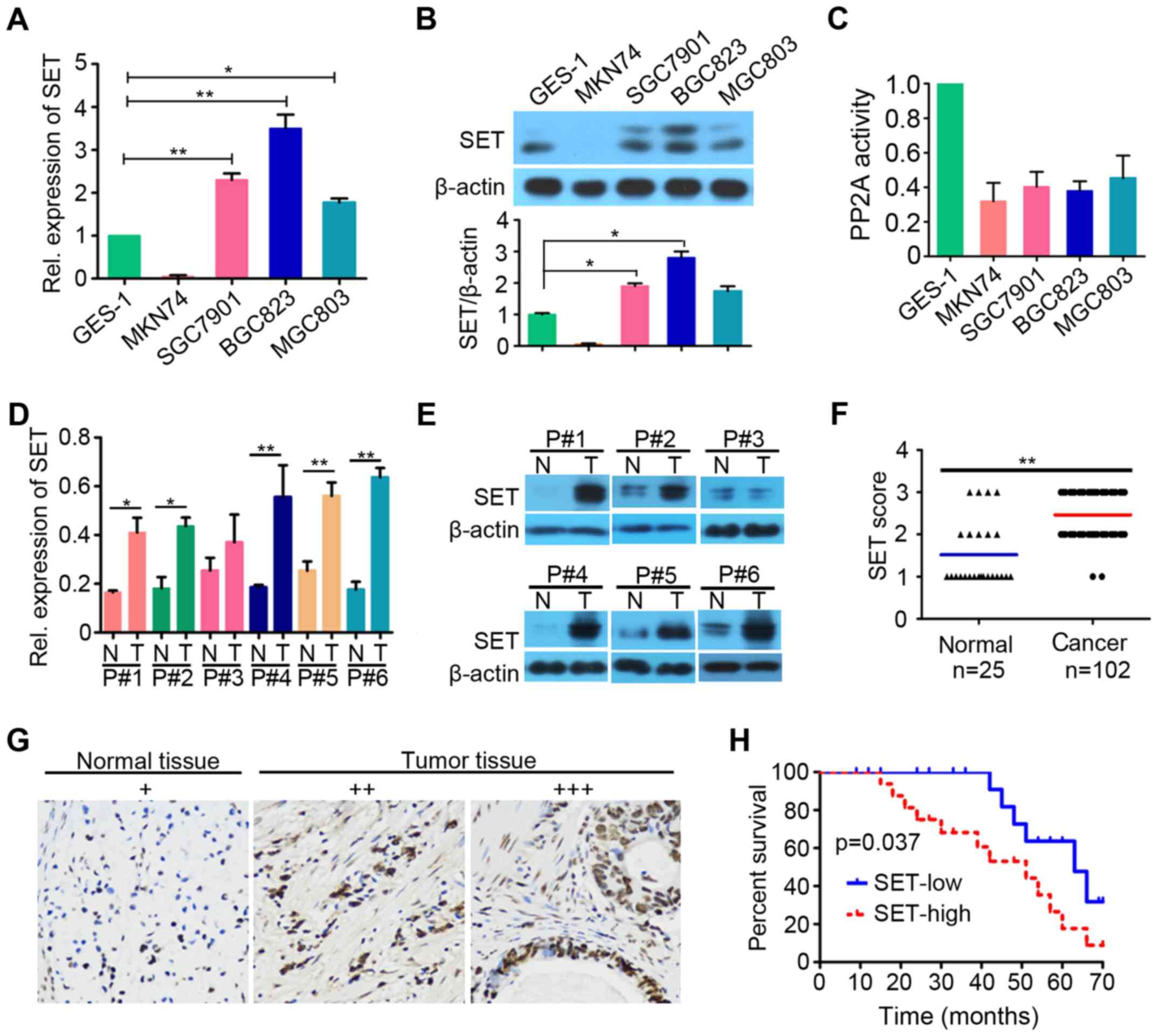
We first investigated SET prevalence in a panel of 4
human GC cell lines (MKN74, SGC7901, BGC823 and MGC803) and 1
normal human gastric epithelial cell line GES-1. Real-time
quantitative PCR (qPCR, Fig. 1A)
and western blot analysis (Fig. 1B)
showed high level of SET in SGC7901, BGC823, and MGC803 cells
compared with that in GES-1 (p<0.05). We investigated the
activity of PP2A in the above cell lines and found that GC lines
have lower PP2A activity than GES-1 (Fig. 1C). We detected SET expression in
clinical samples of human GC by qPCR and western blot analysis. The
results showed that SET was upregulated in the majority of tested
specimens (5 out of 6) compared with that in patient-matched
adjacent normal gastric tissues (p<0.05, Fig. 1D and E). We also analyzed SET
expression in 102 GC specimens and 25 adjacent normal tissues using
immunohistochemistry analysis. To evaluate the role of SET
expression in GC tumorigenesis, we divided the GC patients into SET
high (+++), moderate (++), and low expression (+) groups by
immunohistochemical assay. Immunoreactivity scoring was performed
as described (22). We found that
SET was overexpressed in 46% of tumor samples (47 of 102), and the
adjacent normal tissues exhibited low (or moderate) SET staining
(21 of 25) (Fig. 1F and G). These
results suggested that SET might be a critical molecule in GC
development. Survival analysis revealed that GC patients with high
SET expression exhibited poorer overall survival than those with
low SET expression (p=0.037; Fig.
1H). Furthermore, we analyzed the relationship between SET
expression levels and clinicopathological characteristics. As shown
in Table I, SET expression and sex,
age, clinical stage, or tumor size at diagnosis showed no
statistically significant correlations (p>0.05). Nevertheless,
statistically significant correlations among high levels of SET
expression were found with pathological grade (p=0.002), lymph node
stage (p=0.014), and invasive depth (p=0.022). Altogether, these
data suggested that SET is overexpressed in GC, and high level of
SET expression is a prognostic predictor of progression and poor
prognosis of GC patients.
 | Table I.Characteristics of SET expression in
GC patients. |
Table I.
Characteristics of SET expression in
GC patients.
|
|
| SET expression |
|
|---|
|
|
|
|
|
|---|
|
Characteristics | All (n=102)
No. | Low (n=55) No. | High (n=47)
No. |
P-valuea |
|---|
| Age |
|
<60 | 72 | 37 | 35 | 0.427 |
|
≥60 | 30 | 18 | 12 |
|
| Sex |
|
Male | 73 | 38 | 35 | 0.548 |
|
Female | 29 | 17 | 12 |
|
| Clinical stage |
| Early
(ІA-IIB) | 33 | 16 | 17 | 0.446 |
|
Advanced (IIIA-IV) | 69 | 39 | 30 |
|
| Pathological
grade |
|
І-II | 17 | 15 | 2 | 0.002 |
|
III-IV | 85 | 40 | 45 |
|
| Invasive depth |
|
T1-T2 | 21 | 16 | 5 | 0.022 |
|
T3-T4 | 81 | 39 | 42 |
|
| Lymph node
stage |
| N0 | 27 | 20 | 7 | 0.014 |
|
N1-N3 | 75 | 35 | 40 |
|
| Tumor size |
| <7
cm | 83 | 41 | 42 | 0.055 |
| ≥7
cm | 19 | 14 | 5 |
|
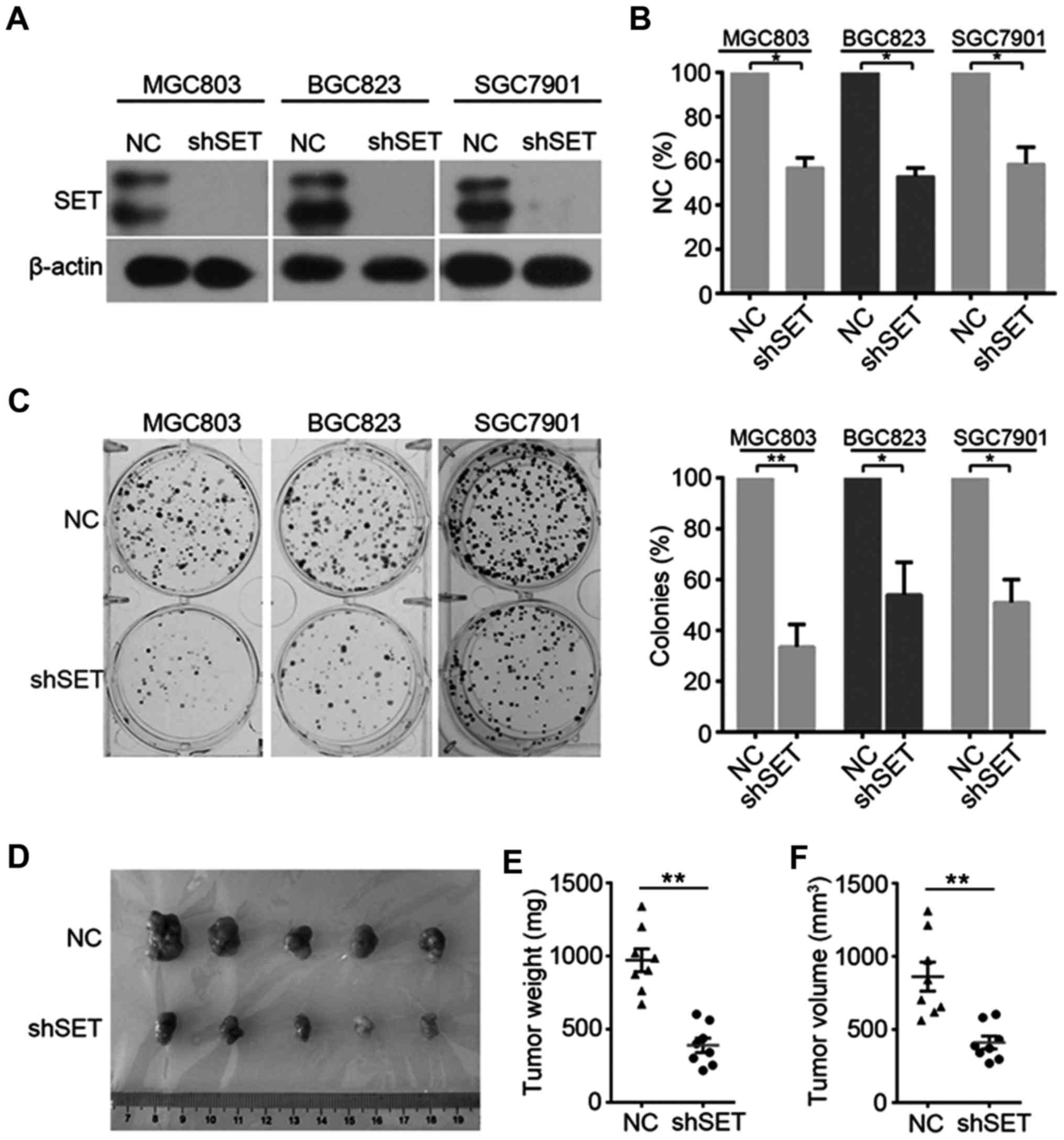
Knockdown of SET inhibits GC cell
proliferation and tumorigenesis
We also evaluated the functional role of SET in GC
proliferation and tumorigenesis. GC cell lines were transfected
with negative control (NC) or SET-specific shRNA (shSET) vectors
(Fig. 2A). MTT and clonogenic assay
were used to investigate the involvement of SET in tumorigenesis.
Knockdown of SET significantly decreased cell viability (Fig. 2B) in MGC803, SGC7901, and BGC823
cells compared with that of the NC group. Furthermore, colony
formation was reduced after transfection with SET shRNA (Fig. 2C). To further determine the
importance of SET in tumorigenesis, we investigated the effect of
SET knockdown on tumor growth in vivo. SGC7901 cells stably
expressing NC or shSET were injected subcutaneously into the right
flanks of nude mice. SET knockdown substantially inhibited in
vivo tumor growth on SGC7901 cells (Fig. 2D-F).
Knockdown of SET suppresses GC cell
invasion and migration
To examine the role of SET in GC cell metastasis, we
evaluated the effect of SET knockdown on GC cell invasion and
migration by Transwell and wound healing assays. As shown in
Fig. 3A, SET knockdown drastically
reduced the invasive ability of MGC803, BGC823, and SGC7901 cells
compared with that of NC. Wound healing assay also showed that
knockdown of SET significantly decreased the migratory rate of
MGC803, BGC823, and SGC7901 cells (Fig.
3B). We also evaluated the effects of SET on the expression of
several proliferative and invasive protein markers. Western blot
analysis revealed that knockdown of SET markedly suppressed the
protein expression of c-Myc and cyclin D1 and decreased the
expression of matrix metalloproteinase 2 (MMP2) (Fig. 3C). c-Myc is a target of PP2A
(23). Moreover, SET participates
in the regulation of cellular molecular processes by inhibiting the
tumor suppressor PP2A (9). Thus, we
analyzed whether SET silence can alter the effects of PP2A in GC
cells. SET depletion significantly upregulated the phosphatase
activity of MGC803, BGC823 and SGC7901 cells (Fig. 3D). In addition, we used western blot
analysis to investigate Akt phosphorylation (pAkt) and found that
SET silence also reduced pAkt, a critical PP2A target (Fig. 3C).
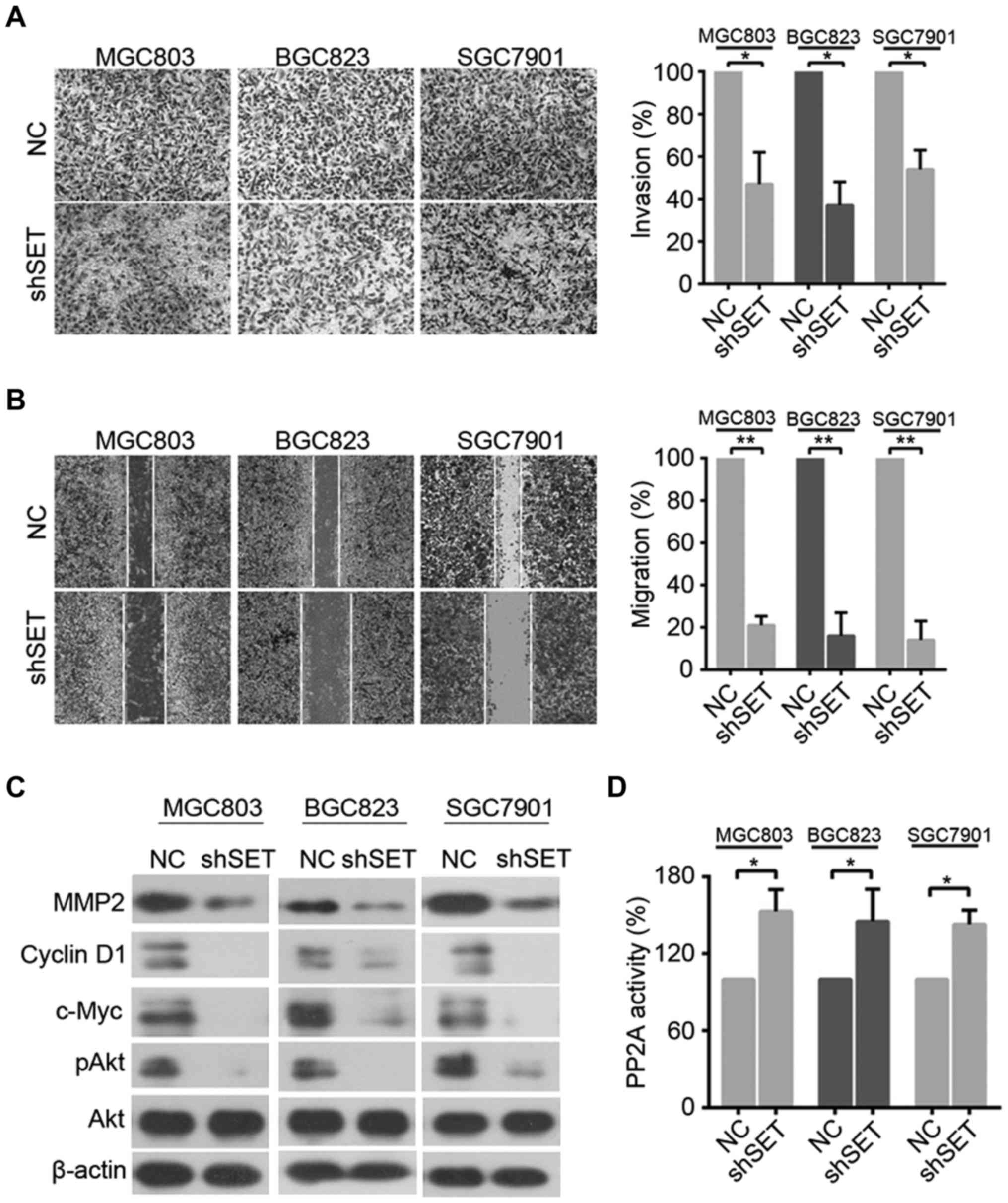 | Figure 3.Knockdown of SET suppresses the
invasion and migration of GC cells. (A) MGC803, BGC823, and SGC7901
cells were transfected with shSET, cell invasive ability was
detected by Matrigel Transwell assays. *p<0.05. (B) The
migratory speed of shSET-expressing MGC803, BGC823, and SGC7901
cells was monitored through a wound healing assay. **p<0.01. (C)
MGC803, BGC823, and SGC7901 cells were transfected with shSET,
western blot analysis was performed using antibodies indicated. (D)
MGC803, BGC823, and SGC7901 cells transfected with shSET for 48 h
and lysed, and cell lysates were prepared for detecting PP2A
activity as described in Materials and methods. *p<0.05. |
Inhibition of PP2A is essential for
SET-induced proliferation and metastasis
To further evaluate whether SET-induced cell
proliferation is caused by the inhibition of PP2A activity, we
compared cell proliferation in shSET-transfected GC cells and
NC-transfected GC cells in the presence and absence of PP2A
inhibitor, that is, okadaic acid (OA). OA significantly reversed
the cell proliferation inhibited by SET silence (Fig. 4A). In addition, we compared cell
proliferation in shSET-transfected GC cells and NC-transfected GC
cells in the presence and absence of PP2A activator FTY720. FTY720
significantly enhances the cell proliferation inhibited by SET
silence (Fig. 4B). Furthermore, OA
also significantly reversed the cell invasion and migration
inhibited by SET silence (Fig. 4C and
D). Hence, inhibition of PP2A is, at least in part, essential
for SET-induced proliferation and metastasis.
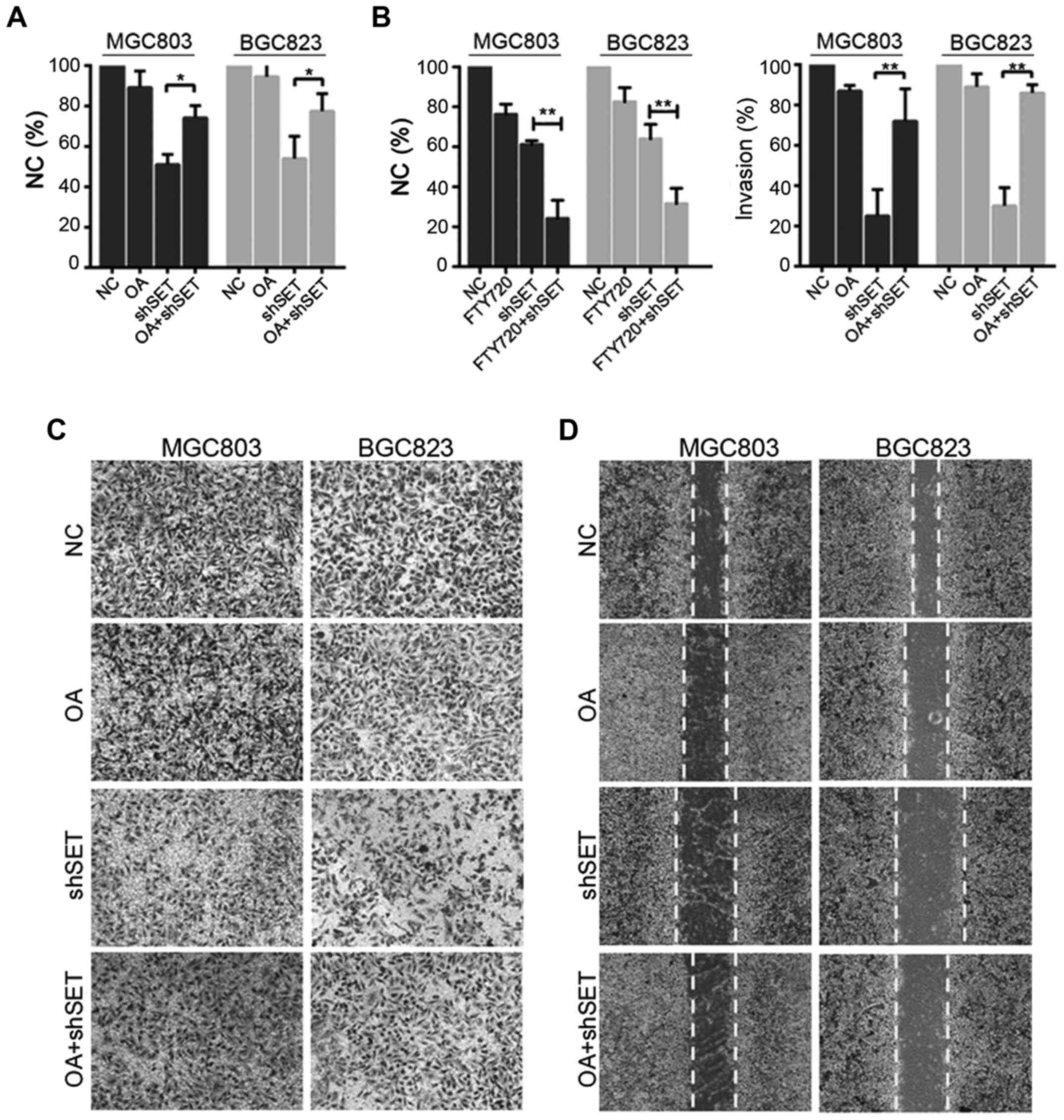 | Figure 4.SET promotes proliferation and
metastasis through inhibiting PP2A. (A) MGC803, or BGC823 cells
were transfected with shSET (or NC), 6 h after transfection, cells
were treated with or without 0.5 nM OA for 48 h, cell viability was
evaluated by MTT assay. *p<0.05. (B) MGC803, or BGC823 cells
were transfected with shSET (or NC), 6 h after transfection, cells
were treated with or without 2.5 µM FTY720 for 48 h, cell viability
was evaluated by MTT assay. **p<0.01. (C) MGC803 or BGC823 cells
were transfected with shSET (or NC), 6 h after transfection, cells
were treated with or without 0.5 nM OA for 48 h, cell invasive
ability was detected by Matrigel Transwell assay. **p<0.01. (D)
MGC803 or BGC823 cells were transfected with shSET (or NC), 6 h
after transfection, cells were treated with or without 0.5 nM OA
for 48 h, cell migration ability was detected by wound healing
assay. |
Discussion
As one of the most common human malignancies in
China, GC remains as a challenging disease. Complete resection is
the primary treatment of localized tumors, but patients who are
newly diagnosed with GC tend to present advanced and often
incurable diseases (24).
Therefore, novel diagnostic markers, molecular pathways implicated
in GC pathogenesis, and targeted therapy should be developed
urgently. This report is the first to demonstrate that SET
expression is significantly upregulated in GC cell lines and cancer
tissues at both protein and mRNA levels in comparison with those of
normal cells and paired adjacent non-cancerous gastric tissues. We
found that high expression of SET was correlated with pathological
grade, lymph node stage, and invasive depth. SET also played an
oncogenic role of promoting cell proliferation, colony formation,
and metastasis. Furthermore, SET was required for tumorigenesis of
GC.
SET is a predictive marker for prognosis in Wilm
tumor (12), pancreatic tumor
(13), prostate cancer (14), ovarian cancer (15), lung tumor (16), acute myelogenous leukemia (17), and chronic lymphocytic leukemia
(18). Overexpression of SET
oncoprotein in these cancer cells decreases PP2A activity and
promotes cell proliferation, survival, drug resistance, invasion,
and metastasis. In this study, SET was overexpressed in all GC cell
lines, but showed low expression in normal gastric epithelial
cells, thereby suggesting that SET has a positive role in GC
formation (Fig. 1A and B). In
addition, GC samples expressed high levels of SET compared with
those of adjacent normal gastric tissues (Fig. 1C-F). Multivariate analysis indicated
that SET was closely associated with tumor progression and poor
prognosis (Fig. 1G and Table I), and SET may represent an
independent prognostic biomarker for GC patients. SET knockdown
attenuated the tumorigenicity of SGC7901 cells in nude mice, which
suggested that SET is required in GC tumorigenesis and progression,
which is consistent with previous findings (Fig. 2D-F).
Invasion and migration are important characteristics
of cancer cell metastasis (25). To
determine the potential mechanisms of GC, we further tested the
effect of SET knockdown on migration and invasion. Wound healing
and Transwell assay data indicated that silencing of SET in GC
cells could efficiently inhibit the invasive capability of GC cells
(Fig. 3A and B). Downregulation of
SET also inhibited the expression of cyclin D1, MMP2, c-Myc, and
pAkt and upregulated the activity of PP2A (Fig. 3C and D). These abnormally expressed
downstream molecules of CIP2A are critical for GC proliferation and
metastasis.
PP2A, one of the four major classes (PP1, PP2A,
PP2B, and PP2C) of eukaryotic serine/threonine phosphoprotein
phosphatases, is a key tumor suppressor that regulates survival,
cell cycle progression, and differentiation-related signaling
pathways with high relevance in human cancer (4,23). SET
oncoprotein is an important binding partner of PP2A with an
inhibitory function, and collected evidence supports the idea that
SET-promoted tumor progression is mediated by PP2A inhibition
(16). We treated OA, the inhibitor
of PP2A in SET-depleted GC cells. The results showed that loss of
PP2A function can reverse the inhibition effect of SET depletion in
GC cell proliferation, migration, and invasion (Fig. 4A-C). These results confirmed that
PP2A inactivation plays a critical role in SET-promoted GC
proliferation and invasion progression. PP2A-activating drugs
represent promising anticancer molecules that can be used in cancer
treatment (16,26,27).
Thus, targeting SET protein is a potential strategy to treat cancer
by reactivation of PP2A.
In conclusion, high expression of SET is a recurrent
event in GC, where it serves as a marker of reduced overall
survival and a poor prognostic factor, as previously reported in
other tumors. Moreover, SET depletion upregulates PP2A activity,
thereby reducing cell proliferation, invasion, and migration. Our
results confirmed that SET behaves as an oncoprotein in GC and
could represent a novel therapeutic target in GC.
Acknowledgements
This study was supported by grants from the National
Natural Sciences Foundation of China (grant no. 81400157); the
Natural Science Foundation of Hubei Province of China (grant no.
2016CFB528); the Foundation of Health and Family planning
Commission of Hubei Province (grant no., WJ2017F065 and
WJ2017F067); the Foundation of Hubei University of Medicine (grant
no. FDFR201605) and the National Training Program of Innovation and
Entrepreneurship for undergraduates (grant nos. 201710929004 and
201710929012).
References
|
1
|
Kanda M and Kodera Y: Recent advances in
the molecular diagnostics of gastric cancer. World J Gastroenterol.
21:9838–9852. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2015. CA Cancer J Clin. 65:5–29. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Jemal A, Tiwari RC, Murray T, Ghafoor A,
Samuels A, Ward E, Feuer EJ, Thun MJ, et al: American Cancer
Society: Cancer statistics, 2004. CA Cancer J Clin. 54:8–29. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Perrotti D and Neviani P: Protein
phosphatase 2A: A target for anticancer therapy. Lancet Oncol.
14:e229–e238. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Junttila MR, Puustinen P, Niemelä M, Ahola
R, Arnold H, Böttzauw T, Ala-aho R, Nielsen C, Ivaska J, Taya Y, et
al: CIP2A inhibits PP2A in human malignancies. Cell. 130:51–62.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Seshacharyulu P, Pandey P, Datta K and
Batra SK: Phosphatase: PP2A structural importance, regulation and
its aberrant expression in cancer. Cancer Lett. 335:9–18. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Schönthal AH: Role of serine/threonine
protein phosphatase 2A in cancer. Cancer Lett. 170:1–13. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lv P, Wang Y, Ma J, Wang Z, Li JL, Hong
CS, Zhuang Z and Zeng YX: Inhibition of protein phosphatase 2A with
a small molecule LB100 radiosensitizes nasopharyngeal carcinoma
xenografts by inducing mitotic catastrophe and blocking DNA damage
repair. Oncotarget. 5:7512–7524. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Li M, Makkinje A and Damuni Z: The myeloid
leukemia-associated protein SET is a potent inhibitor of protein
phosphatase 2A. J Biol Chem. 271:11059–11062. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Adachi Y, Pavlakis GN and Copeland TD:
Identification and characterization of SET, a nuclear
phosphoprotein encoded by the translocation break point in acute
undifferentiated leukemia. J Biol Chem. 269:2258–2262.
1994.PubMed/NCBI
|
|
11
|
Kalousi A, Hoffbeck AS, Selemenakis PN,
Pinder J, Savage KI, Khanna KK, Brino L, Dellaire G, Gorgoulis VG
and Soutoglou E: The nuclear oncogene SET controls DNA repair by
KAP1 and HP1 retention to chromatin. Cell Rep. 11:149–163. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Carlson SG, Eng E, Kim EG, Perlman EJ,
Copeland TD and Ballermann BJ: Expression of SET, an inhibitor of
protein phosphatase 2A, in renal development and Wilms' tumor. J Am
Soc Nephrol. 9:1873–1880. 1998.PubMed/NCBI
|
|
13
|
Bhutia YD, Hung SW, Krentz M, Patel D,
Lovin D, Manoharan R, Thomson JM and Govindarajan R: Differential
processing of let-7a precursors influences RRM2 expression and
chemosensitivity in pancreatic cancer: Role of LIN-28 and SET
oncoprotein. PLoS One. 8:e534362013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Anazawa Y, Nakagawa H, Furihara M, Ashida
S, Tamura K, Yoshioka H, Shuin T, Fujioka T, Katagiri T and
Nakamura Y: PCOTH, a novel gene overexpressed in prostate cancers,
promotes prostate cancer cell growth through phosphorylation of
oncoprotein TAF-Ibeta/SET. Cancer Res. 65:4578–4586. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ouellet V, Page CL, Guyot M-C, Lussier C,
Tonin PN, Provencher DM and Mes-Masson A-M: SET complex in serous
epithelial ovarian cancer. Int J Cancer. 119:2119–2126. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Liu H, Gu Y, Wang H, Yin J, Zheng G, Zhang
Z, Lu M, Wang C and He Z: Overexpression of PP2A inhibitor SET
oncoprotein is associated with tumor progression and poor prognosis
in human non-small cell lung cancer. Oncotarget. 6:14913–14925.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Cristóbal I, Garcia-Orti L, Cirauqui C,
Cortes-Lavaud X, García-Sánchez MA, Calasanz MJ and Odero MD:
Overexpression of SET is a recurrent event associated with poor
outcome and contributes to protein phosphatase 2A inhibition in
acute myeloid leukemia. Haematologica. 97:543–550. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Christensen DJ, Chen Y, Oddo J, Matta KM,
Neil J, Davis ED, Volkheimer AD, Lanasa MC, Friedman DR, Goodman
BK, et al: SET oncoprotein overexpression in B-cell chronic
lymphocytic leukemia and non-Hodgkin lymphoma: A predictor of
aggressive disease and a new treatment target. Blood.
118:4150–4158. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Cao W, Liu Y, Zhang R, Zhang B, Wang T,
Zhu X, Mei L, Chen H, Zhang H, Ming P, et al: Homoharringtonine
induces apoptosis and inhibits STAT3 via IL-6/JAK1/STAT3 signal
pathway in Gefitinib-resistant lung cancer cells. Sci Rep.
5:84772015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Xiao X, He Z, Cao W, Cai F, Zhang L, Huang
Q, Fan C, Duan C, Wang X, Wang J, et al: Oridonin inhibits
gefitinib-resistant lung cancer cells by suppressing
EGFR/ERK/MMP-12 and CIP2A/Akt signaling pathways. Int J Oncol.
48:2608–2618. 2016.PubMed/NCBI
|
|
22
|
Khanna A, Böckelman C, Hemmes A, Junttila
MR, Wiksten JP, Lundin M, Junnila S, Murphy DJ, Evan GI, Haglund C,
et al: MYC-dependent regulation and prognostic role of CIP2A in
gastric cancer. J Natl Cancer Inst. 101:793–805. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Rincón R, Cristóbal I, Zazo S, Arpí O,
Menéndez S, Manso R, Lluch A, Eroles P, Rovira A, Albanell J, et
al: PP2A inhibition determines poor outcome and doxorubicin
resistance in early breast cancer and its activation shows
promising therapeutic effects. Oncotarget. 6:4299–4314. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Li L, Fan B, Zhang LH, Xing XF, Cheng XJ,
Wang XH, Guo T, Du H, Wen XZ and Ji JF: Trichostatin A potentiates
TRAIL-induced antitumor effects via inhibition of ERK/FOXM1 pathway
in gastric cancer. Tumour Biol. 37:10269–10278. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Liu Y, Cao W, Zhang B, Liu YQ, Wang ZY, Wu
YP, Yu XJ, Zhang XD, Ming PH, Zhou GB, et al: The natural compound
magnolol inhibits invasion and exhibits potential in human breast
cancer therapy. Sci Rep. 3:30982013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Pippa R, Dominguez A, Christensen DJ,
Moreno-Miralles I, Blanco-Prieto MJ, Vitek MP and Odero MD: Effect
of FTY720 on the SET-PP2A complex in acute myeloid leukemia; SET
binding drugs have antagonistic activity. Leukemia. 28:1915–1918.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Agarwal A, MacKenzie RJ, Pippa R, Eide CA,
Oddo J, Tyner JW, Sears R, Vitek MP, Odero MD, Christensen DJ, et
al: Antagonism of SET using OP449 enhances the efficacy of tyrosine
kinase inhibitors and overcomes drug resistance in myeloid
leukemia. Clin Cancer Res. 20:2092–2103. 2014. View Article : Google Scholar : PubMed/NCBI
|