Introduction
Hepatocellular carcinoma (HCC) is one of the
deadliest malignancies worldwide. According to the data from the
International Agency for Research on Cancer, more than half of
global incidence (782000 estimated new cases) and mortality (746000
estimated deaths) were in China in 2012 (1). In recent years, the incidence and
mortality of HCC have increased in the United States, while the
incidence and mortality of HCC have declined in China (2,3).
Improvements in management and follow-up protocols seemly mean that
they could provide additional benefit for HCC patients, whereas the
5-year overall survival rate still remained ~30–40% in HCC.
Currently, molecular targeted therapy has been used as a novel
treatment in advanced HCC patients, but the effect is still
suboptimal due to neoplastic heterogeneity. Therefore, it is
necessary to identify optimal biomarkers for clinical treatment and
prognostic assessment of HCC (4,5).
EVI5 (ecotropic viral integration site 5) is an 810
amino acid protein, which belongs to a small subfamily of the
Tre-2/Bub2/Cdc16 (TBC) domain-containing proteins. Earlier studies
showed that EVI5 was expressed in various tissues, including in
liver, brain, thymus and adrenal tissues (6). Many studies supported that EVI5 was a
disease risk gene for multiple sclerosis (7,8). In
the field of cancer research, the studies of EVI5 are gaining
attention. Functional study revealed that EVI5 played significant
roles in cell cycle progression by interacting with Emi1 and Rab11
respectively, and that this regulation was also involved in
vesicular trafficking and cytokinesis (9,10). In
addition, EVI5 protein associates with the INCENP-aurora B
kinase-survivin chromosomal passenger complex, facilitating the
final stages of cell separation at the end of mitosis (11). Furthermore, it has been confirmed
that EVI5 showed a close relationship with disease process in AKXD
T-cell lymphomas (12). More
importantly, EVI5 promoted the development of lymphomas by
cooperating with the human BCL6 gene (13). All of above suggested that EVI5
plays a vital role in tumorigenesis.
During the past two decades, EVI5 has been
recognized as a potential oncogene and a cell cycle regulator
(14). However, it remains largely
unclear between EVI5 and human cancers. In the present study, we
investigated the prognostic significance of EVI5 expression in
HCC.
Materials and methods
Cell lines and cell culture
Human normal liver cell line LO2 and seven
hepatocellular carcinoma cell lines were cultured in this study.
H2M and H2P were gifts from Professor Tiebang Kang laboratory
(State Key Laboratory of Oncology in South China). HuH7, SMMC7721,
BEL7402 and QGY7703 were purchased from the Shanghai Cell Bank
Chinese Academy Sciences. HepG2 was purchased from the American
Type Culture Collection (ATCC; Manassas, VA, USA). Cultured cells
were maintained in Dulbeccos modified Eagles medium ((DMEM;
Invitrogen, Carlsbad, CA, USA) supplemented with 10% fetal bovine
serum (FBS; Invitrogen), and incubated in a humidified atmosphere
of 5% CO2 at 37°C.
Patient information and tissue
specimens
A total of 205 consecutive patients who underwent
radical hepatectomy between January 1, 2002 and December 31, 2004,
at the Affiliated Tumor Hospital of Xinjiang Medical University
were enrolled in the present study. Hepatectomy was carried out
under general anesthesia using a right subcostal incision with a
midline extension. Intraoperative ultrasound was routinely used.
Radical hepatectomy was performed aiming at a surgical margin of at
least 2 cm. The clinicopathological information of the patients is
listed in Table I. Paired fresh
hepatocellular carcinoma tissues and adjacent non-tumor liver
tissues were collected and processed within 30 min after resection.
Each specimen was divided into two parts, one immediately preserved
in liquid nitrogen for protein extraction, the other was immersed
in RNAlater (Ambion, Inc., Austin, TX, USA) at 4°C overnight and
then transferred to liquid nitrogen storage for RNA isolation. All
patients were followed up and monitored prospectively, detailed
methods including physical examination, serum α-fetoprotein (AFP)
and abdomen ultrasonography every 1–3 months in the first year, and
every 3–6 months thereafter for surveillance of recurrence or
metastases. For patients with test results suggestive of
recurrence, contrast computed tomography and/or magnetic resonance
imaging were used to verify whether recurrence had occurred. None
of the patients combined with synchronous cancers, nor had
undergone interventional therapy, chemotherapy or radiotherapy in
the preoperation. Patients were staged according to the 7th edition
of the American Joint Committee on Cancer (AJCC)
tumor-node-metastasis (TNM) classification system. Overall survival
(OS) was calculated from the day of hepatectomy to the day of death
or to the date of the last follow-up. Recurrence-free survival
(RFS) was defined as the interval from primary liver resection to
the first recurrence. A diagnosis of recurrence was based on
typical imaging appearance in computed tomography and/or magnetic
resonance imaging scan and an elevated serum AFP level. For the use
of these clinical materials for research purposes, prior written
consent of the patients and approval from the Ethics Committee of
the Affiliated Tumor Hospital of Xinjiang Medical University were
obtained.
 | Table I.Clinicopathological correlation of
EVI5 expression in HCC patients. |
Table I.
Clinicopathological correlation of
EVI5 expression in HCC patients.
|
|
| EVI5 expression (n,
%) |
|
|---|
|
|
|
|
|
|---|
| Variables | N | Low | High | P-value |
|---|
| All cases | 205 | 112 | 93 |
|
| Age (years) |
|
|
| 0.414 |
|
≤50 | 110 (53.7) | 63 (56.3) | 47 (50.5) |
|
|
>50 | 95 (46.3) | 49 (43.7) | 46 (49.5) |
|
| Sex |
|
|
| 0.698 |
|
Female | 24 (11.7) | 14 (12.5) | 10 (10.8) |
|
|
Male | 181 (88.3) | 98 (87.5) | 83 (89.2) |
|
| HBsAg |
|
|
| 0.737 |
|
Negative | 26 (12.7) | 15 (13.4) | 11 (11.8) |
|
|
Positive | 179 (87.3) | 97 (86.6) | 82 (88.2) |
|
| Liver function |
|
|
| 0.013 |
|
Child-Pugh A | 176 (85.9) | 90 (80.4) | 86 (92.5) |
|
|
Child-Pugh B | 29 (14.1) | 22 (19.6) | 7 (7.5) |
|
| Serum AFP
(ng/ml) |
|
|
| 0.942 |
|
≤25 | 70 (34.1) | 38 (33.9) | 32 (34.4) |
|
|
>25 | 135 (65.9) | 74 (66.1) | 61 (65.6) |
|
| Tumor size
(cm) |
|
|
| 0.632 |
| ≤5 | 160 (78.0) | 86 (76.8) | 74 (79.6) |
|
|
>5 | 45 (22.0) | 26 (23.2) | 19 (20.4) |
|
| Tumor number |
|
|
| 0.384 |
|
Solitary | 188 (91.7) | 101 (90.2) | 87 (93.5) |
|
|
Multiple | 17 (8.3) | 11 (9.8) | 6 (6.5) |
|
| Venous
invasion |
|
|
| 0.015 |
|
Absent | 180 (87.8) | 104 (92.9) | 76 (81.7) |
|
|
Present | 25 (12.2) | 8 (7.1) | 17 (18.3) |
|
| Histological
gradea |
|
|
| 0.592 |
|
I/II | 110 (53.7) | 62 (55.4) | 48 (51.6) |
|
|
III/IV | 95 (46.3) | 50 (44.6) | 45 (48.4) |
|
| TNM
stageb |
|
|
| 0.014 |
| Stage
I | 148 (72.2) | 73 (65.2) | 75 (80.6) |
|
| Stage
II/III | 57 (27.8) | 39 (34.8) | 18 (19.4) |
|
RNA extraction and quantitative
real-time polymerase chain reaction (RT-qPCR)
The detailed protocol of this experiment was
described in a previous study (15). Total RNA was isolated from cells and
tissues using the TRIzol reagent (Invitrogen) according to the
manufacturers instruction, the mean yield rates were 150 µg per
5×106 cells and 38 µg per 10 mg tissues, respectively.
The first strand cDNA was generated using PrimeScript®
RT reagent kit with gDNA Eraser (Takara Bio, Inc., Dalian, China).
Subsequently, qPCR was conducted for detection of EVI5 mRNA using a
SYBR-Green PCR kit (Takara Bio) and the CFX96 sequence detection
system (Bio-Rad Laboratories, Inc., Hercules, CA, USA).
Glyceraldehyde-3-phosphate dehydrogenase (GAPDH; forward: 5-GGAGC
GAGATCCCTCCAAAAT-3 and reverse, 5-GGCTGTTGTC ATACTTCTCATGG-3) was
used as the input control. The sequences of primers for EVI5 were
as follows: forward, 5-AGAAACCCTAGTGGGAAACAGG-3 and reverse, 5-TG
ACTGTATGCGATACTGTGTTC-3. The 2−ΔΔCt method was used to
analyze the relative change in EVI5 expression. The PCR condition
was: 95°C for 10 min, followed by 40 cycles of 95°C for 5 sec and
60°C for 60 sec. Negative non-template control was also run on
PCR.
Western blot assay
Western blotting was performed as previously
described (15). In brief,
specimens were ground to powder in liquid nitrogen, collected and
lysed by RIPA buffer, centrifuged at 13,400 g for 30 min at 4°C,
and then heated at 95°C for 10 min. Proteins (35 µg per lane) were
separated on 10% sodium dodecyl sulfate-polyacrylamide gradient
gels, transferred onto PVDF membranes, and followed by blocking
with 5% non-fat milk for 2 h at room temperature. Membranes were
incubated with primary antibody and horseradish
peroxidase-conjugated secondary antibody, and then detected by
chemiluminesence using the enhanced chemiluminescence (ECL) system.
Antibodies against EVI5 and β-actin were purchased from Santa Cruz
Biotechnology (Santa Cruz, CA, USA; sc-160055, 0.2 mg/ml, 1:200
dilution) and Bioworld Technology (St. Louis Park, MN, USA; AP0060,
1 mg/ml, 1:10,000 dilution), respectively.
Immunohistochemistry (IHC) and IHC
evaluation
After hematoxylin and eosin (H&E) staining, the
degree of tumor differentiation was evaluated in terms of
Edmondson-Steiner histological grading system. IHC was performed on
hepatocellular carcinoma and paratumor tissues as previously
described (15). Paraffin-embedded
sections were deparaffinized, rehydrated and then prepared for
antigen retrieval. Sections were blocked with 10% normal goat
serum, incubated with primary antibody against EVI5 (Sigma-Aldrich,
St. Louis, MO, USA; HPA027339, 1:100 dilution) at 4°C overnight in
a moist chamber, and treated by biotin-labeled secondary antibody
for 1 h at room temperature. Subsequently, the samples were
developed by adding DAB (Genetech, Co., Ltd., Beijing, China) and
counterstained with hematoxylin. The sections were reviewed and
scored independently by two observers who were masked to
clinicopathological materials, based on both the proportion of
positively stained tumor cells and the staining intensity. The
immunoreactions for EVI5 in cytoplasm was quantified, respectively,
using widely accepted German semi-quantitative scoring system
(16). The proportion of positive
tumor cells was scored as follows: 0, no positive tumor cells; 1,
1–25%; 2, 26–50%; 3, 51–75%; and 4, 76–100%. The intensity of
staining was categorized as follows: 0, no staining; 1, weak
staining (light yellow); 2, moderate staining (yellow brown); and
3, strong staining (brown). The staining index (SI) was calculated
as staining intensity score multiplied by positive tumor cells
percentage score (17). The range
of SI was from 0 to 12, and cut-offs were selected by the receiver
operating characteristic (ROC) curve analysis. SI<6 was defined
as low EVI5 expression and SI≥6 was defined as high EVI5
expression. Confirmed immunostaining-positive slides were used as
positive controls. A rabbit polyclonal IgG antibody (ab27478;
Abcam) replaced the primary antibody as a negative control.
Statistical analysis
The SPSS statistical software package (version 16.0;
SPSS, Inc., Chicago, IL, USA) was used. The Pearsons Chi-square
test was utilized to analyze the relationship between the EVI5
expression and the clinicopathological parameters. Survival curves
were generated by the Kaplan-Meier method and compared by the
log-rank test. Univariate analysis identified variables associated
with survival, and than significant variables associated with
survival in univariate analysis were entered into a multivariate
Cox proportional hazard regression model. Finally, independent
prognostic factors were determined. A two-sided P<0.05 was
considered to be statistically significant.
Results
EVI5 is overexpressed in HCC cell
lines and clinical samples
To detect EVI5 expression profile, initially,
multiple HCC cell lines were used to measure mRNA and protein
expression levels of EVI5 by RT-qPCR and western blotting assay,
respectively. Both the mRNA and protein levels of EVI5 were
elevated in the 7 HCC cell lines, including H2M, H2P, HuH7,
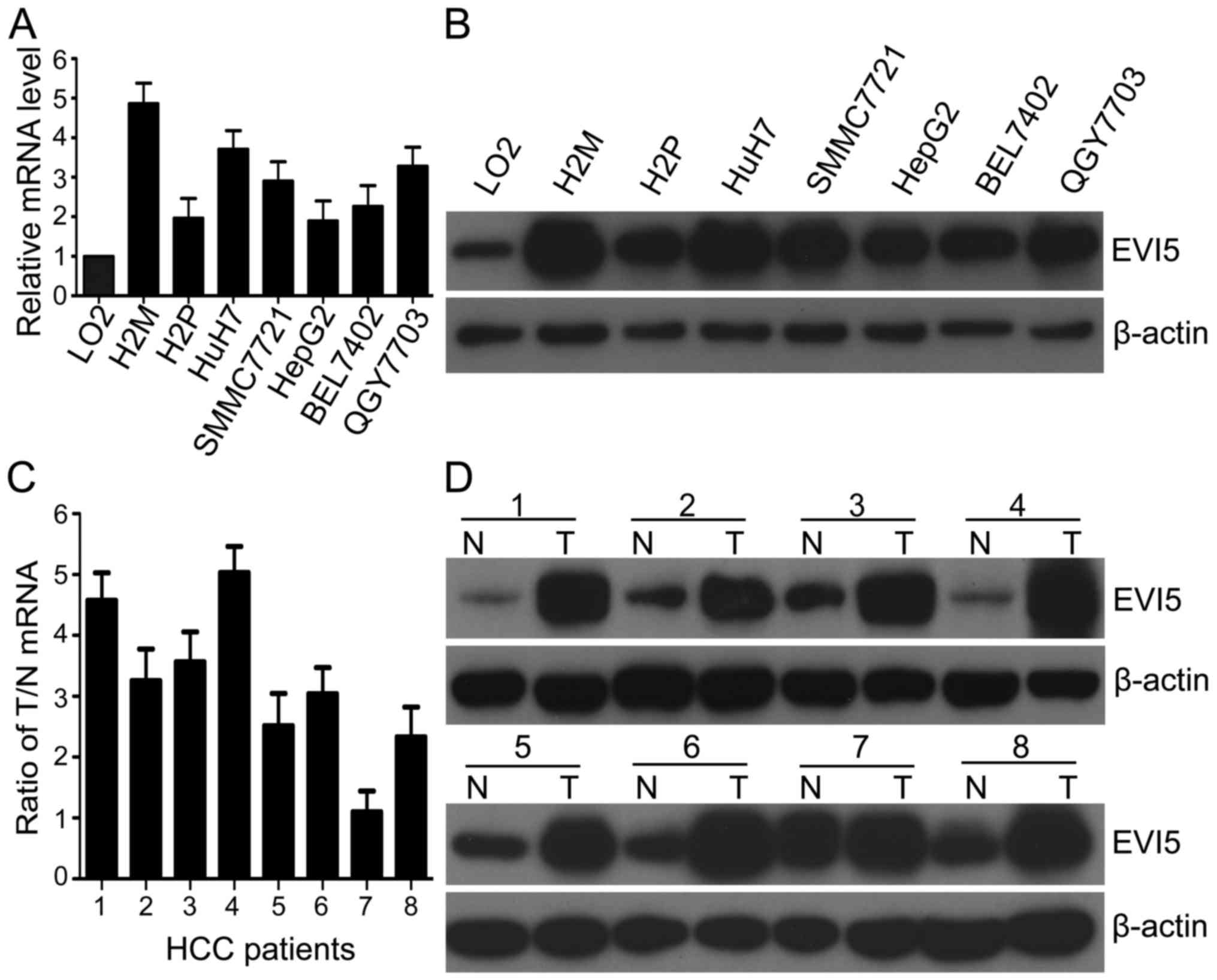
SMMC7721, HepG2, BEL7402 and QGY7703, than those in LO2 (Fig. 1A and B). Among these cancer cell
lines, EVI5 mRNA was almost 5-fold overexpressed in H2M compared to
normal LO2 liver cells. Furthermore, by use of the fresh HCC
specimens, we also observed that the expression of EVI5 mRNA were
remarkably higher in 7 of 8 HCC tissues than that in the matched
adjacent non-tumor liver tissues, which was consistent with
corresponding paired protein levels (Fig. 1C and D). These results show that
EVI5 is upregulated in HCC.
Correlation between EVI5 protein
expression and HCC clinicopathological features
To further evaluate the relationship between the
EVI5 protein expression and the clinicopathological characteristics
of HCC, 205 paraffin-embedded HCC samples were detected by IHC.
According to the cut-off value of 6, all of patients could be
divided into high-expression group (45.4%, 93/205) and
low-expression group (54.6%, 112/205). Chi-square test indicated
that the EVI5 protein level was closely associated with some of the
clinicopathological variables of HCC, including liver function
(P=0.013), venous invasion (P=0.015) and TNM stage (P=0.014).
However, EVI5 protein expression is unrelated to age, sex,
hepatitis B surface antigen (HBs Ag), serum AFP, tumor size, tumor
number and histological grade (Table
I). Moreover, patients in the venous invasion group (68.0%,
17/25) had a greater proportion of high EVI5 expression, compared
with those in the absence of venous invasion group (42.2%,
76/180).
Association between EVI5 expression
and HCC survival
Among this retrospective cohort of 205 HCC cases,
including 148 cases of stage I (72.2%), 32 cases of stage II
(15.6%), 25 cases of stage III (12.2%), IHC was conducted to
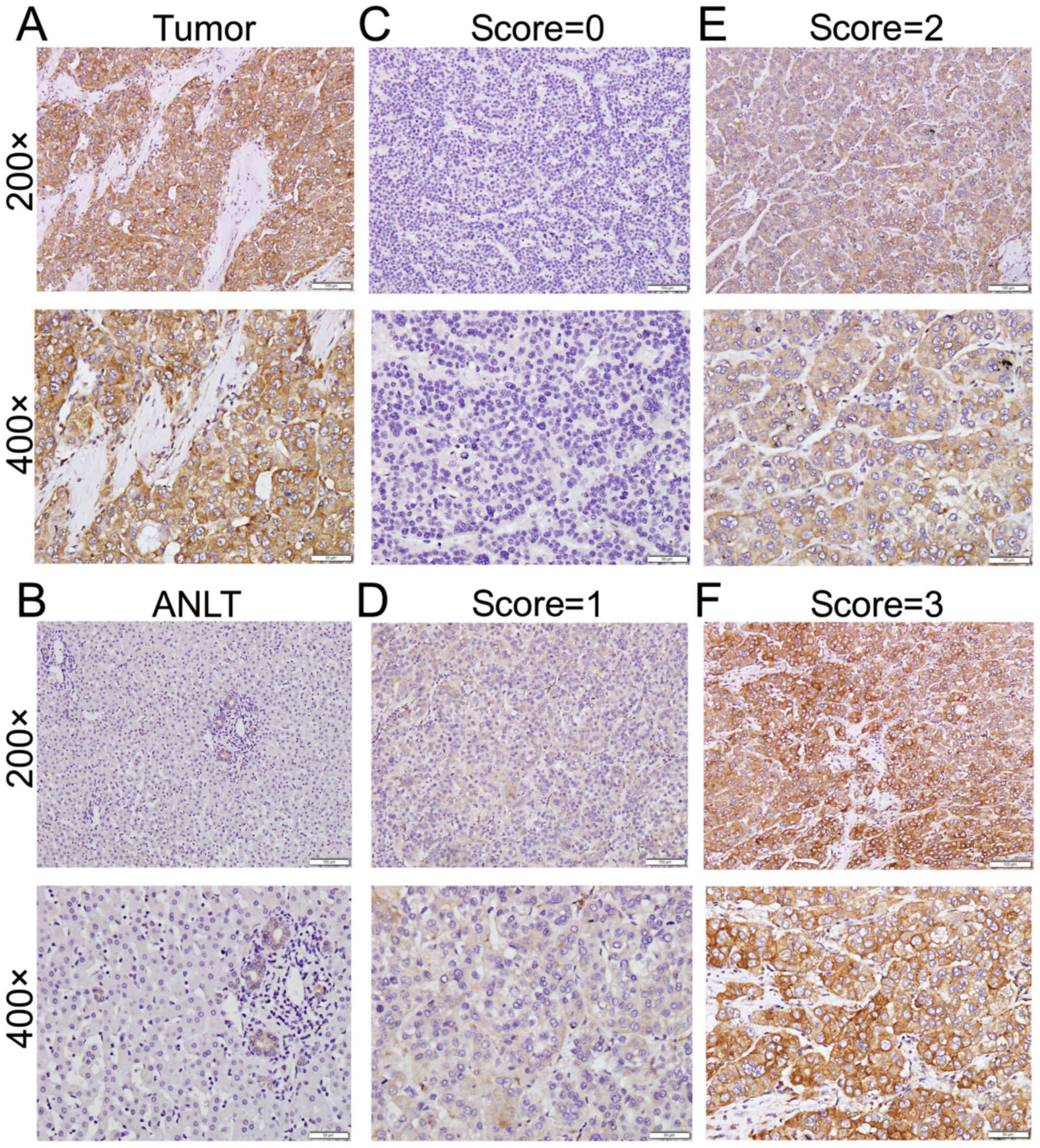
explore the expression pattern of EVI5. The immunostaining of EVI5
was stronger in HCC than in matched adjacent non-tumor tissue
(Fig. 2A and B). The staining
intensity was classified into four grades (negative, score = 0;
weak, score = 1; moderate, score = 2; strong, score = 3) (Fig. 2C-F). The positive staining
predominantly localized in the cytoplasm.
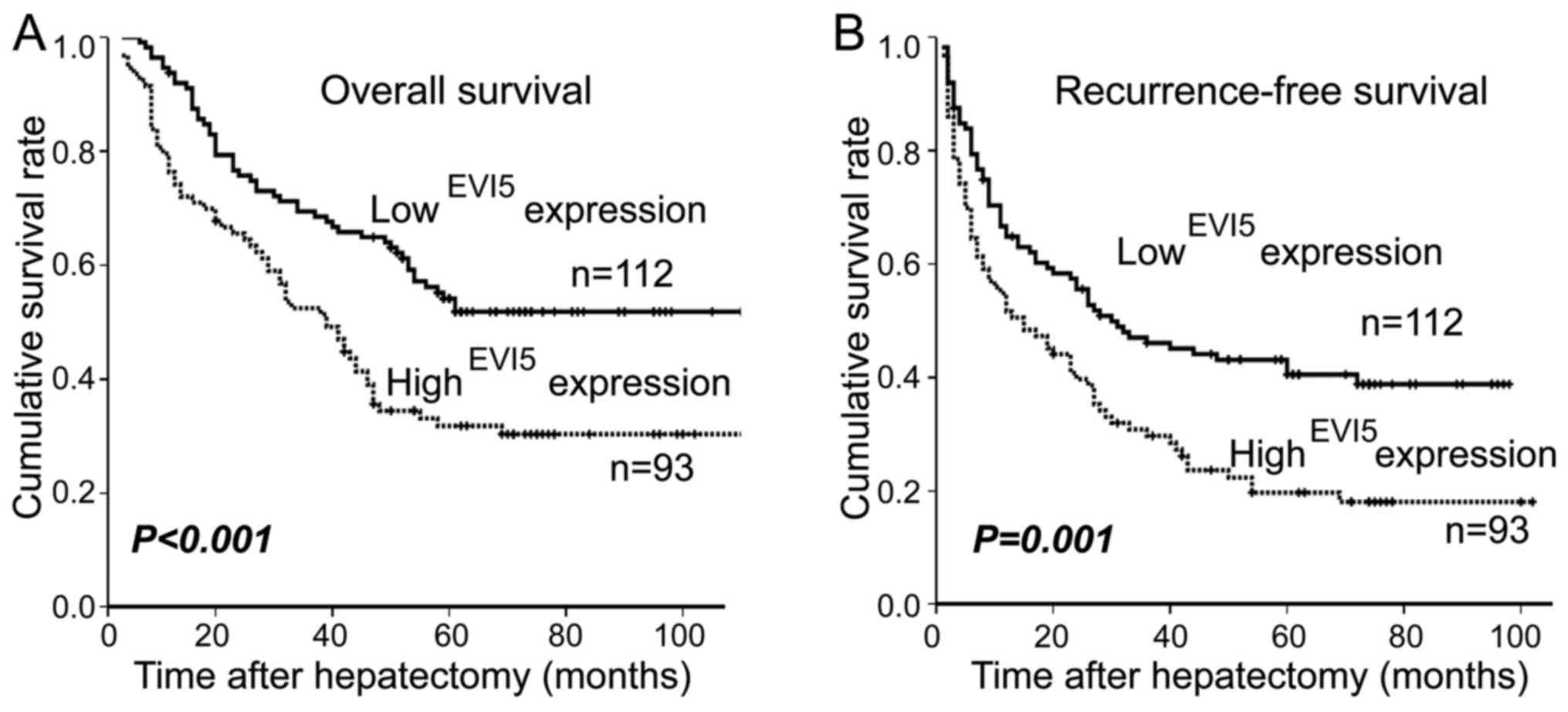
The EVI5 protein level was negatively correlated
with overall survival (OS, P<0.001; Fig. 3A) and recurrence-free survival (RFS,
P=0.001; Fig. 3B). The patients
with low EVI5 expression in HCC tumor tissue had better OS and RFS
rates than those with high EVI5 expression. The overall 3- and
5-year cumulative survival rate of patients with high EVI5
expression were 62.3 and 39.1%. For those with low EVI5 expression,
the rates were 68.5 and 54.1% (Fig.
3A). The recurrence-free survival (RFS) rate at 3- and 5-year
were 61.3 and 38.6% for high EVI5 expression subjects compared with
70.3 and 49.9% for low EVI5 expression ones, respectively (Fig. 3B). According to the following-up
data analysis, EVI5 high-expression group had a shorter overall
survival time (median OS, 39.0 months) and recurrence-free survival
time (median RFS, 15.0 months) compared with the low-expression
group (median OS, 63.0 months; median RFS, 30.0 months).
Univariate analysis revealed there were 7 risk
factors associated with OS and RFS, respectively (Table II). Moreover, Cox regression
analysis demonstrated that EVI5 expression was an independent
prognostic factor for OS in HCC patients as well as tumor number.
Independent prognostic factors for RFS including EVI5 expression,
tumor number and serum AFP (Table
III).
 | Table II.Univariate prognostic analysis of
overall survival and recurrence-free survival rates for 205
surgical HCC patients. |
Table II.
Univariate prognostic analysis of
overall survival and recurrence-free survival rates for 205
surgical HCC patients.
|
| Overall
survival | Recurrence-free
survival |
|---|
|
|
|
|
|---|
| Variables | HR (95% CI) | P-value | HR (95% CI) | P-value |
|---|
| Age, years (≤50 vs.
>50) | 1.359
(0.943–1.960) | 0.100 | 1.267
(0.908–1.767) | 0.163 |
| Sex (female vs.
male) | 1.122
(0.630–1.998) | 0.696 | 1.368
(0.787–2.378) | 0.267 |
| HBsAg (negative vs.
positive) | 1.095
(0.626–1.916) | 0.750 | 1.183
(0.703–1.994) | 0.527 |
| Serum AFP, ng/ml
(≤25 vs. >25) | 1.548
(1.032–2.320) | 0.034 | 1.771
(1.221–2.568) | 0.003 |
| Liver function
(Child-Pugh A vs. B) | 1.008
(0.699–1.454) | 0.965 | 0.891
(0.638–1.242) | 0.495 |
| Histological
gradea (I/II vs.
III/IV) | 1.511
(1.048–2.179) | 0.027 | 1.669
(1.195–2.331) | 0.003 |
| Tumor number
(solitary vs. multiple) | 2.190
(1.201–3.994) | 0.011 | 2.143
(1.223–3.755) | 0.008 |
| Tumor size, cm (≤5
vs. >5) | 1.655
(1.090–2.513) | 0.018 | 1.740
(1.189–2.546) | 0.004 |
| Venous invasion
(absent vs. present) | 2.366
(1.467–3.814) | 0.000 | 2.341
(1.474–3.718) | 0.000 |
| TNM
stageb (I vs.
II/III) | 1.550
(1.205–1.992) | 0.001 | 1.505
(1.040–2.170) | 0.028 |
| EVI5 expression
(low vs. high) | 1.903
(1.316–2.751) | 0.001 | 1.706
(1.221–2.383) | 0.002 |
 | Table III.Cox multivariate regression analysis
on overall survival and recurrence-free survival in 205 HCC
patients after hepatectomy. |
Table III.
Cox multivariate regression analysis
on overall survival and recurrence-free survival in 205 HCC
patients after hepatectomy.
|
| Overall
survival | Recurrence-free
survival |
|---|
|
|
|
|
|---|
| Variables | HR (95% CI) | P-value | HR (95% CI) | P-value |
|---|
| Serum AFP ng/ml
(≤25 vs. >25) | 1.378
(0.892–2.130) | 0.149 | 1.577
(1.066–2.333) | 0.023 |
| Histological
gradea (I/II vs.
III/IV) | 1.356
(0.917–2.005) | 0.127 | 1.570
(0.826–2.984) | 0.169 |
| Tumor number
(solitary vs. multiple) | 1.992
(1.011–3.925) | 0.046 | 1.467
(1.026–2.099) | 0.036 |
| Tumor size, cm (≤5
vs. >5) | 1.324
(0.764–2.294) | 0.318 | 1.512
(0.921–2.483) | 0.102 |
| Venous invasion
(absent vs. present) | 1.255
(0.668–2.360) | 0.480 | 1.191
(0.671–2.111) | 0.551 |
| TNM
stageb (I vs. II
/III) | 1.155
(0.673–1.983) | 0.600 | 1.236
(0.754–2.027) | 0.401 |
| EVI5 expression
(low vs. high) | 2.141
(1.437–3.190) | 0.000 | 1.903
(1.334–2.715) | 0.000 |
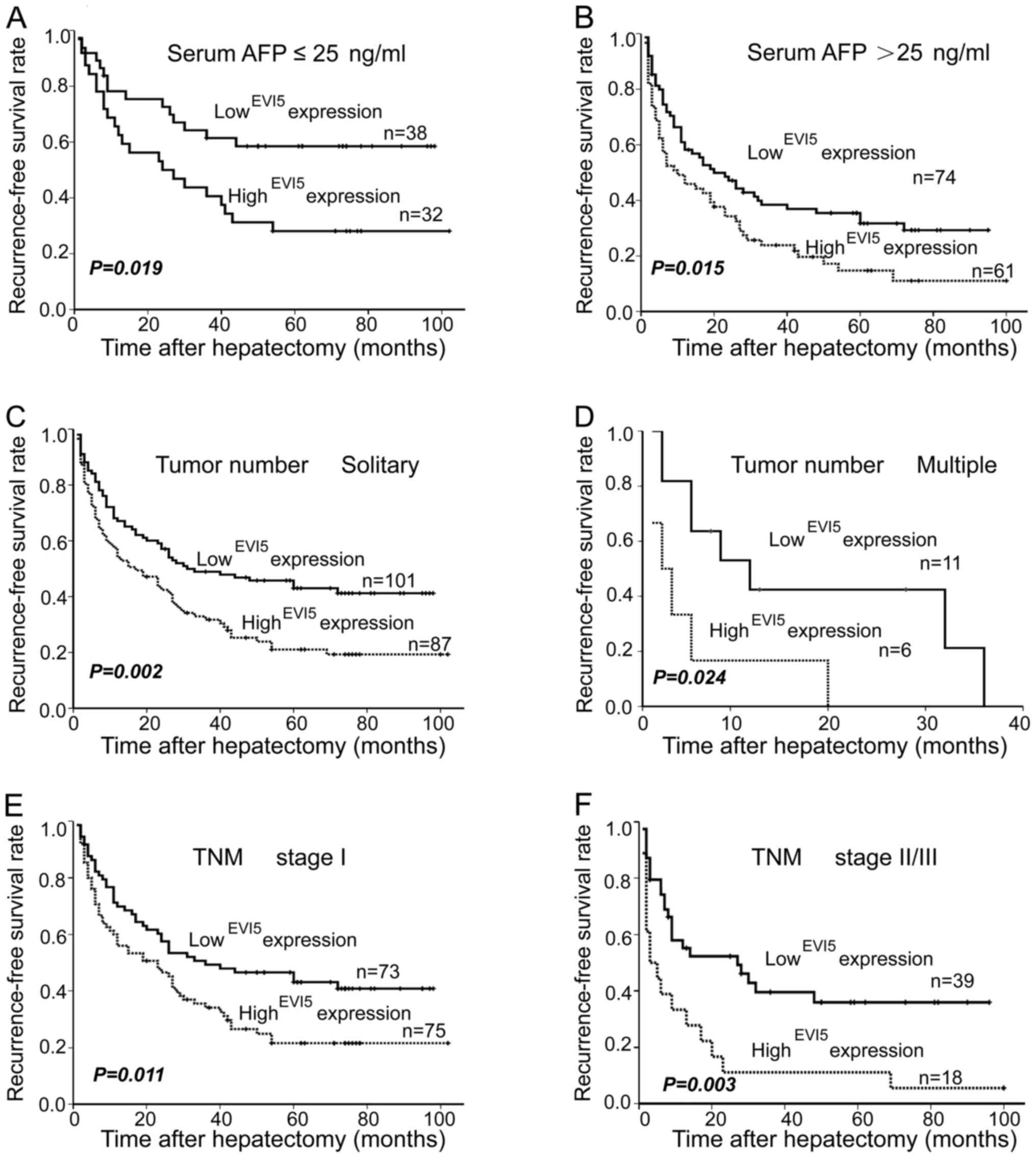
Furthermore, based on the analysis of the 3
subgroups, the influence of EVI5 expression on postoperative
recurrence was further revealed by stratified analysis. As shown in
Fig. 4, high EVI5 expression
exhibited short-term recurrence in the serum AFP subgroup (Fig. 4A, P=0.019; Fig. 4B, P=0.015). Similarly, whether the
patients had a solitary tumor or multiple tumors, the patients with
high EVI5 expression were associated with significantly worse RFS
(Fig. 4C, P=0.002; Fig. 4D, P=0.024). Additionally, in TNM
subgroup, high EVI5 expression was confirmed as an unfavorable
factor once again, and this meant that high EVI5 expression
contributed to shorter RFS under the same TNM stage (Fig. 4E, P=0.011; Fig. 4F, P=0.003). Thus, EVI5 is a valuable
prognostic marker for HCC patients.
Discussion
Hepatocellular carcinoma (HCC) which is one of the
most common malignant tumors worldwide, it is a serious threat to
human health and survival. In recent years, the studies focusing on
tumor molecular biomarkers seemed to provide new insight into
diagnosis, treatment and prognostic assessment for precise
medicine. Therefore, it is significant to identify sensitive and
specific biomarkers for HCC. Here, we identified and validated EVI5
as a novel prognostic biomarker for HCC.
Previous studies show that EVI5 was located in the
nucleus and cytoplasm (18,19). But interestingly, we failed to
observe a clear and specific nuclear staining within tumor cells.
Thus, we scored only for cytoplastic staining status. In recent
years, an increasing number of reports have linked EVI5 to various
human neoplasms, such as HCC (20),
bladder cancer (21), melanoma
(22), leukemogenesis (23) and lymphomas (13).
In the initial studies, EVI5 participated in
regulation of cell cycle in different compartments of cell,
functioned by stabilizing the Emi1 protein and promoting cyclin-A
accumulation, EVI5 misregulation led to abnormity of cell cycle and
cell division (24). Investigation
found that EVI5 levels were significantly increased during the
formation of embryo limb regeneration in newts (25). Furthermore, EVI5 widely participated
in various regulatory functions, including cell fate determination
(26), autophagy (27), membrane trafficking and cytokinesis
(28), cell migration (29,30,32),
as well as growth and differentiation of cells (31). In general, prior studies indicated
that EVI5 can function as an onco-protein which might promote
malignant phenotype of cancer cells.
Our results support the above previous findings. We
showed that EVI5 protein overexpressed in HCC cell lines and tumor
tissues compared with normal liver cell line (LO2) and
corresponding paratumor tissues. Furthermore, we validated the
differential expression by RT-qPCR and IHC. In particular, a pair
of cell models, from heptic portal vein tumor thrombus (H2M) and
primary tumor (H2P) (33), helped
explore metastatic mechanism in HCC. Notably, we found that the
expression of EVI5 in H2M was much higher than that in H2P, which
supported that EVI5 was involved in the metastasis of HCC. However,
much work should be done to unveil the detailed mechanism in
future. The study of Li et al (20) have revealed that
HSF1/miR-135b/RECK&EVI5 axis may be related to the mechanism of
metastasis in HCC. These results indicated that EVI5 behaved as a
regulator in tumorigenesis and metastasis.
Chi-square analysis demonstrated that EVI5
expression level was correlated with serum AFP, tumor number and
TNM stage. Among TNM stages, the lower percentage of high EVI5 in
advanced stage II/III (19.4%) than stage I (80.6%), may be due to
fewer subjects with stage II/III (57/205, 27.8%). Interestingly, we
still found the high EVI5 expression tended to occur in sufferers
with more advanced stage (stage II, 25.0% vs. stage III, 40.0%),
suggesting that EVI5 might be involved in tumor progression.
Kaplan-Meier analysis showed that the OS and RFS curves were
significantly separated between two cohorts with different EVI5
expression. The high expression of EVI5 was associated with shorter
OS and RFS, low expression group showed contrary results. Tumor
relapse is a key cause for therapeutic failure. Therefore, we
investigated the prognostic factors that attributed to recurrence
by stratified analysis. Cox regression analysis showed that serum
AFP, tumor number and EVI5 expression were independent prognostic
factors. TNM stage was failed to be selected, which may be due to
fewer advanced patients (TNM stage II/III, 27.8%). Subgroup
analysis demonstrated that among these patients with lower serum
AFP, solitary or TNM stage I, high EVI5 expression was consistently
associated with shorter RFS, which means that these patients could
be distinguished for early intervention to prevent tumor
recurrence. To those patients with EVI5 high expression,
accompanying with high serum AFP, multiple or advanced stages,
comprehensive treatment strategy should be considered as soon as
possible.
In conclusion, our findings showed that the EVI5 was
upregulated in HCC. The high expression of EVI5 indicated a worse
prognosis in postoperative HCC subjects. EVI5 is a novel predictive
biomarker that may help guide the clinical therapy for HCC
patients.
Acknowledgements
The present study was supported by grants from the
National Natural Science Foundation in China (grant no. 81460360 to
B.W. and grant no. 81560403 to J.T.); the Natural Science
Foundation of Xinjiang Uygur Autonomous Region, (grant no.
2015211C126 to B.W. and grant no. 2016D01C374 to J.T.). The authors
thank professor Tiebang Kang of Sun Yat-sen University Cancer
Center for providing experimental platform and expert opinions.
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2015. CA Cancer J Clin. 65:5–29. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chen S, Ling Q, Yu K, Huang C, Li N, Zheng
J, Bao S, Cheng Q, Zhu M and Chen M: Dual oxidase 1: A predictive
tool for the prognosis of hepatocellular carcinoma patients. Oncol
Rep. 35:3198–3208. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wu Y, Lin X, Di X, Chen Y, Zhao H and Wang
X: Oncogenic function of Plac1 on the proliferation and metastasis
in hepatocellular carcinoma cells. Oncol Rep. 37:465–473. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Roberts T, Chernova O and Cowell JK: NB4S,
a member of the TBC1 domain family of genes, is truncated as a
result of a constitutional t(1;10)(p22;q21) chromosome
translocation in a patient with stage 4S neuroblastoma. Hum Mol
Genet. 7:1169–1178. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Didonna A, Isobe N, Caillier SJ, Li KH,
Burlingame AL, Hauser SL, Baranzini SE, Patsopoulos NA and
Oksenberg JR: A non-synonymous single-nucleotide polymorphism
associated with multiple sclerosis risk affects the EVI5
interactome. Hum Mol Genet. 24:7151–7158. 2015.PubMed/NCBI
|
|
8
|
Liu J, Liu X, Liu Y, Deng S, Huang H, Chen
Q, Liu W and Huang Z: Association of EVI5 rs11808092, CD58
rs2300747, and CIITA rs3087456 polymorphisms with multiple
sclerosis risk: A meta-analysis. Meta Gene. 9:97–103. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Westlake CJ, Junutula JR, Simon GC, Pilli
M, Prekeris R, Scheller RH, Jackson PK and Eldridge AG:
Identification of Rab11 as a small GTPase binding protein for the
Evi5 oncogene. Proc Natl Acad Sci USA. 104:1236–1241. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Eldridge AG, Loktev AV, Hansen DV,
Verschuren EW, Reimann JD and Jackson PK: The evi5 oncogene
regulates cyclin accumulation by stabilizing the anaphase-promoting
complex inhibitor emi1. Cell. 124:367–380. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Faitar SL, Sossey-Alaoui K, Ranalli TA and
Cowell JK: EVI5 protein associates with the INCENP-aurora B
kinase-survivin chromosomal passenger complex and is involved in
the completion of cytokinesis. Exp Cell Res. 312:2325–2335. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Liao X, Buchberg AM, Jenkins NA and
Copeland NG: Evi-5, a common site of retroviral integration in AKXD
T-cell lymphomas, maps near Gfi-1 on mouse chromosome 5. J Virol.
69:7132–7137. 1995.PubMed/NCBI
|
|
13
|
Baron BW, Anastasi J, Bies J, Reddy PL,
Joseph L, Thirman MJ, Wroblewski K, Wolff L and Baron JM: GFI1B,
EVI5, MYB - additional genes that cooperate with the human BCL6
gene to promote the development of lymphomas. Blood Cells Mol Dis.
52:68–75. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Heber-Katz E, Zhang Y, Bedelbaeva K, Song
F, Chen X and Stocum DL: Cell cycle regulation and regeneration.
Curr Top Microbiol Immunol. 367:253–276. 2013.PubMed/NCBI
|
|
15
|
Wang B, Tang J, Liao D, Wang G, Zhang M,
Sang Y, Cao J, Wu Y, Zhang R, Li S, et al: Chromobox homolog 4 is
correlated with prognosis and tumor cell growth in hepatocellular
carcinoma. Ann Surg Oncol. 20 Suppl 3:S684–S692. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Henriksen KL, Rasmussen BB, Lykkesfeldt
AE, Møller S, Ejlertsen B and Mouridsen HT: Semi-quantitative
scoring of potentially predictive markers for endocrine treatment
of breast cancer: A comparison between whole sections and tissue
microarrays. J Clin Pathol. 60:397–404. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Liao WT, Guo L, Zhong Y, Wu YH, Li J and
Song LB: Astrocyte elevated gene-1 (AEG-1) is a marker for
aggressive salivary gland carcinoma. J Transl Med. 9:2052011.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Faitar SL, Dabbeekeh JT, Ranalli TA and
Cowell JK: EVI5 is a novel centrosomal protein that binds to alpha-
and gamma-tubulin. Genomics. 86:594–605. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Fuchs E, Haas AK, Spooner RA, Yoshimura S,
Lord JM and Barr FA: Specific Rab GTPase-activating proteins define
the Shiga toxin and epidermal growth factor uptake pathways. J Cell
Biol. 177:1133–1143. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Li Y, Xu D, Bao C, Zhang Y, Chen D, Zhao
F, Ding J, Liang L, Wang Q, Liu L, et al: MicroRNA-135b, a HSF1
target, promotes tumor invasion and metastasis by regulating RECK
and EVI5 in hepatocellular carcinoma. Oncotarget. 6:2421–2433.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Scaravilli M, Asero P, Tammela TL,
Visakorpi T and Saramäki OR: Mapping of the chromosomal
amplification 1p21-22 in bladder cancer. BMC Res Notes. 7:5472014.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Varghese S, Xu H, Bartlett D, Hughes M,
Pingpank JF, Beresnev T and Alexander HR Jr: Isolated hepatic
perfusion with high-dose melphalan results in immediate alterations
in tumor gene expression in patients with metastatic ocular
melanoma. Ann Surg Oncol. 17:1870–1877. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Jacob B, Osato M, Yamashita N, Wang CQ,
Taniuchi I, Littman DR, Asou N and Ito Y: Stem cell exhaustion due
to Runx1 deficiency is prevented by Evi5 activation in
leukemogenesis. Blood. 115:1610–1620. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Verschuren EW, Ban KH, Masek MA, Lehman NL
and Jackson PK: Loss of Emi1-dependent anaphase-promoting
complex/cyclosome inhibition deregulates E2F target expression and
elicits DNA damage-induced senescence. Mol Cell Biol. 27:7955–7965.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Rao N, Jhamb D, Milner DJ, Li B, Song F,
Wang M, Voss SR, Palakal M, King MW, Saranjami B, et al: Proteomic
analysis of blastema formation in regenerating axolotl limbs. BMC
Biol. 7:832009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Emery G, Hutterer A, Berdnik D, Mayer B,
Wirtz-Peitz F, Gaitan MG and Knoblich JA: Asymmetric Rab 11
endosomes regulate delta recycling and specify cell fate in the
Drosophila nervous system. Cell. 122:763–773. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Longatti A, Lamb CA, Razi M, Yoshimura S,
Barr FA and Tooze SA: TBC1D14 regulates autophagosome formation via
Rab11- and ULK1-positive recycling endosomes. J Cell Biol.
197:659–675. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Giansanti MG, Belloni G and Gatti M: Rab11
is required for membrane trafficking and actomyosin ring
constriction in meiotic cytokinesis of Drosophila males. Mol
Biol Cell. 18:5034–5047. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kawauchi T, Sekine K, Shikanai M, Chihama
K, Tomita K, Kubo K, Nakajima K, Nabeshima Y and Hoshino M: Rab
GTPases-dependent endocytic pathways regulate neuronal migration
and maturation through N-cadherin trafficking. Neuron. 67:588–602.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Kessler D, Gruen GC, Heider D, Morgner J,
Reis H, Schmid KW and Jendrossek V: The action of small GTPases
Rab11 and Rab25 in vesicle trafficking during cell migration. Cell
Physiol Biochem. 29:647–656. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Liao X, Du Y, Morse HC III, Jenkins NA and
Copeland NG: Proviral integrations at the Evi5 locus disrupt a
novel 90 kDa protein with homology to the Tre2 oncogene and
cell-cycle regulatory proteins. Oncogene. 14:1023–1029. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Laflamme C, Assaker G, Ramel D, Dorn JF,
She D, Maddox PS and Emery G: Evi5 promotes collective cell
migration through its Rab-GAP activity. J Cell Biol. 198:57–67.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Hu L, Wen JM, Sham JS, Wang W, Xie D, Tjia
WM, Huang JF, Zhang M, Zeng WF and Guan XY: Establishment of cell
lines from a primary hepatocellular carcinoma and its metastatis.
Cancer Genet Cytogenet. 148:80–84. 2004. View Article : Google Scholar : PubMed/NCBI
|