Introduction
Hepatocellular carcinoma (HCC), mainly induced by
chronic hepatitis B virus (HBV) or hepatitis C virus (HCV)
infection, hepatic cirrhosis or alcoholic liver diseases, is one of
the most common malignancies with a rise in new cases worldwide
each year. HCC has a higher rate in developing countries partly
East Asia as compared to developed countries (1). Accumulating evidence has demonstrated
that abnormal expression and mutation of genes are involved in the
carcinogenesis and progression of HCC, including cyclin D1 (CCND1),
epidermal growth factor receptor (EGFR), c-myc and Ras, as well as
mutations of tumor-suppressor genes. It was found that the G870A
polymorphism in exon 4 of the CCND1 gene may increase the risk of
HBV-related HCC in the Chinese population (2). The chronic stimulation of EGFR plays a
key role in the neoplastic conversion and development of HCC
(3). A progressive increase in
c-myc mRNA and protein was noted during the different steps of
malignancy of HCC (4). Aberrant
activation of different levels of the Ras pathway could play
important roles in HCC. In addition, overexpression of H-ras, DNA
copy number gains of B-Raf and hypermethylation of Ras binding
proteins were found to be involved in the poor prognosis of HCC
patients (5). However, due to the
lack of effective diagnostic methods at the early stage of the
disease, the mortality rate of HCC remains high. Therefore, it is
crucial to understand the precise molecular mechanisms involved in
the carcinogenesis, proliferation and recurrence of HCC and thus
develop effective diagnostic and therapeutic strategies.
During the last decades, microarray technology and
bioinformatic analysis have been widely used to screen genetic
alterations at the genome level, which have helped us identify the
differentially expressed genes (DEGs) and functional pathways
involved in the carcinogenesis and progression of HCC. However,
false-positive rates in independent microarray analysis make it
difficult to obtain reliable results. Thus, in the present study, 3
mRNA microarray datasets from Gene Expression Omnibus (GEO) were
downloaded and analyzed to obtain DEGs between liver cancer tissues
and non-cancerous tissues. Subsequently, Gene Ontology (GO), Kyoto
Encyclopedia of Genes and Genomes (KEGG) pathway enrichment
analysis and protein-protein interaction (PPI) network analyses
were performed to help us understand the molecular mechanisms
underlying carcinogenesis and progression. In conclusion, a total
of 273 DEGs and 16 hub genes were identified, which may be
candidate biomarkers for HCC.
Materials and methods
Microarray data
GEO (http://www.ncbi.nlm.nih.gov/geo) (6) is a public functional genomics data
repository of high throughout gene expression data, chips and
microarrays. Three gene expression datasets [GSE36668 (7), GSE18520 (8) and GSE14407 (9)] were downloaded from GEO (Affymetrix
GPL570 platform, Affymetrix Human Genome U133 Plus 2.0 Array). The
probes were converted into the corresponding gene symbol according
to the annotation information in the platform. The GSE19665 dataset
contained 10 HCC tissue samples and 10 non-cancerous samples.
GSE33006 contained 3 HCC samples and 3 non-cancerous samples.
GSE41804 contained 20 HCC samples and 20 non-cancerous samples.
Identification of DEGs
The DEGs between HCC and non-cancerous samples were
screened using GEO2R (http://www.ncbi.nlm.nih.gov/geo/geo2r). GEO2R is an
interactive web tool that allows users to compare two or more
datasets in a GEO series in order to identify DEGs across
experimental conditions. The adjusted P-values (adj. P) and
Benjamini and Hochberg false discovery rate were applied to provide
a balance between discovery of statistically significant genes and
limitations of false-positives. Probe sets without corresponding
gene symbols or genes with more than one probe set were removed or
averaged, respectively. logFC (fold change) >1 and adj. P-value
<0.01 were considered statistically significant.
KEGG and GO enrichment analyses of
DEGs
The Database for Annotation, Visualization and
Integrated Discovery (DAVID; http://david.ncifcrf.gov) (version 6.7) (10) is an online biological information
database that integrates biological data and analysis tools, and
provides a comprehensive set of functional annotation information
of genes and proteins for users to extract biological information.
KEGG is a database resource for understanding high-level functions
and biological systems from large-scale molecular datasets
generated by high-throughput experimental technologies (11). GO is a major bioinformatics tool to
annotate genes and analyze biological process of these genes
(12). To analyze the function of
DEGs, biological analyses were performed using DAVID online
database. P<0.05 was considered statistically significant.
PPI network construction and module
analysis
The PPI network was predicted using Search Tool for
the Retrieval of Interacting Genes (STRING; http://string-db.org) (version 10.0) (13) online database. Analyzing the
functional interactions between proteins may provide insights into
the mechanisms of generation or development of diseases. In the
present study, PPI network of DEGs was constructed using STRING
database, and an interaction with a combined score >0.4 was
considered statistically significant. Cytoscape (version 3.4.0) is
an open source bioinformatics software platform for visualizing
molecular interaction networks (14). The plug-in Molecular Complex
Detection (MCODE) (version 1.4.2) of Cytoscape is an APP for
clustering a given network based on topology to find densely
connected regions (15). The PPI
networks were drawn using Cytoscape and the most significant module
in the PPI networks was identified using MCODE. The criteria for
selection were as follows: MCODE scores >5, degree cut-off=2,
node score cut-off=0.2, Max depth=100 and k-score=2. Subsequently,
the KEGG and GO analyses for genes in this module were performed
using DAVID.
Hub genes selection and analysis
The hub genes were selected with degrees ≥10. A
network of the genes and their co-expression genes was analyzed
using cBioPortal (http://www.cbioportal.org) (16,17)
online platform. The biological process analysis of hub genes was
performed and visualized using Biological Networks Gene Oncology
tool (BiNGO) (version 3.0.3) plugin of Cytoscape (18). Hierarchical clustering of hub genes
was constructed using UCSC Cancer Genomics Browser (http://genome-cancer.ucsc.edu) (19). The overall survival and disease-free
survival analyses of hub genes were performed using Kaplan-Meier
curve in cBioPortal. The expression profiles of TOP2A and CDK1 were
analyzed and displayed using online database Serial Analysis of
Gene Expression (SAGE; http://www.ncbi.nlm.nih.gov/SAGE). The relationship
between expression patterns and tumor grades, hepatitis virus
infection status, satellites and vascular invasion were analyzed
using online database Oncomine (http://www.oncomine.com) (20–22).
Results
Identification of DEGs in HCC
After standardization of the microarray results,
DEGs (1,558 in GSE19665, 1,330 in GSE33006 and 717 in GSE41804)
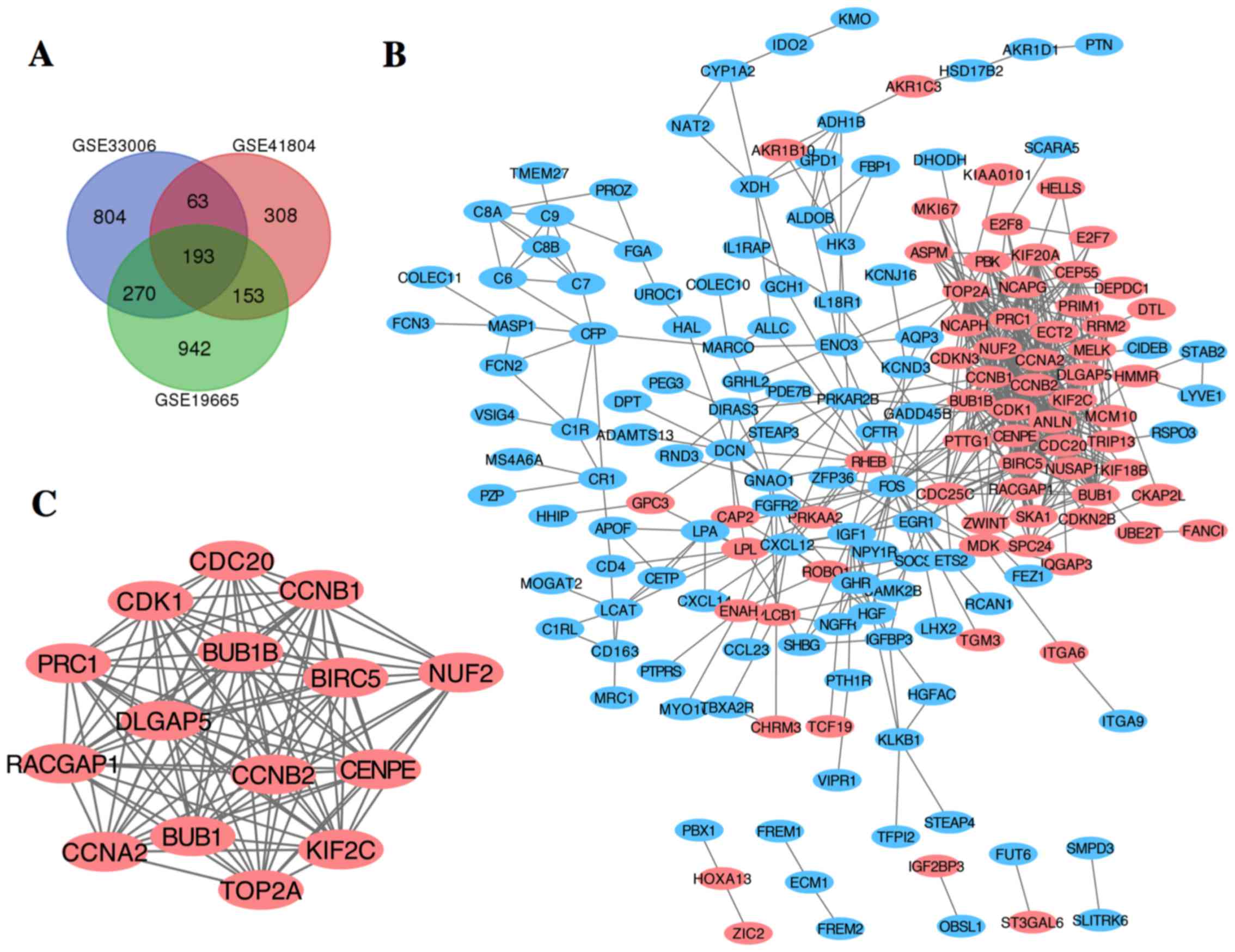
were identified. The overlap among the 3 datasets contained 273
genes as shown in the Venn diagram (Fig. 1A), consisting of 189 downregulated
genes and 84 upregulated genes between liver cancer tissues and
non-cancerous tissues.
KEGG and GO enrichment analyses of
DEGs
To analyze the biological classification of DEGs,
functional and pathway enrichment analyses were performed using
DAVID. GO analysis results showed that changes in biological
processes (BP) of DEGs were significantly enriched in protein
activation cascade, complement activation, defense response,
mitotic cell cycle and cell cycle process (Table I). Changes in molecular function
(MF) were mainly enriched in carbohydrate binding, oxidoreductase
activity, mannose-binding, scavenger receptor activity and
monosaccharide binding (Table I).
Changes in cell component (CC) of DEGs were mainly enriched in the
extracellular region, membrane attack complex and chromosome
(Table I). KEGG pathway analysis
revealed that the downregulated DEGs were mainly enriched in
complement and coagulation cascades, glycolysis/gluconeogenesis and
metabolic pathways, while the upregulated DEGs were mainly enriched
in oocyte meiosis, cell cycle and progesterone-mediated oocyte
maturation.
 | Table I.GO and KEGG pathway enrichment
analysis of DEGs in HCC samples. |
Table I.
GO and KEGG pathway enrichment
analysis of DEGs in HCC samples.
| Term | Description | Count in gene
set | P-value |
|---|
| Downregulated |
|
|
|
|
GO:0072376 | Protein activation
cascade | 16 | 5.27E-15 |
|
GO:0006956 | Complement
activation | 14 | 5.47E-15 |
|
GO:0006952 | Defense
response | 41 | 2.24E-09 |
|
GO:0030246 | Carbohydrate
binding | 13 | 0.00398 |
|
GO:0005537 |
Mannose-binding | 4 | 0.0216 |
|
GO:0016491 | Oxidoreductase
activity | 18 | 0.0216 |
|
GO:0005615 | Extracellular
space | 42 | 1.89E-11 |
|
GO:0005576 | Extracellular
region | 80 | 2.41E-10 |
|
GO:0044421 | Extracellular
region part | 69 | 6.95E-09 |
|
Hsa04610 | Complement and
coagulation cascades | 10 | 1.37E-07 |
|
Hsa05020 | Prion diseases | 6 | 0.000106 |
|
Hsa00232 | Caffeine
metabolism | 3 | 0.000665 |
|
Hsa01100 | Metabolic
pathways | 25 | 0.00296 |
|
Hsa00010 |
Glycolysis/gluconeogenesis | 5 | 0.00958 |
| Upregulated |
|
|
|
|
GO:0022402 | Cell cycle
process | 45 | 4.36E-34 |
|
GO:0007049 | Cell cycle | 48 | 9.01E-34 |
|
GO:0000278 | Mitotic cell
cycle | 41 | 6.08E-33 |
|
GO:0000793 | Condensed
chromosome | 15 | 3.67E-13 |
|
GO:0005819 | Spindle | 17 | 3.67E-13 |
|
GO:0005694 | Chromosome | 23 | 1.05E-12 |
|
Hsa04110 | Cell cycle | 9 | 3.84E-07 |
|
Hsa04114 | Oocyte meiosis | 6 | 0.00058 |
|
Hsa04914 |
Progesterone-mediated oocyte
maturation | 5 | 0.00156 |
PPI network construction and module
analysis
The PPI network of DEGs was constructed (Fig. 1B) and the most significant module
was obtained using Cytoscape (Fig.
1C). The functional analyses of genes involved in this module
were analyzed using DAVID. Results showed that genes in this module
were mainly enriched in cell division, mitotic nuclear division and
cell cycle (Table II).
 | Table II.GO and KEGG pathway enrichment
analysis of DEGs in the most significant module. |
Table II.
GO and KEGG pathway enrichment
analysis of DEGs in the most significant module.
| Pathway ID | Pathway
description | Count in gene
set | FDR |
|---|
| GO:0051301 | Cell division | 14 | 9.99E-19 |
| GO:0000280 | Nuclear
division | 13 | 4.42E-17 |
| GO:0007067 | Mitotic nuclear
division | 12 | 3.42E-16 |
| GO:1903047 | Mitotic cell cycle
process | 13 | 1.75E-14 |
| GO:0000278 | Mitotic cell
cycle | 13 | 6.11E-14 |
| GO:0005819 | Spindle | 9 | 7.60E-11 |
| GO:0000793 | Condensed
chromosome | 7 | 1.36E-08 |
| GO:0000777 | Condensed
chromosome kinetochore | 6 | 1.98E-08 |
| GO:0000779 | Condensed
chromosome, centromeric region | 6 | 2.18E-08 |
| GO:0015630 | Microtubule
cytoskeleton | 10 | 3.12E-08 |
| Hsa04110 | Cell cycle | 6 | 5.20E-08 |
| Hsa04914 |
Progesterone-mediated oocyte
maturation | 4 | 3.91E-05 |
| Hsa04114 | Oocyte meiosis | 4 | 8.19E-05 |
Hub gene selection and analysis
A total of 16 genes were identified as hub genes
with degrees ≥10. The names, abbreviations and functions for these
hub genes are shown in Table III.
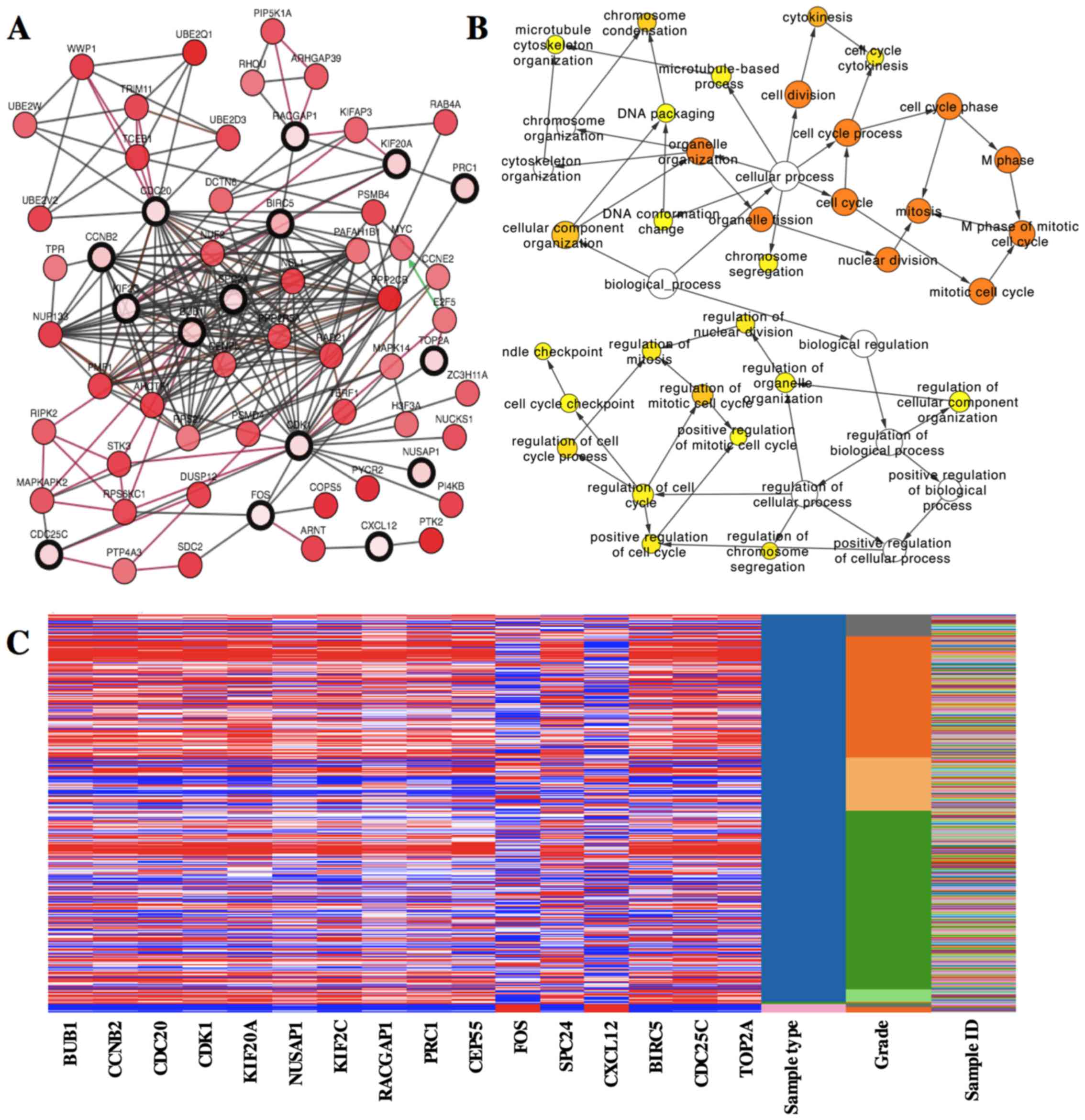
A network of the hub genes and their co-expression genes was
analyzed using cBioPortal online platform (Fig. 2A). The biological process analysis
of the hub genes is shown in Fig.
2B. Hierarchical clustering showed that the hub genes could
basically differentiate the liver cancer samples from the
non-cancerous samples (Fig. 2C).
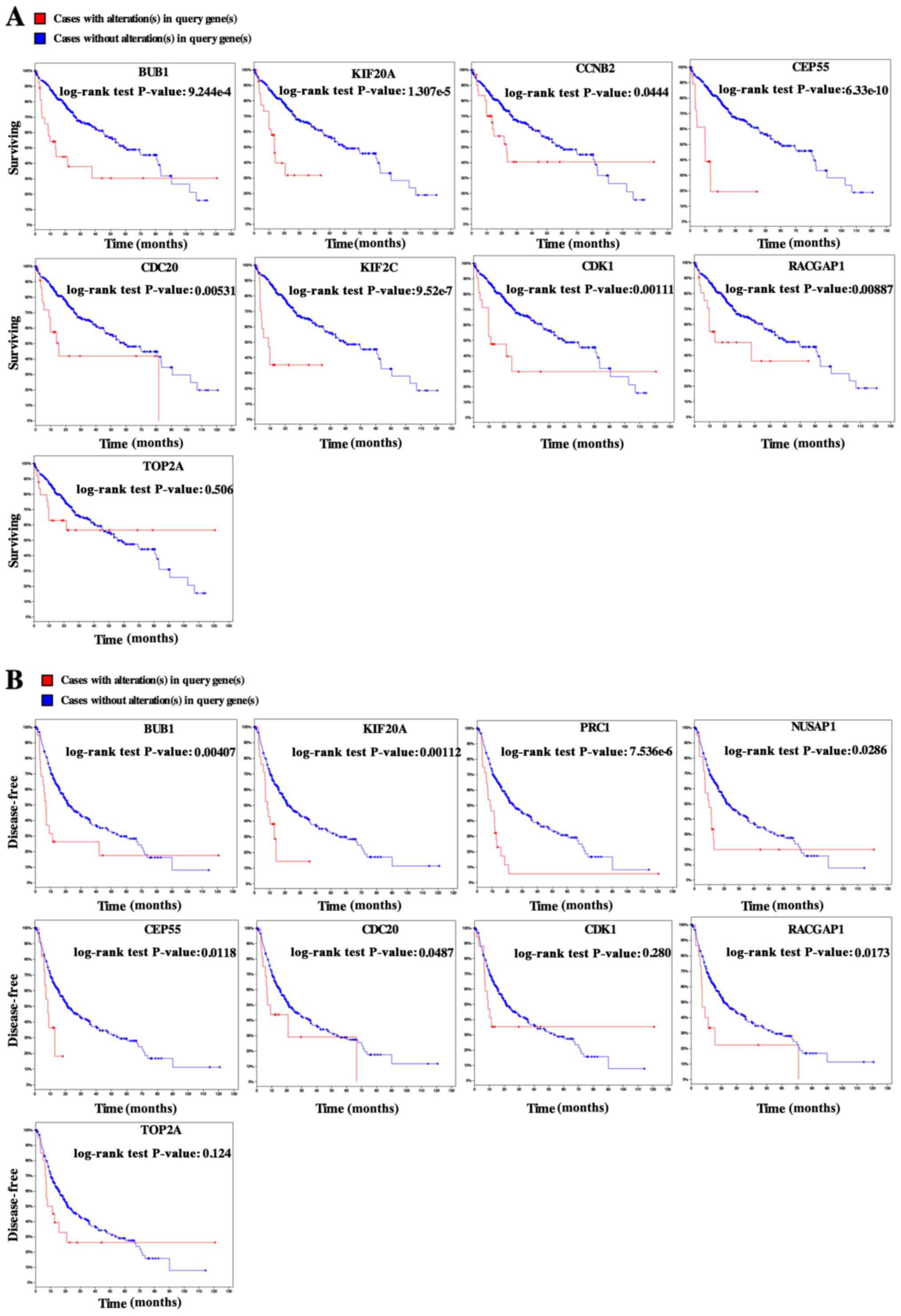
Subsequently, the overall survival analysis of the hub genes was
performed using Kaplan-Meier curve. HCC patients with BUB1, CCNB2,
CDC20, CDK1, KIF20A, KIF2C, RACGAP1 and CEP55 alteration showed
worse overall survival (Fig. 3A).
Nonetheless, HCC patients with BUB1, CDC20, KIF20A, NUSAP1,
RACGAP1, PRC1 and CEP55 alteration showed worse disease-free
survival (Fig. 3B).
 | Table III.Functional roles of 16 hub genes with
degree ≥10. |
Table III.
Functional roles of 16 hub genes with
degree ≥10.
| No. | Gene symbol | Full name | Function |
|---|
| 1 | BIRC5 | Baculoviral IAP
repeat containing 5 | BIRC5 may prevent
apoptotic cell death and is highly expressed in most tumors |
| 2 | BUB1 | BUB1 mitotic
checkpoint serine/threonine kinase | BUB1 promotes the
progression of breast cancer |
| 3 | CCNB2 | Cyclin B2 | CCNB2 (cyclin B2)
is associated with invasion, metastasis and poor prognosis of
several cancers |
| 4 | CDC20 | Cell division cycle
20 | High expression of
CDC20 is associated with development and progression of HCC |
| 5 | CDC25C | Cell division cycle
25C | CDC25C can regulate
the G2/M transition in HCC cells |
| 6 | CDK1 | Cyclin-dependent
kinase 1 | CDK1 can regulate
the cell cycle progression, apoptosis and carcinogenesis of tumor
cells |
| 7 | CEP55 | Centrosomal protein
55 | High expression of
CEP55 can promote the proliferation of lung, breast and thyroid
cancers |
| 8 | CXCL12 | C-X-C motif
chemokine ligand 12 | High expression of
CXCL12 in tumor cells may impede tumor spread |
| 9 | FOS | FBJ murine
osteosarcoma viral oncogene homolog | FOS has been
implicated as a regulator of cell proliferation, differentiation
and transformation |
| 10 | KIF20A | Kinesin family
member 20A | High expression of
KIF20A is involved in the development and progression of various
cancers |
| 11 | NUSAP1 | Nucleolar and
spindle associated protein 1 | High expression of
NUSAP1 is involved in the progression of prostate cancer |
| 12 | KIF2C | Kinesin family
member 2C | KIF2C is
overexpressed in various cancers and may be associated with the
chemoresistance of ovarian cancer |
| 13 | RACGAP1 | Rac GTPase
activating protein 1 | RACGAP1 plays a
regulatory role in cytokinesis, cell growth and
differentiation |
| 14 | PRC1 | Protein regulator
of cytokinesis 1 | PRC1 may be a novel
regulator of early HCC recurrence |
| 15 | SPC24 | SPC24, NDC80
kinetochore complex component | High expression of
SPC24 is associated with worse disease-free survival and overall
survival in HCC |
| 16 | TOP2A | Topoisomerase (DNA)
II α | TOP2A acts as a
target for several anticancer agents and mutations of this gene
have been associated with drug resistance |
Among these genes, TOP2A and CDK1 showed the highest
node degrees with 33, suggesting that they may play important roles
in the carcinogenesis or progression of HCC. Using the data from
cBioPortal, we noted that HCC patients who had an association of
genomic alterations in TOP2A showed reductions in overall and
disease-free survival. However, those observations were not
statistically significant (P=0.506 for overall survival and P=0.124
for disease-free survival). In addition, the CDK1 alteration was
significantly associated with worse overall survival but not
disease-free survival (P=0.00111 for overall survival and P=0.280
for disease-free survival) (Fig.
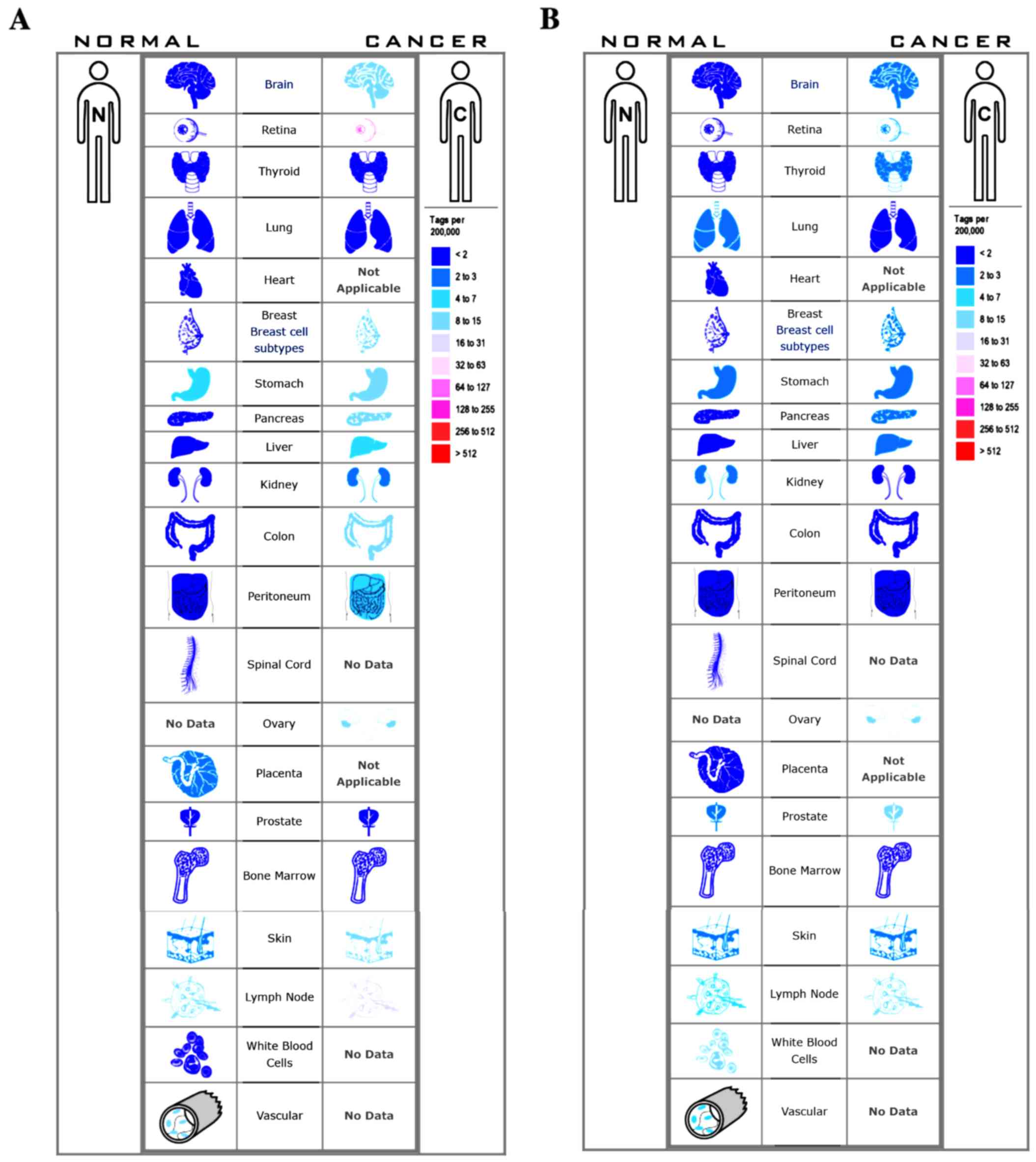
3B). The expression profile of TOP2A and CDK1 in human tissue
was displayed using SAGE. We found that TOP2A mRNA in brain,
retina, breast, pancreas, liver, kidney, colon and peritoneum
displayed higher levels as compared with the matched normal tissues
(Fig. 4A). CDK1 mRNA in brain,
retina, thyroid, breast, pancreas, liver and prostate displayed
higher levels as compared with the matched normal tissues (Fig. 4B). Oncomine analysis of cancer vs.
normal tissue showed that TOP2A and CDK1 were significantly
overexpressed in HCC in the different datasets (Fig. 5A and B). In the Wurmbach Liver
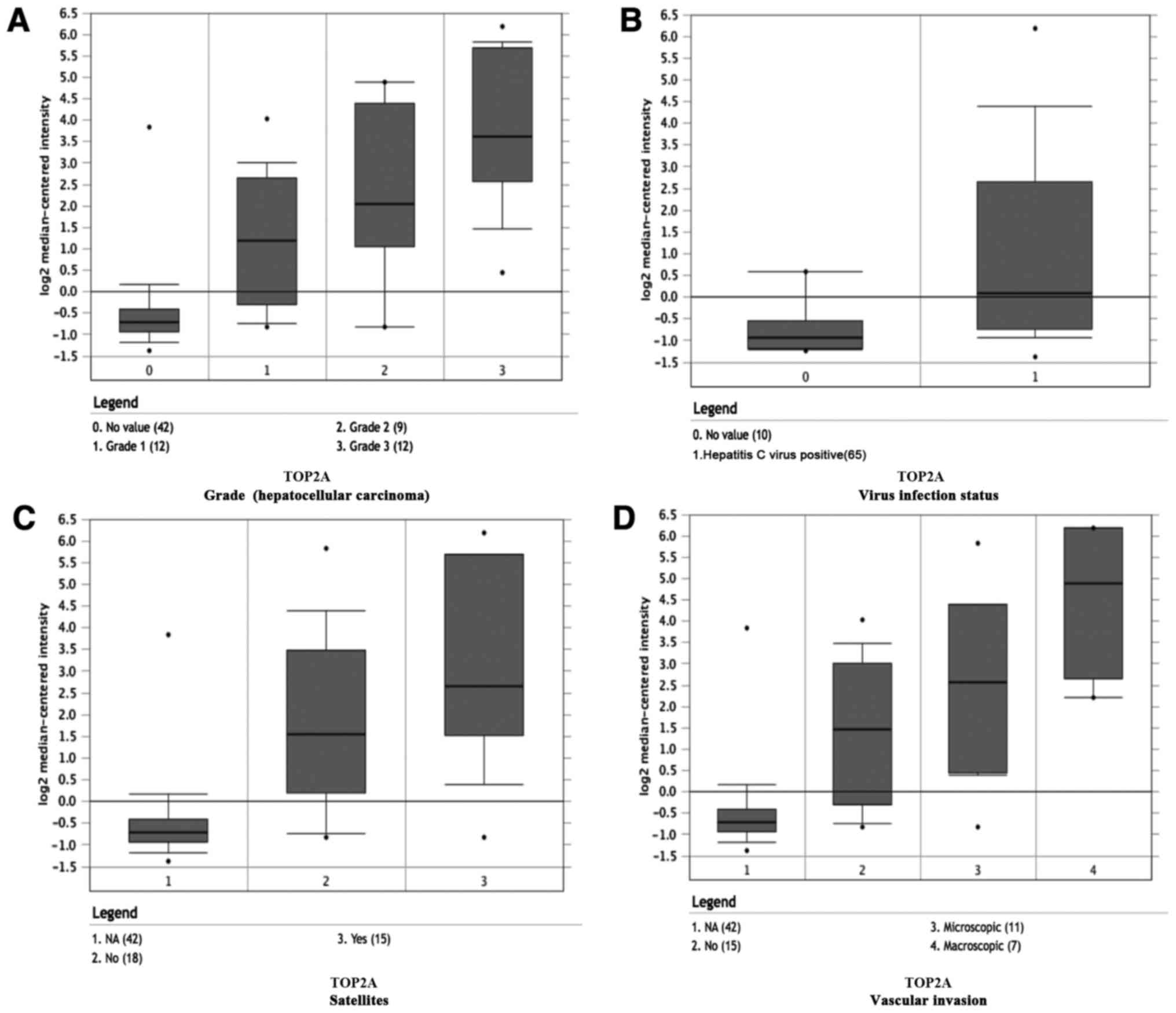
dataset, higher mRNA levels of TOP2A and CDK1 were associated with
tumor grade, hepatitis virus infection status, satellites and
vascular invasion (Fig. 6A-H).
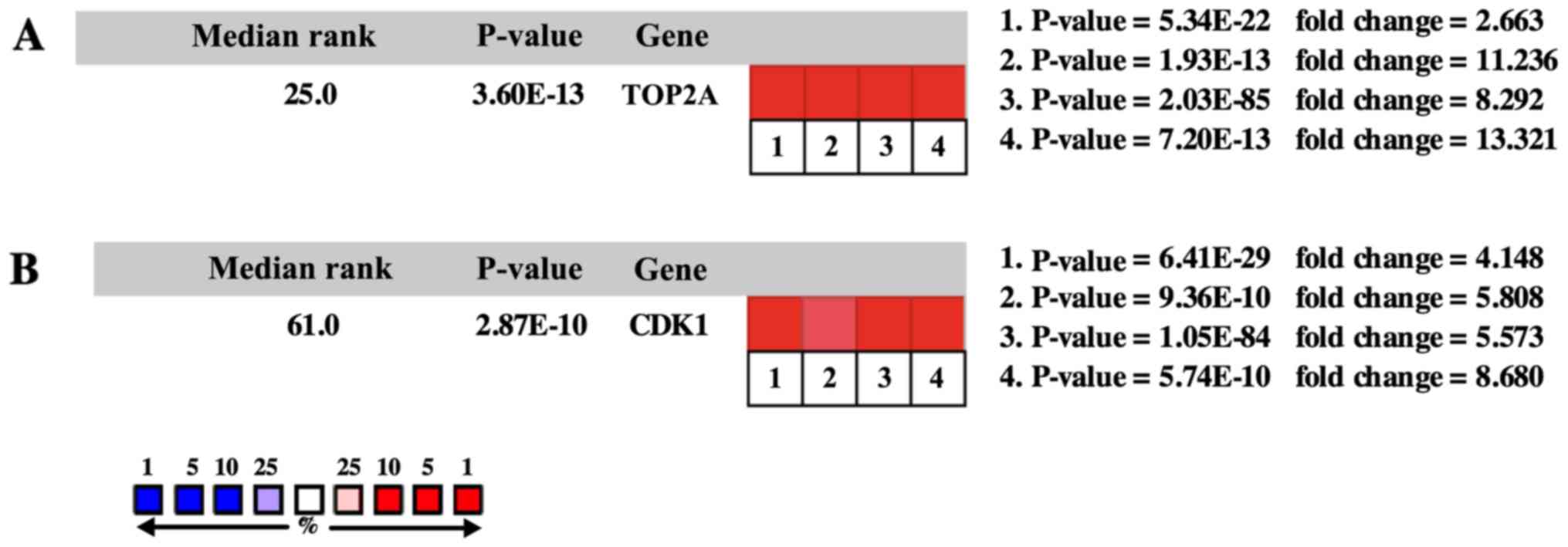 | Figure 5.Oncomine analysis of cancer vs.
normal tissue of (A) TOP2A and (B) CDK1. Heat maps of TOP2A and
CDK1 gene expression in clinical hepatocellular carcinoma samples
vs. normal tissues. 1. Hepatocellular carcinoma vs. normal liver,
Chen, Mol Biol Cell, 2002 (20). 2.
Hepatocellular carcinoma vs. normal liver, Roessler, Cancer Res,
2010 (21). 3. Hepatocellular
carcinoma vs. normal liver, Roessler, Cancer Res, 2010 (21). 4. Hepatocellular carcinoma vs.
normal liver, Wurmbach, Hepatology, 2007 (22). |
Discussion
Hepatocellular carcinoma (HCC) is the fifth most
common malignant tumor worldwide and its mortality rate has
increased in recent years (1). The
main etiology factors of HCC include chronic infection with
hepatitis viruses, gene mutations, cell damage, alcoholic liver
diseases and aflatoxin poisoning (23). The most common cause is chronic
infection with hepatitis B virus (HBV) or hepatitis C virus (HCV),
accounting for over 80% of HCC cases. However, the molecular
mechanisms of HCC remain poorly understood. Cell cycle regulators
play important roles in HCC. Mutation of cyclin D1 (CCND1), c-myc
or Ras, hypermethylation of cyclin D2 (CCND2) promoter and aberrant
expression of p53 or p21 have been reported to be involved in HCC
(24,25). In addition, splicing alterations of
NT5E, Sulf1 or SLC39A14 have been reported to be associated with
HCC (26–28). Most cases of HCC without an early
finding are not candidates for curative therapies, which may be one
of the reasons for the poor patient prognosis. Thus, potential
markers for diagnosis and treatment with high efficiency are
urgently demanded. Microarray technology enables us to explore the
genetic alterations in HCC, and has been proved to be a useful
approach to identify new biomarkers in other diseases.
In the present study, 3 mRNA microarray datasets
were analyzed to obtain DEGs between liver cancer tissues and
non-cancerous tissues. A total of 273 DEGs were identified among
the 3 datasets, including 189 downregulated genes and 84
upregulated genes. GO and KEGG enrichment analyses were performed
to explore interactions among the DEGs. The upregulated genes were
mainly enriched in oocyte meiosis, mitotic cell cycle and cell
cycle, while the downregulated genes were mainly enriched in
protein activation cascade, complement activation and complement
and coagulation cascades. Previous studies have reported that
dysregulation of the cell cycle process and mitotic cell cycle play
important roles in the carcinogenesis or progression of tumors
(24,25,29).
In addition, recent studies have brought forward a tumor-promoting
role for complement activation (30). Moreover, oxidoreductase activity
often plays a major role in antioxidant defense and can encode
tumor suppressors that are frequently altered in tumors (31,32).
In a word, all these theories are consistent with our results. GO
enrichment analysis revealed that changes in the most significant
modules were mainly enriched in cell division, nuclear division and
mitotic cell cycle process, while changes in KEGG were mainly
enriched in cell cycle, progesterone-mediated oocyte maturation and
oocyte meiosis.
We selected 16 DEGs as hub genes with degrees ≥10.
Among these hub genes, TOP2A and CDK1 showed the highest node
degrees with 33. TOP2A, which forms breaks in double-stranded DNA
and alters DNA structure during transcription, has been shown to be
correlated with early age onset, microvascular invasion, shorter
patient survival, chemoresistance and recurrence in HCC (33,34).
Thus, it is regarded as a target for anticancer agents, such as
epirubicin, doxorubicin, etoposide and temozolomide (35–37).
In HER2-amplified breast cancer, HER2 and TOP2A genes are
frequently co-amplified (38).
However, TOP2A overexpression in HCC is independent from HER2
amplification or overexpression (39). In addition, TOP2A overexpression has
also been found in lung, colon and ovarian cancers, and may be
regarded as a valuable biomarker for diagnosis, treatment and
prognosis of tumors (40–42). In the present study, PPI network
showed that TOP2A directly interacts with CDK1, RACGAP1, BIRC5 and
PRC1, indicating a key role of TOP2A in HCC. Cyclin-dependent
kinase 1 (CDK1), a serine/threonine kinase, regulates cell cycle
progression by binding to cyclin B to form a complex called cyclin
B-CDK1. miR-582-5p was found to regulate the progression of HCC by
inhibiting the expression of CDK1 (43). CDK1 overexpression has also been
found in lung, pancreas and other cancers. In addition to its role
in cell cycle progression, cyclin B-CDK1 could interact with
apoptin and regulate the subcellular localization of apoptin,
leading to apoptosis and carcinogenesis (44). Survivin inactivates and blocks CDK1
by increasing the level of Wee1 and thus, inactivates and blocks
the pro-apoptotic activity of CDK1 (45). Cyclin B-CDK1 is also involved in
connecting mitotic arrest and apoptosis by mediating the
phosphorylation of anti-apoptotic Bcl-2 (46). We assessed the expression of TOP2A
and CDK1 in relation to overall and disease-free survival. Gene
alteration in TOP2A showed reductions in overall and disease-free
survival. However, in the present study, those observations were
not statistically significant. In addition, the CDK1 alteration was
significantly associated with worse overall survival, but not
disease-free survival. Nevertheless, several clinical studies have
shown that overexpression of TOP2A is significantly associated with
shorter survival times (37,39).
We speculate that the reason may be that survival analyses in
cBioPortal were performed on the basis of the relationship between
gene mutation and prognosis, while gene overexpression usually
arises from mutation or amplification. Thus, overexpression of
TOP2A in HCC may arise from gene amplification rather than
mutation, and further research is needed to confirm our hypothesis.
Oncomine analysis showed that higher mRNA levels of TOP2A and CDK1
were associated with tumor grade, hepatitis virus infection status,
satellites and vascular invasion, indicating vital roles of TOP2A
and CDK1 in the carcinogenesis or progression of HCC.
RACGAP1, a Rac- and Cdc42-specific GAP, can suppress
Rac and activate RhoA, leading to the promotion of pseudopod
extension and invasion (47).
Moreover, RACGAP1 shows a relationship with AURKA, a negative
prognostic indicator in gastric cancer (48). BIRC5, also called survivin, is
overexpressed and plays important roles in cell division and
proliferation in a majority of cancers including HCC. PRC1 is
involved in the microtubule organization in eukaryotes and is
upregulated in breast tumors (49).
Recent research has found that it can be a novel regulator of early
HCC recurrence via potentiating Wnt signaling (50). High expression of CDC20 is
associated with sex, differentiation, tumor-node-metastasis (TNM)
stage, and p53 expression of HCC (51). The protein kinase CDC25C acts as an
activator of cyclin B-CDK1 that regulates the G2/M transition in
HCC cells (52). HCC patients with
relatively low CXCL12 mRNA levels exhibit worse overall survival
(53). A recent study found that
the transcriptional activity of c-FOS could be induced by membrane
melanoma cell adhesion molecule (MCAM), and it is important for
MCAM-induced liver tumorigenesis (54). High expression of SPC24 is
associated with worse disease-free and overall survival in HCC
patients (55). High expression of
KIF20A is involved in the development and progression of various
cancers such as pancreatic, bladder and breast cancer. A recent
study found that the glioma-associated oncogene 2/KIF20A axis is
crucial for the proliferation of human HCC cells (56).
Literature retrieval results showed that the
interaction among HCC and hub genes NUSAP1, CEP55, BUB1, CCNB2,
KIF2C and RACGAP1 has not been widely reported. NUSAP1 regulates
mitosis, and high expression of NUSAP1 is involved in the
progression of prostate cancer (57). CEP55 is a centrosome-associated
protein, which plays an important role in regulating the cell
cycle. Overexpression of CEP55 was found to promote the
proliferation of several cancers, such as pulmonary adenocarcinoma,
breast cancer and anaplastic thyroid carcinoma (58–60).
BUB1, a component of the spindle assembly checkpoint, is
overexpressed and plays important roles in the progression of
breast cancer (61). However, BUB1
has been reported to have a controversial role in spindle assembly
checkpoint activation (62,63), which needs further investigation.
CCNB2 is overexpressed in bladder, lung and colorectal cancers, and
is associated with invasion, metastasis and poor prognosis of
cancers (64,65). KIF2C plays an important role in the
segregation of chromosomes in mitosis, and is overexpressed in
various cancers and may be associated with the chemoresistance of
ovarian cancer (66,67). In addition, we also performed
hierarchical clustering for hub genes. Results showed that these
hub genes differentiated HCC samples from non-cancerous samples,
and may be candidates for diagnostic biomarkers. Moreover,
alteration of BUB1, CDC20, KIF20A, RACGAP1 and CEP55 is involved in
worse overall and disease-free survival, indicating that these
genes may play important roles in the carcinogenesis, progression,
invasion or recurrence of HCC.
In conclusion, the present study was designed to
identify DEGs that may be involved in the carcinogenesis or
progression of HCC. A total of 273 DEGs and 16 hub genes were
identified and may be regarded as diagnostic biomarkers for HCC.
However, further studies are needed to elucidate the biological
function of these genes in HCC.
Acknowledgements
The present study was supported by a grant (no.
81271838) from the National Science Foundation of China.
References
|
1
|
El-Serag HB and Rudolph KL: Hepatocellular
carcinoma: Epidemiology and molecular carcinogenesis.
Gastroenterology. 132:2557–2576. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Hu Z, Zhou Z, Xiong G, Wang Y, Lai Y, Deng
L and Yang J: Cyclin D1 G870A polymorphism and the risk of
hepatocellular carcinoma in a Chinese population. Tumour Biol.
35:5607–5612. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Berasain C, Castillo J, Prieto J and Avila
MA: New molecular targets for hepatocellular carcinoma: The ErbB1
signaling system. Liver Int. 27:174–185. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Gan FY, Gesell MS, Alousi M and Luk GD:
Analysis of ODC and c-myc gene expression in hepatocellular
carcinoma by in situ hybridization and immunohistochemistry. J
Histochem Cytochem. 41:1185–1196. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Newell P, Toffanin S, Villanueva A, Chiang
DY, Minguez B, Cabellos L, Savic R, Hoshida Y, Lim KH,
Melgar-Lesmes P, et al: Ras pathway activation in hepatocellular
carcinoma and anti-tumoral effect of combined sorafenib and
rapamycin in vivo. J Hepatol. 51:725–733. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Edgar R, Domrachev M and Lash AE: Gene
Expression Omnibus: NCBI gene expression and hybridization array
data repository. Nucleic Acids Res. 30:207–210. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Elgaaen BV, Olstad OK, Sandvik L, Odegaard
E, Sauer T, Staff AC and Gautvik KM: ZNF385B and VEGFA are strongly
differentially expressed in serous ovarian carcinomas and correlate
with survival. PLoS One. 7:e463172012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Mok SC, Bonome T, Vathipadiekal V, Bell A,
Johnson ME, Wong KK, Park DC, Hao K, Yip DK, Donninger H, et al: A
gene signature predictive for outcome in advanced ovarian cancer
identifies a survival factor: Microfibril-associated glycoprotein
2. Cancer Cell. 16:521–532. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bowen NJ, Walker LD, Matyunina LV, Logani
S, Totten KA, Benigno BB and McDonald JF: Gene expression profiling
supports the hypothesis that human ovarian surface epithelia are
multipotent and capable of serving as ovarian cancer initiating
cells. BMC Med Genomics. 2:712009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Huang DW, Sherman BT, Tan Q, Collins JR,
Alvord WG, Roayaei J, Stephens R, Baseler MW, Lane HC and Lempicki
RA: The DAVID Gene Functional Classification Tool: A novel
biological module-centric algorithm to functionally analyze large
gene lists. Genome Biol. 8:R1832007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kanehisa M: The KEGG database. Novartis
Found Symp. 247:91–252. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ashburner M, Ball CA, Blake JA, Botstein
D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT,
et al: Gene ontology: Tool for the unification of biology. The Gene
Ontology Consortium. Nat Genet. 25:25–29. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Franceschini A, Szklarczyk D, Frankild S,
Kuhn M, Simonovic M, Roth A, Lin J, Minguez P, Bork P, von Mering
C, et al: STRING v9.1: Protein-protein interaction networks, with
increased coverage and integration. Nucleic Acids Res.
41:D808–D815. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Smoot ME, Ono K, Ruscheinski J, Wang PL
and Ideker T: Cytoscape 2.8: New features for data integration and
network visualization. Bioinformatics. 27:431–432. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Bandettini WP, Kellman P, Mancini C,
Booker OJ, Vasu S, Leung SW, Wilson JR, Shanbhag SM, Chen MY and
Arai AE: MultiContrast Delayed Enhancement (MCODE) improves
detection of subendocardial myocardial infarction by late
gadolinium enhancement cardiovascular magnetic resonance: A
clinical validation study. J Cardiovasc Magn Reson. 14:832012.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Gao J, Aksoy BA, Dogrusoz U, Dresdner G,
Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al:
Integrative analysis of complex cancer genomics and clinical
profiles using the cBioPortal. Sci Signal. 6:pl12013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Cerami E, Gao J, Dogrusoz U, Gross BE,
Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et
al: The cBio cancer genomics portal: An open platform for exploring
multidimensional cancer genomics data. Cancer Discov. 2:401–404.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Maere S, Heymans K and Kuiper M: BiNGO: A
Cytoscape plugin to assess overrepresentation of gene ontology
categories in biological networks. Bioinformatics. 21:3448–3449.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kent WJ, Sugnet CW, Furey TS, Roskin KM,
Pringle TH, Zahler AM and Haussler D: The human genome browser at
UCSC. Genome Res. 12:996–1006. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chen X, Cheung ST, So S, Fan ST, Barry C,
Higgins J, Lai KM, Ji J, Dudoit S, Ng IO, et al: Gene expression
patterns in human liver cancers. Mol Biol Cell. 13:1929–1939. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Roessler S, Jia HL, Budhu A, Forgues M, Ye
QH, Lee JS, Thorgeirsson SS, Sun Z, Tang ZY, Qin LX, et al: A
unique metastasis gene signature enables prediction of tumor
relapse in early-stage hepatocellular carcinoma patients. Cancer
Res. 70:10202–10212. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wurmbach E, Chen YB, Khitrov G, Zhang W,
Roayaie S, Schwartz M, Fiel I, Thung S, Mazzaferro V, Bruix J, et
al: Genome-wide molecular profiles of HCV-induced dysplasia and
hepatocellular carcinoma. Hepatology. 45:938–947. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Turner PC, Sylla A, Diallo MS, Castegnaro
JJ, Hall AJ and Wild CP: The role of aflatoxins and hepatitis
viruses in the etiopathogenesis of hepatocellular carcinoma: A
basis for primary prevention in Guinea-Conakry, West Africa. J
Gastroenterol Hepatol. 17 Suppl:S441–S448. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wang Y, Cheng J, Xu C, Liu S, Jiang S, Xu
Q, Chen X, Zhuang H and Lu F: Quantitative methylation analysis
reveals gender and age differences in p16INK4a hypermethylation in
hepatitis B virus-related hepatocellular carcinoma. Liver Int.
32:420–428. 2012.PubMed/NCBI
|
|
25
|
Choi YL, Park SH, Jang JJ and Park CK:
Expression of the G1-S modulators in hepatitis B virus-related
hepatocellular carcinoma and dysplastic nodule: Association of
cyclin D1 and p53 proteins with the progression of hepatocellular
carcinoma. J Korean Med Sci. 16:424–432. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Snider NT, Altshuler PJ, Wan S, Welling
TH, Cavalcoli J and Omary MB: Alternative splicing of human NT5E in
cirrhosis and hepatocellular carcinoma produces a negative
regulator of ecto-5-nucleotidase (CD73). Mol Biol Cell.
25:4024–4033. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Gill RBS, Day A, Barstow A, Zaman G, Chenu
C and Dhoot GK: Mammalian Sulf1 RNA alternative splicing and its
significance to tumour growth regulation. Tumour Biol.
33:1669–1680. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Franklin RB, Levy BA, Zou J, Hanna N,
Desouki MM, Bagasra O, Johnson LA and Costello LC: ZIP14 zinc
transporter downregulation and zinc depletion in the development
and progression of hepatocellular cancer. J Gastrointest Cancer.
43:249–257. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Tripathi V, Shen Z, Chakraborty A, Giri S,
Freier SM, Wu X, Zhang Y, Gorospe M, Prasanth SG, Lal A, et al:
Long noncoding RNA MALAT1 controls cell cycle progression by
regulating the expression of oncogenic transcription factor B-MYB.
PLoS Genet. 9:e10033682013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Markiewski MM and Lambris JD: Unwelcome
complement. Cancer Res. 69:6367–6370. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Abu-Remaileh M and Aqeilan RI: The tumor
suppressor WW domain-containing oxidoreductase modulates cell
metabolism. Exp Biol Med. 240:345–350. 2015. View Article : Google Scholar
|
|
32
|
Roszczenko P, Radomska KA, Wywial E,
Collet JF and Jagusztyn-Krynicka EK: A novel insight into the
oxidoreductase activity of Helicobacter pylori HP0231 protein. PLoS
One. 7:e465632012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Watanuki A, Ohwada S, Fukusato T, Makita
F, Yamada T, Kikuchi A and Morishita Y: Prognostic significance of
DNA topoisomerase IIalpha expression in human hepatocellular
carcinoma. Anticancer Res. 22:1113–1119. 2002.PubMed/NCBI
|
|
34
|
Nakajima T, Yasui K, Zen K, Inagaki Y,
Fujii H, Minami M, Tanaka S, Taniwaki M, Itoh Y, Arii S, et al:
Activation of B-Myb by E2F1 in hepatocellular carcinoma. Hepatol
Res. 38:886–895. 2008.PubMed/NCBI
|
|
35
|
Chan HH, Chu TH, Chien HF, Sun CK, Wang
EM, Pan HB, Kuo HM, Hu TH, Lai KH, Cheng JT, et al: Rapid induction
of orthotopic hepatocellular carcinoma in immune-competent rats by
non-invasive ultrasound-guided cells implantation. BMC
Gastroenterol. 10:832010. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wang N, Zhu M, Tsao SW, Man K, Zhang Z and
Feng Y: MiR-23a-mediated inhibition of topoisomerase 1 expression
potentiates cell response to etoposide in human hepatocellular
carcinoma. Mol Cancer. 12:1192013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wong N, Yeo W, Wong WL, Wong NL, Chan KY,
Mo FK, Koh J, Chan SL, Chan AT, Lai PB, et al: TOP2A overexpression
in hepatocellular carcinoma correlates with early age onset,
shorter patients survival and chemoresistance. Int J Cancer.
124:644–652. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Fritz P, Cabrera CM, Dippon J, Gerteis A,
Simon W, Aulitzky WE and van der Kuip H: c-erbB2 and topoisomerase
IIalpha protein expression independently predict poor survival in
primary human breast cancer: A retrospective study. Breast Cancer
Res. 7:R374–R384. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Panvichian R, Tantiwetrueangdet A,
Angkathunyakul N and Leelaudomlipi S: TOP2A amplification and
overexpression in hepatocellular carcinoma tissues. Biomed Res Int.
2015:3816022015. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Dingemans AM, Witlox MA, Stallaert RA, van
der Valk P, Postmus PE and Giaccone G: Expression of DNA
topoisomerase IIalpha and topoisomerase IIbeta genes predicts
survival and response to chemotherapy in patients with small cell
lung cancer. Clin Cancer Res. 5:2048–2058. 1999.PubMed/NCBI
|
|
41
|
Lazaris AC, Kavantzas NG, Zorzos HS,
Tsavaris NV and Davaris PS: Markers of drug resistance in relapsing
colon cancer. J Cancer Res Clin Oncol. 128:114–118. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Costa MJ, Hansen CL, Holden JA and Guinee
D Jr: Topoisomerase II alpha: Prognostic predictor and cell cycle
marker in surface epithelial neoplasms of the ovary and peritoneum.
Int J Gynecol Pathol. 19:248–257. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Zhang Y, Huang W, Ran Y, Xiong Y, Zhong Z,
Fan X, Wang Z and Ye Q: miR-582-5p inhibits proliferation of
hepatocellular carcinoma by targeting CDK1 and AKT3. Tumour Biol.
36:8309–8316. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Zhao J, Han SX, Ma JL, Ying X, Liu P, Li
J, Wang L, Zhang Y, Ma J, Zhang L, et al: The role of CDK1 in
apoptin-induced apoptosis in hepatocellular carcinoma cells. Oncol
Rep. 30:253–259. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Guzman JR, Fukuda S and Pelus LM:
Inhibition of caspase-3 by Survivin prevents Wee1 Kinase
degradation and promotes cell survival by maintaining
phosphorylation of p34Cdc2. Gene Ther Mol Biol. 13B:1–273.
2009.
|
|
46
|
Terrano DT, Upreti M and Chambers TC:
Cyclin-dependent kinase 1-mediated Bcl-xL/Bcl-2
phosphorylation acts as a functional link coupling mitotic arrest
and apoptosis. Mol Cell Biol. 30:640–656. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Jacquemet G, Green DM, Bridgewater RE, von
Kriegsheim A, Humphries MJ, Norman JC and Caswell PT: RCP-driven
α5β1 recycling suppresses Rac and promotes RhoA activity via the
RacGAP1-IQGAP1 complex. J Cell Biol. 202:917–935. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Bornschein J, Nielitz J, Drozdov I,
Selgrad M, Wex T, Jechorek D, Link A, Vieth M and Malfertheiner P:
Expression of aurora kinase A correlates with the Wnt-modulator
RACGAP1 in gastric cancer. Cancer Med. 5:516–526. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Subramanian R, Ti SC, Tan L, Darst SA and
Kapoor TM: Marking and measuring single microtubules by PRC1 and
kinesin-4. Cell. 154:377–390. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Chen J, Rajasekaran M, Xia H, Zhang X,
Kong SN, Sekar K, Seshachalam VP, Deivasigamani A, Goh BK, Ooi LL,
et al: The microtubule-associated protein PRC1 promotes early
recurrence of hepatocellular carcinoma in association with the
Wnt/β-catenin signalling pathway. Gut. 65:1522–1534. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Li J, Gao JZ, Du JL, Huang ZX and Wei LX:
Increased CDC20 expression is associated with development and
progression of hepatocellular carcinoma. Int J Oncol. 45:1547–1555.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Deng LJ, Peng QL, Wang LH, Xu J, Liu JS,
Li YJ, Zhuo ZJ, Bai LL, Hu LP, Chen WM, et al: Arenobufagin
intercalates with DNA leading to G2 cell cycle arrest
via ATM/ATR pathway. Oncotarget. 6:34258–34275. 2015.PubMed/NCBI
|
|
53
|
Semaan A, Dietrich D, Bergheim D, Dietrich
J, Kalff JC, Branchi V, Matthaei H, Kristiansen G, Fischer HP and
Goltz D: CXCL12 expression and PD-L1 expression serve as prognostic
biomarkers in HCC and are induced by hypoxia. Virchows Arch.
470:185–196. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Wang J, Tang X, Weng W, Qiao Y, Lin J, Liu
W, Liu R, Ma L, Yu W, Yu Y, et al: The membrane protein melanoma
cell adhesion molecule (MCAM) is a novel tumor marker that
stimulates tumorigenesis in hepatocellular carcinoma. Oncogene.
34:5781–5795. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Zhu P, Jin J, Liao Y, Li J, Yu XZ, Liao W
and He S: A novel prognostic biomarker SPC24 up-regulated in
hepatocellular carcinoma. Oncotarget. 6:41383–41397. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Shi C, Huang D, Lu N, Chen D, Zhang M, Yan
Y, Deng L, Lu Q, Lu H and Luo S: Aberrantly activated Gli2-KIF20A
axis is crucial for growth of hepatocellular carcinoma and predicts
poor prognosis. Oncotarget. 7:26206–26219. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Gordon CA, Gulzar ZG and Brooks JD: NUSAP1
expression is upregulated by loss of RB1 in prostate cancer cells.
Prostate. 75:517–526. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Jiang W, Wang Z, Chen G and Jia Y:
Prognostic significance of centrosomal protein 55 in stage I
pulmonary adenocarcinoma after radical resection. Thorac Cancer.
7:316–322. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Wang Y, Jin T, Dai X and Xu J:
Lentivirus-mediated knockdown of CEP55 suppresses cell
proliferation of breast cancer cells. Biosci Trends. 10:67–73.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Weinberger P, Ponny SR, Xu H, et al: Cell
cycle M-phase genes are highly upregulated in anaplastic thyroid
carcinoma. Thyroid. 27:236–252. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Wang Z, Katsaros D, Shen Y, Fu Y, Canuto
EM, Benedetto C, Lu L, Chu WM, Risch HA and Yu H: Biological and
clinical significance of MAD2L1 and BUB1, genes frequently
appearing in expression signatures for breast cancer prognosis.
PLoS One. 10:e01362462015. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Kawashima SA, Yamagishi Y, Honda T,
Ishiguro K and Watanabe Y: Phosphorylation of H2A by Bub1 prevents
chromosomal instability through localizing shugoshin. Science.
327:172–177. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
London N and Biggins S: Mad1 kinetochore
recruitment by Mps1-mediated phosphorylation of Bub1 signals the
spindle checkpoint. Genes Dev. 28:140–152. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Takashima S, Saito H, Takahashi N, Imai K,
Kudo S, Atari M, Saito Y, Motoyama S and Minamiya Y: Strong
expression of cyclin B2 mRNA correlates with a poor prognosis in
patients with non-small cell lung cancer. Tumour Biol.
35:4257–4265. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Lei CY, Wang W, Zhu YT, Fang WY and Tan
WL: The decrease of cyclin B2 expression inhibits invasion and
metastasis of bladder cancer. Urol Oncol. 34:237.e1–237.e10. 2016.
View Article : Google Scholar
|
|
66
|
Zhao F, Siu MK, Jiang L, Tam KF, Ngan HY,
Le XF, Wong OG, Wong ES, Gomes AR, Bella L, et al: Overexpression
of forkhead box protein M1 (FOXM1) in ovarian cancer correlates
with poor patient survival and contributes to paclitaxel
resistance. PLoS One. 9:e1134782014. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Shimo A, Tanikawa C, Nishidate T, Lin ML,
Matsuda K, Park JH, Ueki T, Ohta T, Hirata K, Fukuda M, et al:
Involvement of kinesin family member 2C/mitotic
centromere-associated kinesin overexpression in mammary
carcinogenesis. Cancer Sci. 99:62–70. 2008.PubMed/NCBI
|