Introduction
Lung cancer is the most common cause of cancer
related mortality worldwide (1).
Lung adenocarcinoma (LAC) is currently the predominant histological
subtype of lung cancer and accounts for 38% of all newly diagnosed
lung cancers (2). Despite recent
advancement in targeted therapies such as EGFR tyrosine kinase
inhibitors, the average 5-year survival rate is only 15%,
highlighting the desperate need for additional therapeutic
strategies (3,4). This poor outcome is largely due to the
delay of early diagnosis and lack of effective treatment against
metastasis (5). Evidence suggests
once they have developed metastatic disease, only 15% patients
survive for 1 year after surgery with almost no long-term survivors
(6). Therefore, identification of
previously unrecognized molecular events that propel cancer
development, particularly metastasis, would not just be of
biological relevance, but also be of clinical significance allowing
development of novel biomarkers or drug targets for the management
of lung cancer patients.
14-3-3ζ, belongs to scaffold 14-3-3 proteins family,
regulates multiple signal transduction pathways and acts as an
oncogene in cancer. 14-3-3ζ was initially identified as metastasis
enhancer genes through comparing lung cancer cell lines with
different metastatic potentials (7). Furthermore, 14-3-3ζ was demonstrated
to promote invasiveness of cancer cells by activating
epithelial-mesenchymal transition (EMT) pathway (8–10). It
is of note that 14-3-3ζ has been recently identified as a
clinically relevant prognostic marker for breast cancer, prostate
cancer, gastric cancer, and head and neck cancer (11–14).
Given the important role of 14-3-3ζ in carcinogenesis, particularly
tumor metastasis, 14-3-3ζ may serve as a promising biomarker or
therapeutic target for lung adenocarcinoma. However, the oncogenic
function and clinical relevance of 14-3-3ζ in lung adenocarcinoma
remains largely unexplored.
Based upon this important gap in knowledge in the
literature, we envisaged this study to interrogate the molecular
contributions of 14-3-3ζ in lung adenocarcinoma, identify novel
downstream target of 14-3-3ζ, and decipher whether these genes may
have translational relevance as clinically-relevant disease
biomarkers or therapeutic target.
Materials and methods
Cell cultures
The cell lines A549 and H1299 were purchased from
the American Type Culture Collection (ATCC). Cells were cultured in
Dulbecco's modified Eagle's medium with 10% FBS and 1%
penicillin-streptomycin at 37°C in a humidified atmosphere of 5%
CO2. These cell lines were periodically authenticated
using a panel of genetic and epigenetic biomarkers.
Plasmid, siRNA and transfection
Validated siRNA for 14-3-3ζ and its negative control
were purchased from Ambion. LAC cells were seeded in 6-well plate
and transfected with siRNA at a final concentration of 50 nmol/l
using Lipofectamine RNAi MAX (Invitrogen) and Opti-MEM (Gibco)
according to the manufacturer's instructions when the cell density
reached 30–50% confluence. A549 and H1299 cells were also stably
transfected with a control pHR-CMV vector or one expressing MUC1-C
according to a previous study (15). The transfection efficiency was
evaluated at 48 and 72 h time points for evaluating corresponding
changes in the mRNA and protein expression, respectively.
Cell proliferation assays
Cells with different treatment were seeded at 2000
cells per well in 96-well plates following which 20 µl MTT reagent
(Sigma) was added to each well with 200 µl culture medium. The MTT
mixture medium were replaced by 150 µl dimethyl sulfoxide after
being incubated for 4 h at 37°C in a 5%CO2 incubator.
The time points were set at 0, 24, 48 and 72 h. Optical densities
were determined using the Infinite 200 Pro multi-readers and
i-control 1.10 software (Tecan).
Migration and invasion
Cell migration and invasion assays were performed
using Boyden chambers (Corning) using 8 µm-size pore membrane
coated with Matrigel (for invasion assays) or without Matrigel (for
migration assays). The Transfected cells were seeded onto inserts
at 2×105 cells in 200 µl serum-free medium and
transferred to lower wells with 600 µl culture medium containing
10% FBS. After 24 h incubation at 37°C in a 5% CO2
incubator, non-invading cells were removed by scraping the top
surface of the membrane. Invaded cells on the bottom of the
membrane were thereafter fixed and stained by using Diff-Quick
Stain kit (Thermo Scientific) according to the manufacturer's
instructions. The stained cells were counted from 10 different
fields randomly using a light microscope.
RNA extraction and qPCR
Cells were harvest at 24 h after transfection. Total
RNA was extracted using TRIzol (Invitrogen) reagent according to
the manufacturer's protocol. The qRT-PCR assays were performed
using QuantStudio 6 Flex Real-time PCR System (Applied Biosystems).
For gene-expression analysis, 2 µg of total RNA was synthesized to
cDNA by using GoScript™ Reverse Transcription System (Promega) and
the Fast SYBR Green Master Mix (Applied Biosystems). The relative
expression of target genes was normalized against β-actin using
2−∆∆ct method. The sequences of all primers were shown
as follows: E-cadherin: forward, AGCCCCGCCTTATGATTCTCTG, reverse,
TGCCCCA TTCGTTCAAGTAGTCAT; ZEB1: forward, AGCGCTT
CTCACACTCTGGGTCTT, reverse, CCTGCCTCTTCCTG CTCTGTGC; and β-actin:
forward, AGAGCTACGA GCTGCCTGAC, reverse, AGCACTGTGTTGGCGTACAG. For
miRNA analysis, qRT-PCR was conducted using TaqMan®
MicroRNA Reverse Transcription kit and TaqMan Universal PCR Master
Mix kit (Applied Biosystems) according to manufacturer's
instructions. Relative expression of miR-200c was determined by
2-∆∆ct method using U6 as normalizer. Taqman primers for
miR-200c and U6 were purchased from Ambion.
Immunoblot analysis
Cellular proteins were separated by SDS-PAGE gel and
transferred onto a polyvinylidene difluoride(PVDF) membrane
(Millipore) using Bio-Rad electrophoresis system. The PVDF membrane
was blocked in 5% non-fat milk for 1 h at room temperature and the
diluted primary antibodies (1–1000)
were incubated with the target protein overnight at 4°C.
Precipitates and lysates not subjected to precipitation were
analyzed by immunoblotting with the following antibodies:
anti-14-3-3ζ (sc-1019) and anti-aPKC antibodies (sc-216) were from
Santa Cruz Biotechnology; and anti-P-aPKCThr560 (CG1453)
antibody was purchased from Cell Applications. Anti-IκBα (4814),
anti-P-p65 (3303), anti-NF-κB p65 (8242), and anti-β-actin (4970)
antibodies were from Cell Signaling Technology. Immunoreactive
complexes were detected using horseradish peroxidase-conjugated
secondary antibodies (1:2000) and an enhanced chemiluminescence
(ECL) detection system.
Immunoprecipitation
Cells were lysed after transfection for 48 h using
NP-40 lysis buffer containing protease inhibitor cocktail (Thermo
Scientific). Soluble proteins were immunoprecipitated with
anti-MUC1, then 5 µg antibody was incubated with 500 µl cell lysate
overnight at 4°C. The solution was incubated with 100 µl
pre-cleared Protein G beads (Thermo Scientific) for 2 h at room
temperature. The bead lysate was prepared for western blotting
after being washed and boiled with 1X SDS-PAGE buffer (Beyotime
Biotechnology).
Luciferase
HEK293T cells were cultured in a 96-well plate and
transfected with an empty pGL3 luciferase reporter vector, a
pMUC1-Luc vector, and SV-40-Renilla-Luc as an internal control in
the presence of Lipofectamine 3000 reagent (Invitrogen). After 24
h, the transfected cells were lysed using passive lysis buffer
according to the instruction of Promega Dual-Luciferase reporter
system kit and the lysates were analyzed using the dual luciferase
assay system (Promega).
Statistical analysis
All statistical analysis was performed using the
GraphPad Prism Ver. 6.0 programs (GraphPad Software Inc. San Diego,
CA). All data were expressed as mean ± SD Statistical differences
between groups were determined by Wilcoxon's signed rank test, the
χ2 test or Mann-Whitney U test. Kaplan-Meier analysis
and log-rank test were used to estimate and compare survival rates
of LAC patients with high and low 14-3-3ζ or MUC1 expression. The
cut-off values were established by X-tile program (X-tile software
version 3.6.1, Yale University School of Medicine, New Haven, CT,
USA) to calculate cut-off value (16). All P-values were 2-sided, and those
<0.05 were considered statistically significant.
Results
14-3-3ζ promotes cell proliferation,
migration and invasion in lung adenocarcinoma
Since the role of 14-3-3ζ remains largely unexplored
in lung adenocarcinoma, we initially investigated whether knockdown
of 14-3-3ζ could lead to the alteration of biological function in
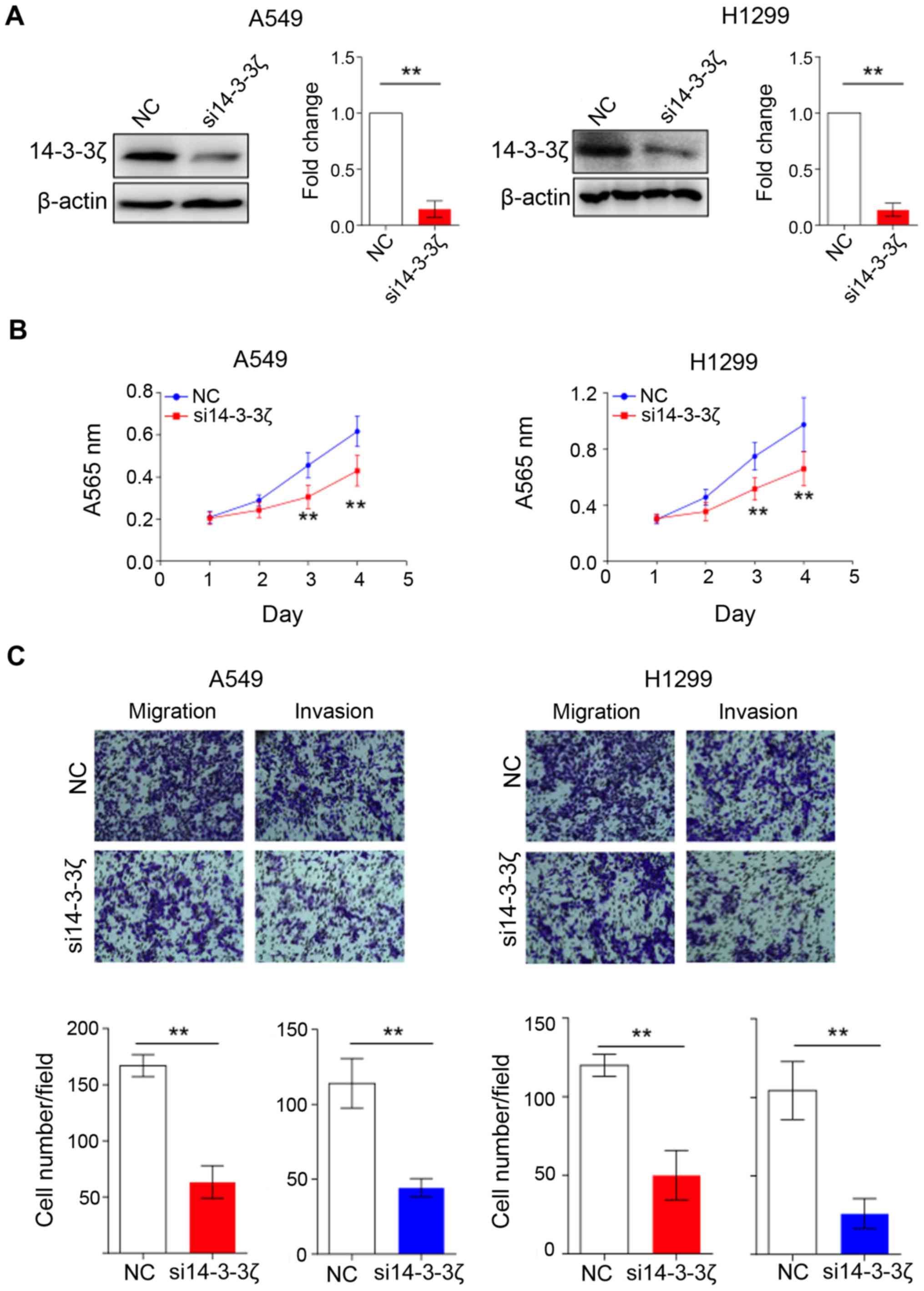
lung cancer cell lines. As shown in Fig. 1A, we successfully reduced 14-3-3ζ
expression in A549 and H1299 by using RNA interference (RNAi)
method according to a previously reported study (9). We subsequently performed MTT assay to
evaluate the growth effect of 14-3-3ζ knockdown on lung cancer
cells. The cell growth of A549 and H1299 cells was significantly
suppressed by 14-3-3ζ siRNA treatment compared to the control group
(P<0.01, Fig. 1B), suggesting
14-3-3ζ protein plays a critical role in promoting cell
proliferation in lung cancer. Furthermore, we also performed
Transwell and Matrigel invasion assays to investigate the
functional role of 14-3-3ζ on cell migration and invasion. As
expected, the decreased expression of 14-3-3ζ resulted in an
impairment of the invasive ability of both A549 and H1299 cells
(Fig. 1C). Collectively, we showed
interference with 14-3-3ζ expression caused by suppression of cell
growth as well as inhibition of cell migration and invasion in lung
cancer cells.
MUC1 is identified as a novel target
of 14-3-3ζ in lung adenocarcinoma
Owing to its oncogenic role in lung cancer, 14-3-3ζ
may affect certain signaling pathways and promote the development
of lung cancer. Several important surface receptors have been found
upregulated in lung adenocarcinoma for sustaining proliferative or
metastatic signaling. We hypothesized that 14-3-3ζ may have impact
on the expression of such surface receptors. Based on our
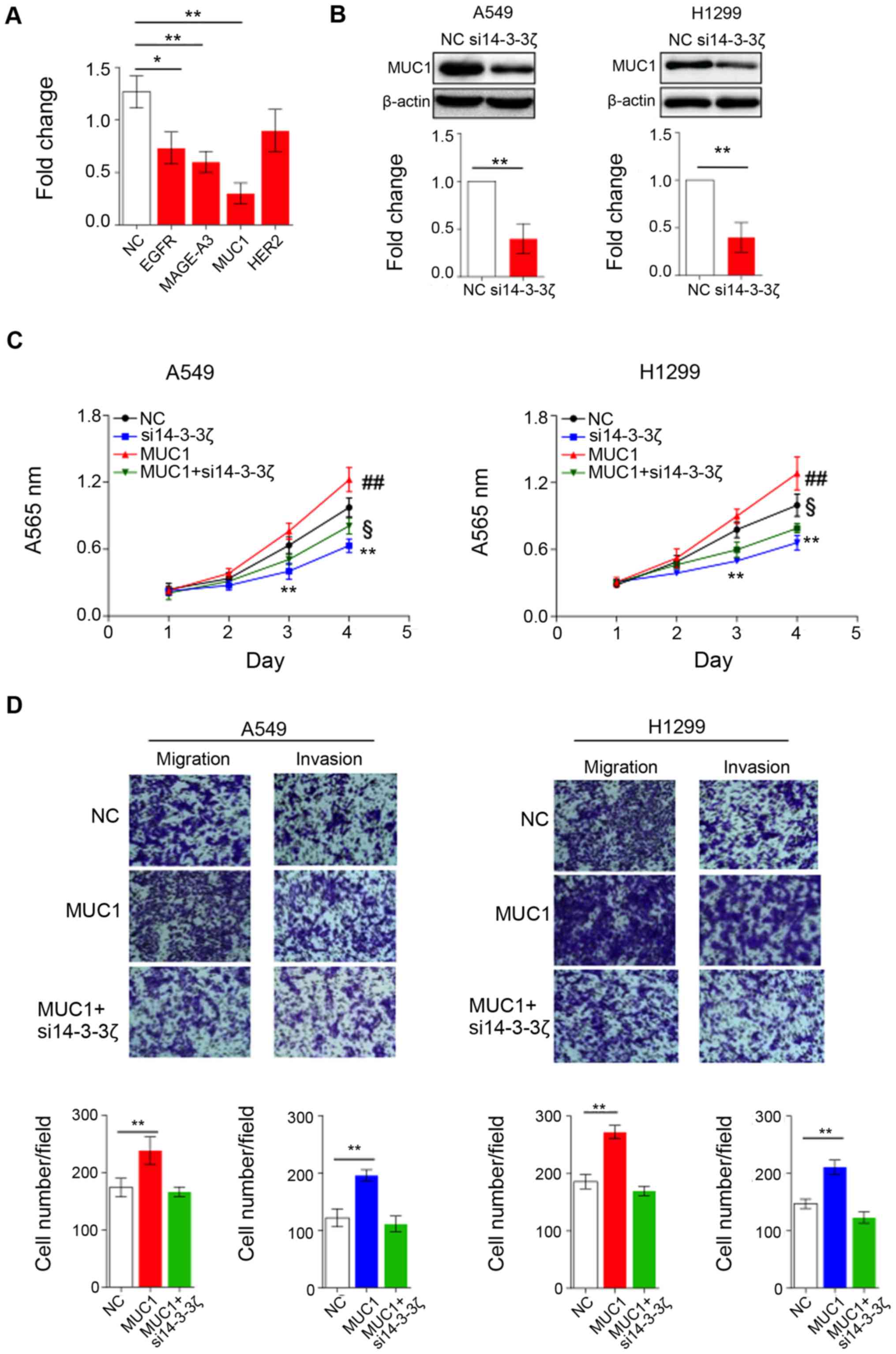
assumption, we first evaluated the expression level of a panel of
receptors (EGFR, MAGE-A3, MUC1 and HER2), which were reported
previously as activators of lung cancer, in 14-3-3ζ siRNA treated
and untreated H1299 cells. The results showed the expression of
EGFR, MAGE-A3 and MUC1 was significantly reduced, while HER2
remains unchanged upon 14-3-3ζ siRNA treatment (Fig. 2A). Notably, the expression of MUC1
was severely decreased by 70% compared to EGFR (30%) and MAGE-A3
(40%). To confirm that the expression of MUC1 can be regulated by
14-3-3ζ, we also treated A549 and H1299 cells with 14-3-3ζ siRNA
and then measured protein level of MUC1 by western blotting.
Consistently, MUC1 protein level is markedly downregulated when
14-3-3ζ expression is decreased in A549 and H1299 cells (Fig. 2B).
Since 14-3-3ζ is able to affect expression of MUC1,
we hypothesized that 14-3-3ζ may also have impact on biological
function of MUC1. As depicted in Fig.
2C, overexpression of MUC1 significantly enhanced proliferation
of both A549 and H1299 cells compared to control cells; however,
14-3-3ζ knockdown effectively inhibits MUC1 function and results in
the suppression of cell growth. Similar to MTT assay, MUC1
overexpression promotes cell migration and invasion, while
knockdown of 14-3-3ζ completely neutralized function of MUC1
(Fig. 2D). Our data clearly showed
MUC1 is a novel target of 14-3-3ζ in lung cancer. 14-3-3ζ plays a
crucial role for MUC1-mediated phenotypic characteristics of lung
cancer cells.
14-3-3ζ upregulates MUC1 expression
through enhancing MUC1/NFκB feedback loop
Recently, MUC1 COOH-terminal subunit (MUC1-C)
cytoplasmic domain was reported to bind directly to p65, trigger
p65 release from its inhibitor IκBα, leading to the constitutive
activation of the NF-κB pathway. Furthermore, the MUC1-C/p65
complex can bind to NF-κB target genes, including the promoter of
the MUC1 gene itself, thus resulting in auto-inductive regulatory
loop (17). Noteworthy, 14-3-3ζ was
demonstrated to activate NF-κB signal transduction pathway through
atypical protein kinase C (aPKC) activation (18,19),
suggesting 14-3-3ζ may affect MUC1 expression through MUC1/NF-κB
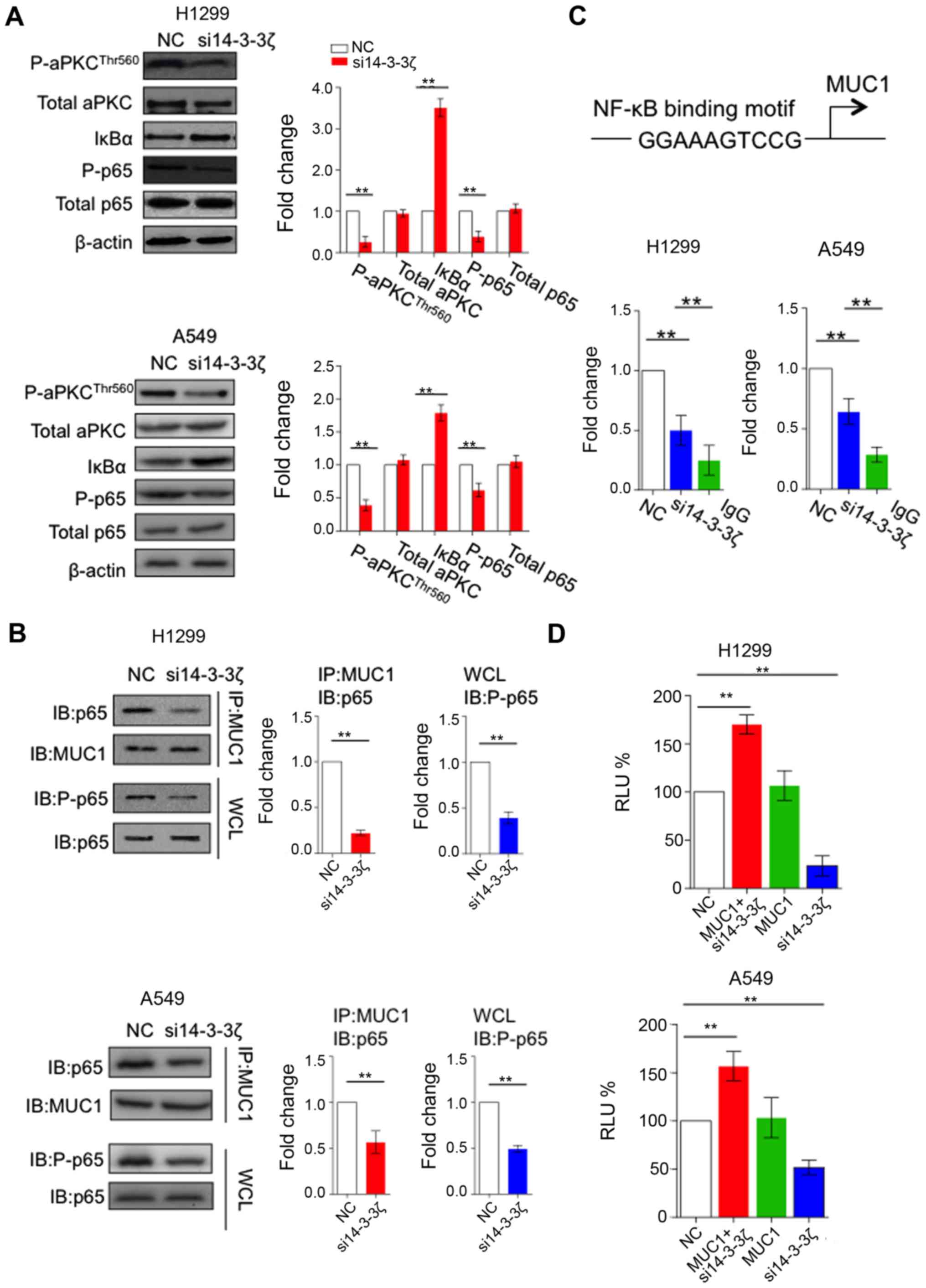
feedback loop. We therefore first confirmed that the activity of
aPKC is inhibited by the knockdown of 14-3-3ζ in H1299 and A549
cells, as measured by the phosphorylation of Thr-560. We further
observed that 14-3-3ζ knockdown caused increased level of IκBα
subunit and decreased phosphorylation of p65, implicating that
14-3-3ζ knockdown inhibited aPKC activity and its target NF-κB
signaling pathway (Fig. 3A).
Since 14-3-3ζ knockdown inhibits the NF-κB signaling
pathway, we assume 14-3-3ζ may affect the formation of MUC1-C/p65
complex. As expected, knockdown of 14-3-3ζ attenuates binding of
MUC1-C and p65 (Fig. 3B). Previous
study showed MUC1-C/p65 complex is able to bind to the promoter
region (NF-κB responsive element) of MUC1 (Fig. 3C) and induce its mRNA expression
(17). We thereafter performed ChIP
assay with MUC1-C antibody to determine whether occupancy of MUC1
promoter by MUC1-C was decreased by silencing 14-3-3ζ. Consistent
with our hypothesis, the binding of MUC1 to the NF-κB responsive
element was significantly decreased when silencing 14-3-3ζ in H1299
and A549 cells (Fig. 3C). To
determine whether 14-3-3ζ affects activation of MUC1 transcription,
we first overexpressed MUC1 in H1299 and A549 cells. In line with
previous studies, MUC1 overexpression can successfully promote the
activity of MUC1-promoter luciferase reporter. However, such
positive effect was significantly attenuated by 14-3-3ζ knockdown
(Fig. 3D), suggesting 14-3-3ζ
regulates MUC1 expression in a promoter-dependent manner.
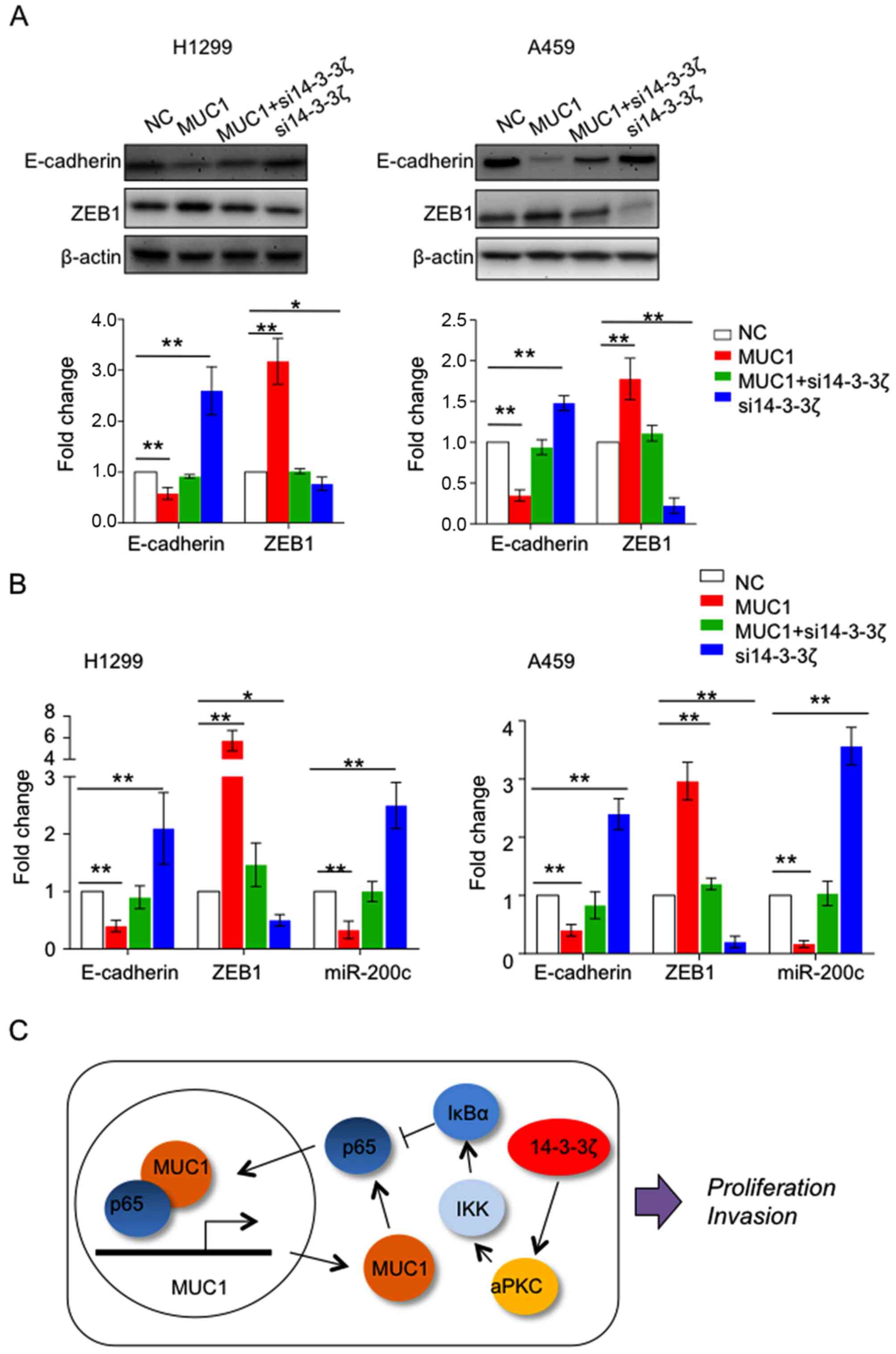
14-3-3ζ and MUC1 was reported to drive
epithelial-mesenchymal transition (EMT) in different cancer types
(8,20–22).
We observed that overexpression of MUC1 in H1299 and A549 cells is
able to upregulate mesenchymal transcription factor ZEB1, while
downregulated epithelial markers E-cadherin and inducer of
epithelial differentiation miR-200c compared to the control group.
In contrast, reduction of 14-3-3ζ expression upregulated ZEB1,
while decreased expression of E-cadherin and miR-200c, suggesting
both 14-3-3ζ and MUC1 contribute to EMT in lung cancer (Fig. 4A and B). Furthermore, we noted that
downregulation of 14-3-3ζ suppressed function of MUC1 to induce
EMT, suggesting the oncogenic function of MUC1 requires the
expression of 14-3-3ζ (Fig.
4C).
High expression of 14-3-3ζ and MUC1 is
associated with poor survival in lung adenocarcinoma patients
To explore the clinical relevance of 14-3-3ζ and
MUC1 in lung cancer, we used public available gene expression
omnibus (GEO) to analyze their expression levels in association
with clinicopathological characteristics and survival. GSE68465
datasets were used since it was a large and multi-site lung
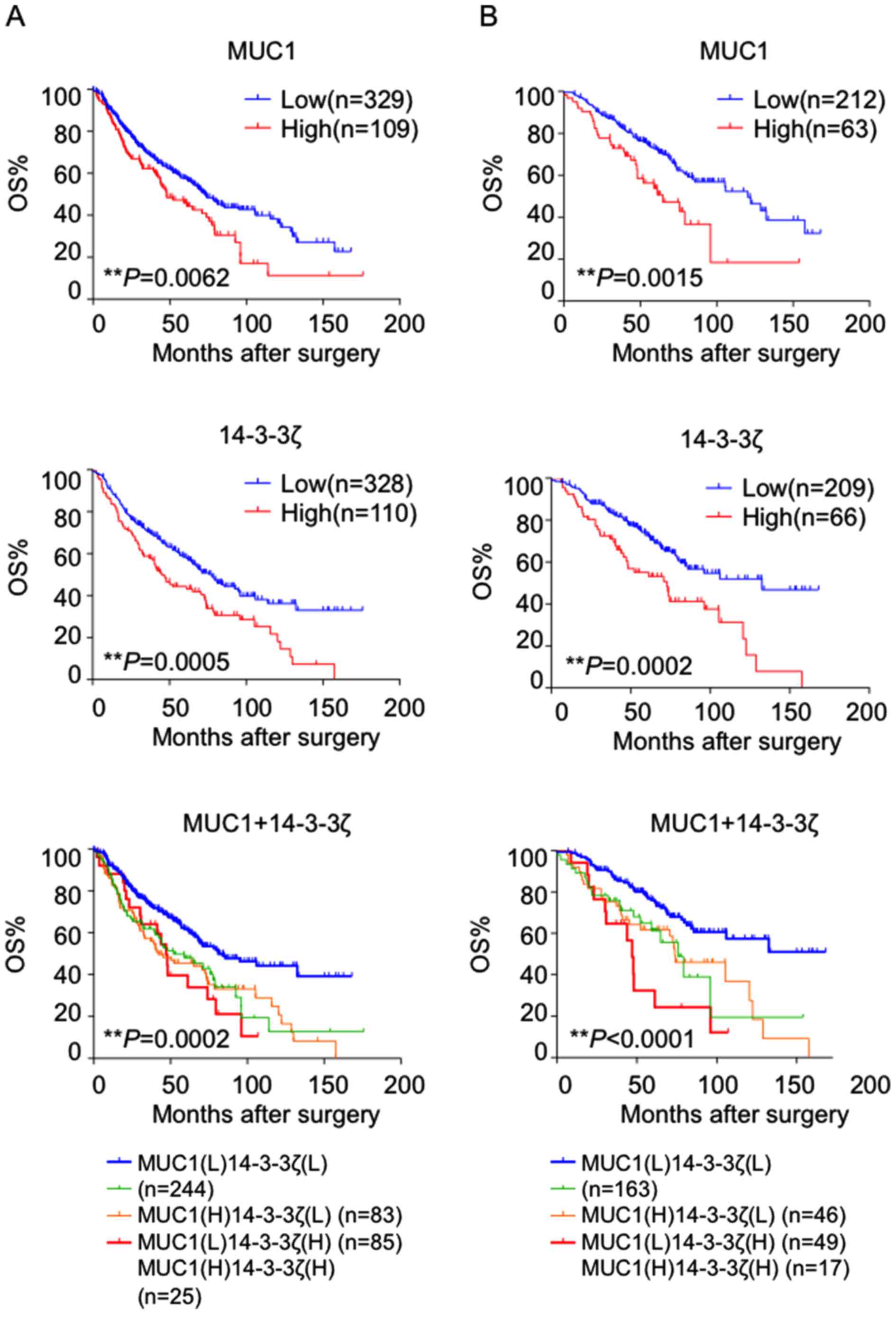
adenocarcinoma cohort with complete follow-up information (23). The correlation between 14-3-3ζ and
MUC1 expression and clinicopathological characteristics is shown in
Table I. Kaplan-Meier survival
analysis showed the group with high expression level of 14-3-3ζ and
MUC1 showed remarkably poor prognosis compared to the low
expression group (P=0.0062, P=0.0005, respectively). Importantly,
when we segregated patients into 4 groups based on 14-3-3ζ levels
and MUC1 expression, we discovered that the group with both high
14-3-3ζ and high MUC1 expression showed worst prognosis compared to
other 3 groups (Fig. 5A).
 | Table I.The correlation between
clinicopathological characteristics, MUC1 and 14-3-3ζ expression in
lung adenocarcinomas. |
Table I.
The correlation between
clinicopathological characteristics, MUC1 and 14-3-3ζ expression in
lung adenocarcinomas.
|
| MUC1 |
| 14-3-3ζ |
|
|---|
|
|
|
|
|
|
|---|
| Characteristics | Low | High | P-value | Low | High | P-value |
|---|
| Gender |
|
|
|
|
|
|
|
Female | 160 | 57 | 0.305 | 169 | 48 | 0.1863 |
| Male | 169 | 52 |
| 159 | 62 |
|
| Age |
|
|
|
|
|
|
| ≥65 | 156 | 56 | 0.368 | 168 | 44 | 0.0539 |
|
<65 | 173 | 53 |
| 160 | 66 |
|
| Smoking |
|
|
|
|
|
|
|
Never | 31 | 17 | 0.0585 | 35 | 13 | 0.8187 |
| Past | 202 | 63 |
| 204 | 61 |
|
|
Current | 28 | 4 |
| 24 | 8 |
|
|
Unknown | 68 | 25 |
| 65 | 28 |
|
| Grade |
|
|
|
|
|
|
| Well | 45 | 15 | 0.8522 | 50 | 10 | 0.3415 |
|
Moderate | 155 | 53 |
| 156 | 52 |
|
| Poor | 126 | 39 |
| 118 | 47 |
|
|
Unknown | 3 | 2 |
| 4 | 1 |
|
| Lymph node
metastasis |
|
|
|
|
|
|
|
Negative | 232 | 66 | 0.0695 | 222 | 76 | 0.8761 |
|
Positive | 97 | 43 |
| 106 | 34 |
|
| Stage |
|
|
|
|
|
|
| I | 212 | 63 | 0.4419 | 209 | 66 | 0.6536 |
| II | 69 | 26 |
| 71 | 24 |
|
|
III | 48 | 20 |
| 48 | 20 |
|
It was reported that a subset of patients with stage
I disease have poorer prognosis and may benefit significantly from
adjuvant chemotherapy (24,25). When we analyzed stage I patients
only, 14-3-3ζ and MUC1 still showed to be a highly predictive
prognostic indicator, wherein patients in the group with high
14-3-3ζ or MUC1 expression were more likely to have a poor outcome
vs. that with low expression (P=0.0002, P=0.0002 respectively).
Consistently, when we segregated patients into 4 groups based on
14-3-3ζ levels and MUC1 expression in stage I patients, we also
observed that the group with both high 14-3-3ζ and high MUC1
expression group showed worst prognosis compared to the other 3
groups (Fig. 5B). Collectively,
these results indicate that 14-3-3ζ and MUC1 could successfully
segregate high vs. low-risk patients with stage I disease.
Discussion
In this study, we for the first time showed Mucin 1
(MUC1) is a novel downstream target of 14-3-3ζ in lung
adenocarcinoma. Second, we unraveled a novel mechanism that 14-3-3ζ
affects MUC1 expression through MUC1/NF-κB feedback loop. Third,
from a biological perspective, we demonstrated that both 14-3-3ζ
and MUC1 are involved in activation of EMT pathway and promoted
aggressiveness of lung cancer cells, highlighting the therapeutic
importance of 14-3-3ζ and MUC1 in lung adenocarcinoma. Fourth, high
expression of 14-3-3ζ and MUC1 correlated with poor patient
outcomes, highlighting its applicability as a promising prognostic
biomarker in lung adenocarcinoma.
Recently, 14-3-3ζ protein has aroused considerable
interest since it was overexpressed in different types of cancer
and reported to activate EMT-associated genes to promote
metastasis. Our data clearly showed interference with 14-3-3ζ
expression in lung cancer cell lines caused suppression of cell
growth, migration and invasion, suggesting its oncogenic role in
the development of lung cancer.
To further investigate the molecular event mediated
by 14-3-3ζ, we screened several important cell surface receptors
which may be affected by 14-3-3ζ. Of note, we observed that the
expression of MUC1 was dramatically downregulated by silencing
14-3-3ζ in lung cancer cells, suggesting MUC1 is a potential target
of 14-3-3ζ. MUC1 is a trans-membrane heterodimeric protein that is
aberrantly expressed in non-small cell lung cancer. It was reported
that over 80% of lung adenocarcinoma express high level of MUC1
(26). Of importance, MUC1
undergoes autocleavage into two subunits. The MUC1 N-terminal
subunit (MUC1-N) contains glycosylated tandem repeats, while
C-terminal subunit (MUC1-C) can interact with receptor tyrosine
kinases, such as EGFR (27).
Moreover, MUC1-C is able to bind directly to NF-κB p65 and promotes
NF-κB-mediated gene transcription including MUC1 gene itself. Our
study revealed a new mechanism that 14-3-3ζ activated NF-κB
signaling pathway through aPKC, the hyperactivated p65 can bind
more MUC1-C, and MUC1-C/p65 complex therefore persistently
activates the promoter of MUC1, resulting in a hyperactive
MUC1/NF-κB feedback loop.
Not only MUC1 gene per se, but other NF-κB target
genes, ZEB1, for instance, could be affected by 14-3-3ζ. The
available evidence showed that MUC1-C occupies the ZEB1 promoter
with NF-κB and thereby promotes ZEB1 transcription. Furthermore,
MUC1-C associates with ZEB1 and the MUC1-C/ZEB1 complex suppresses
transcription of miR-200c, an inducer of epithelial differentiation
(20). Herein, our data
successfully validated that overexpression of MUC1 leads to
upregulation of ZEB1, while suppressed the expression of miR-200c.
More importantly, we found 14-3-3ζ knockdown could abolish the
impact of MUC1 overexpression on ZEB1 and miR-200c, implicating
that the function of MUC1 to induce EMT is dependent on 14-3-3ζ
protein and 14-3-3ζ promotes metastasis, at least in part, through
MUC1/NF-κB feedback loop.
To better appreciate the oncogenic role of 14-3-3ζ
and MUC1 for its contribution to lung carcinogenesis, we also
investigated their correlation in association with clinical
outcome. The GSE68465 datasets showed that the group with high
expression level of 14-3-3ζ and/or MUC1 showed remarkably poor
prognosis compared to the low expression group. Furthermore, when
we analyzed stage I patients only, 14-3-3ζ and/or MUC1 still showed
to be a highly predictive prognostic indicator, wherein patients in
the high expression group were more likely to have a poor outcome
vs. those with low-risk, suggesting that 14-3-3ζ and MUC1 could
successfully segregate high vs. low-risk patients with stage I
disease.
In summary, ours is the first study to
systematically interrogate the functional and clinical significance
of 14-3-3ζ and its novel target MUC1 in lung adenocarcinoma, and we
provide comprehensive evidence that 14-3-3ζ can regulate MUC1
through MUC1/NF-κB feedback loop. We conclude that 14-3-3ζ and/or
MUC1 is a promising prognostic biomarker for lung cancer patients;
therapeutic targeting of 14-3-3ζ and/or MUC1 may be a potential
treatment option for patients with lung adenocarcinoma.
References
|
1
|
Jemal A, Siegel R, Ward E, Hao Y, Xu J,
Murray T and Thun MJ: Cancer statistics, 2008. CA Cancer J Clin.
58:71–96. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Travis WD: Pathology of lung cancer. Clin
Chest Med. 32:669–692. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Yang X, Yang K and Kuang K: The efficacy
and safety of EGFR inhibitor monotherapy in non-small cell lung
cancer: A systematic review. Curr Oncol Rep. 16:3902014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Rossi A, Maione P, Sacco PC, Sgambato A,
Casaluce F, Ferrara ML, Palazzolo G, Ciardiello F and Gridelli C:
ALK inhibitors and advanced non-small cell lung cancer (Review).
Int J Oncol. 45:499–508. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Inamura K: Diagnostic and therapeutic
potential of microRNAs in lung cancer. Cancers (Basel). 9:92017.
View Article : Google Scholar :
|
|
6
|
Groome PA, Bolejack V, Crowley JJ, Kennedy
C, Krasnik M, Sobin LH and Goldstraw P; IASLC International Staging
Committee; Cancer Research and Biostatistics; Observers to the
Committee, : Participating Institutions: The IASLC Lung Cancer
Staging Project: Validation of the proposals for revision of the
TN, and M descriptors and consequent stage groupings in the
forthcoming (seventh) edition of the TNM classification of
malignant tumours. J Thorac Oncol. 2:694–705. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Chen JJ, Peck K, Hong TM, Yang SC, Sher
YP, Shih JY, Wu R, Cheng JL, Roffler SR, Wu CW, et al: Global
analysis of gene expression in invasion by a lung cancer model.
Cancer Res. 61:5223–5230. 2001.PubMed/NCBI
|
|
8
|
Yang Y, Liu Y, He JC, Wang JM, Schemmer P,
Ma CQ, Qian YW, Yao W, Zhang J, Qi WP, et al: 14-3-3ζ and aPKC-ι
synergistically facilitate epithelial-mesenchymal transition of
cholangiocarcinoma via GSK-3β/Snail signaling pathway. Oncotarget.
7:55191–55210. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Huang XY, Ke AW, Shi GM, Zhang X, Zhang C,
Shi YH, Wang XY, Ding ZB, Xiao YS, Yan J, et al: αB-crystallin
complexes with 14-3-3ζ to induce epithelial-mesenchymal transition
and resistance to sorafenib in hepatocellular carcinoma.
Hepatology. 57:2235–2247. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Tang Y, Liu S, Li N, Guo W, Shi J, Yu H,
Zhang L, Wang K, Liu S and Cheng S: 14-3-3ζ promotes hepatocellular
carcinoma venous metastasis by modulating hypoxia-inducible
factor-1α. Oncotarget. 7:15854–15867. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Rüenauver K, Menon R, Svensson MA,
Carlsson J, Vogel W, Andrén O, Nowak M and Perner S: Prognostic
significance of YWHAZ expression in localized prostate cancer.
Prostate Cancer Prostatic Dis. 17:310–314. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Nishimura Y, Komatsu S, Ichikawa D, Nagata
H, Hirajima S, Takeshita H, Kawaguchi T, Arita T, Konishi H,
Kashimoto K, et al: Overexpression of YWHAZ relates to tumor cell
proliferation and malignant outcome of gastric carcinoma. Br J
Cancer. 108:1324–1331. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Matta A, DeSouza LV, Shukla NK, Gupta SD,
Ralhan R and Siu KW: Prognostic significance of head-and-neck
cancer biomarkers previously discovered and identified using
iTRAQ-labeling and multidimensional liquid chromatography-tandem
mass spectrometry. J Proteome Res. 7:2078–2087. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Neal CL, Yao J, Yang W, Zhou X, Nguyen NT,
Lu J, Danes CG, Guo H, Lan KH, Ensor J, et al: 14-3-3zeta
overexpression defines high risk for breast cancer recurrence and
promotes cancer cell survival. Cancer Res. 69:3425–3432. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Rajabi H, Ahmad R, Jin C, Joshi MD, Guha
M, Alam M, Kharbanda S and Kufe D: MUC1-C oncoprotein confers
androgen-independent growth of human prostate cancer cells.
Prostate. 72:1659–1668. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Camp RL, Dolled-Filhart M and Rimm DL:
X-tile: a new bio-informatics tool for biomarker assessment and
outcome-based cut-point optimization. Clin Cancer Res.
10:7252–7259. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ahmad R, Raina D, Joshi MD, Kawano T, Ren
J, Kharbanda S and Kufe D: MUC1-C oncoprotein functions as a direct
activator of the nuclear factor-kappaB p65 transcription factor.
Cancer Res. 69:7013–7021. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Tang Y, Lv P, Sun Z, Han L, Luo B and Zhou
W: 14-3-3ζ up-regulates hypoxia-inducible factor-1α in
hepatocellular carcinoma via activation of PI3K/Akt/NF-κB signal
transduction pathway. Int J Clin Exp Pathol. 8:15845–15853.
2015.PubMed/NCBI
|
|
19
|
Tong S, Xia T, Fan K, Jiang K, Zhai W, Li
JS, Wang SH and Wang JJ: 14-3-3ζ promotes lung cancer cell invasion
by increasing the Snail protein expression through atypical protein
kinase C (aPKC)/NF-κB signaling. Exp Cell Res. 348:1–9. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Rajabi H, Alam M, Takahashi H, Kharbanda
A, Guha M, Ahmad R and Kufe D: MUC1-C oncoprotein activates the
ZEB1/miR-200c regulatory loop and epithelial-mesenchymal
transition. Oncogene. 33:1680–1689. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Roy LD, Sahraei M, Subramani DB, Besmer D,
Nath S, Tinder TL, Bajaj E, Shanmugam K, Lee YY, Hwang SI, et al:
MUC1 enhances invasiveness of pancreatic cancer cells by inducing
epithelial to mesenchymal transition. Oncogene. 30:1449–1459. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chen CH, Chuang SM, Yang MF, Liao JW, Yu
SL and Chen JJ: A novel function of YWHAZ/β-catenin axis in
promoting epithelial-mesenchymal transition and lung cancer
metastasis. Mol Cancer Res. 10:1319–1331. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Shedden K, Taylor JM, Enkemann SA, Tsao
MS, Yeatman TJ, Gerald WL, Eschrich S, Jurisica I, Giordano TJ,
Misek DE, et al: Director's Challenge Consortium for the Molecular
Classification of Lung Adenocarcinoma: Gene expression-based
survival prediction in lung adenocarcinoma: A multi-site, blinded
validation study. Nat Med. 14:822–827. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Booth CM and Shepherd FA: Adjuvant
chemotherapy for resected non-small cell lung cancer. J Thorac
Oncol. 1:180–187. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Gandara DR, Wakelee H, Calhoun R and
Jablons D: Adjuvant chemotherapy of stage I non-small cell lung
cancer in North America. J Thorac Oncol. 2 Suppl 3:S125–S127. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Situ D, Wang J, Ma Y, Zhu Z, Hu Y, Long H
and Rong T: Expression and prognostic relevance of MUC1 in stage IB
non-small cell lung cancer. Med Oncol. 28 Suppl 1:S596–S604. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kufe DW: Mucins in cancer: Function,
prognosis and therapy. Nat Rev Cancer. 9:874–885. 2009. View Article : Google Scholar : PubMed/NCBI
|